Abstract
The thermodynamic hypothesis of Anfinsen postulates that structures and stabilities of globular proteins are determined by their amino acid sequences. Chain topology, however, is known to influence the folding reaction, in that motifs with a preponderance of local interactions typically fold more rapidly than those with a larger fraction of non-local interactions. Together, the topology and sequence can modulate the energy landscape and influence the rate at which the protein folds to the native conformation. To explore the relationship of sequence and topology in the folding of βα–repeat proteins, which are dominated by local interactions, a combined experimental and simulation analysis was performed on two members of the flavodoxin-like, α/β/α sandwich fold. Spo0F and the N-terminal receiver domain of NtrC (NT-NtrC) have similar topologies but low sequence identity, enabling a test of the effects of sequence on folding. Experimental results demonstrated that both response-regulator proteins fold via parallel channels through highly structured sub-millisecond intermediates before accessing their cis prolyl peptide bond-containing native conformations. Global analysis of the experimental results preferentially places these intermediates off the productive folding pathway. Sequence-sensitive Gō-model simulations conclude that frustration in the folding in Spo0F, corresponding to the appearance of the off-pathway intermediate, reflects competition for intra-subdomain van der Waals contacts between its N- and C-terminal subdomains. The extent of transient, premature structure appears to correlate with the number of isoleucine, leucine and valine (ILV) side-chains that form a large sequence-local cluster involving the central β-sheet and helices α2, α3 and α4. The failure to detect the off-pathway species in the simulations of NT-NtrC may reflect the reduced number of ILV side-chains in its corresponding hydrophobic cluster. The location of the hydrophobic clusters in the structure may also be related to the differing functional properties of these response regulators. Comparison with the results of previous experimental and simulation analyses on the homologous CheY argues that prematurely-folded unproductive intermediates are a common property of the βα-repeat motif.
Keywords: response regulators, protein folding landscape, global analysis, coarse-grained molecular dynamics, topological frustration
Introduction
Although it is well accepted that the native conformation of a protein represents its global free energy minimum,1 an understanding of the dynamic process by which the sequence is decoded into its three-dimensional structure on a biologically-feasible time scale remains elusive. Landscape theory2 posits the view that sequences have evolved not only to be stable but also to have the capacity to rapidly and efficiently access the native conformation via a funnel-shaped energy surface biased towards the formation of the native structure. The explicit role for conformational entropy in determining the shape of the energy surface and correlations between folding rate constants and metrics for chain topology3–5 argue for the importance of topology in folding reactions. However, the role of the sequence remains evident in the results of mutational analyses on dozens of proteins,6 where single amino acid replacements can significantly alter the stability and the folding kinetics. Also, structural homologs with diverse sequences have been observed to fold at very different rates7,8 or via different mechanisms.9 Thus, deciphering the folding information contained in the amino acid sequence of a protein remains a major challenge in biophysics.
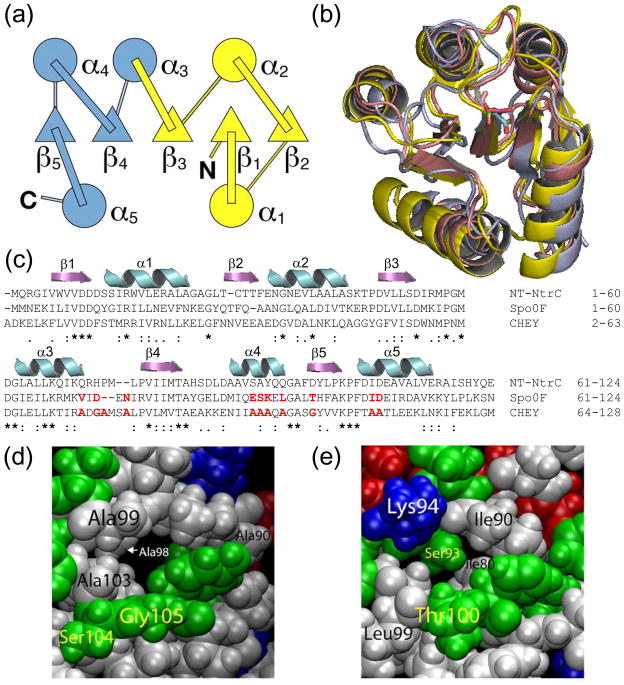
We have adopted a combined experimental and computational approach towards the elucidation of the relative contributions of the sequence and the topology to the folding mechanisms of three members of the CheY-like family of response-regulator proteins: the bacterial chemotaxis protein CheY from Escherichia coli,10 the N-terminal receiver domain of nitrogen regulation protein NtrC from Salmonella typhimurium (NT-NtrC)11 and the sporulation response regulatory protein Spo0F from Bacillus subtilis.12 These small repeat-structure proteins, (βα)5, typically contain ~125 amino acids arranged as a α/β/α sandwich. The five β-strands form a central parallel β-sheet, β2β1β3β4β5, with helices α1 and α5 docking on one face of the β-sheet and helices α2, α3 and α4 docking on the opposing face (Fig. 1a). The pair-wise RMSD values for CheY:NT-NtrC, CheY:Spo0F and NT-NtrC:Spo0F are 2.57 Å, 1.85 Å and 2.44 Å (Fig. 1b), respectively, and the pair-wise sequence alignment scores calculated using ClustalW13 are 30%, 25% and 33%, respectively (Fig. 1c). Based upon the results of previous studies on other folds with similar structures but very different sequences, 7–9,14–17 we hypothesize that common features in the folding mechanisms will reflect the topology while the differences in the mechanisms and the perturbations in the kinetic and thermodynamic properties will reflect the variable sequences.
Figure 1.
(a) Topology of CheY-like proteins. The central β-sheet comprises 5 parallel β strands in the order β2β1β3β4β5 and forms an α/β/α sandwich with helices α1 and α5 on one face of the sheet and helices α2, α3 and α4 on the other. The N-terminal (yellow) and C-terminal (blue) folding subdomains of CheY21 are comprised of β1α1β2α2β3 and α3β4α4β5α5, respectively. (b) Structural alignment of NT-NtrC (blue), Spo0F (red) and CheY (yellow). The pair-wise RMSD values for CheY:NT-NtrC, CheY:Spo0F and NT-NtrC:Spo0F are 2.57 Å, 1.85 Å and 2.44 Å respectively. The catalytic aspartic acid residue, 54 in NT-NtrC, 54 in Spo0F and 57 in CheY, is at the beginning of the loop connecting the two subdomains and is shown as sticks. The PDB codes used were NT-NtrC: 1DC7, 11 Spo0F: 1SRR12 and CheY: 3CHY.10 (c) Sequence alignment of NT-NtrC, Spo0F and CheY using ClustalW.13 The pair-wise sequence alignment scores for CheY:NT-NtrC, CheY:Spo0F and NT-NtrC:Spo0F are 30%, 25% and 33%, respectively. The elements of secondary structure are indicated above the aligned sequences, and the sequence conservation is indicated below, (*) identical, (:) conserved and (.) semi-conserved. Residues highlighted in red font denote either C-terminal alanines and glycines in CheY or corresponding residues in Spo0F with bulkier side-chains. (d) An alanine-rich cavity resides between α4 and β4β5 in the inactive CheY structure. (e) The same region in Spo0F is filled with bulkier residues.
As a basis for this comparative analysis, a recent experimental study of CheY18 found that folding initiates with the appearance of an off-pathway partially-folded state in the sub-millisecond time range. This kinetically-trapped species must at least partially unfold before the protein can access the productive transition state ensemble (TSE) and fold to the native conformation. The companion coarse-grained Gō-simulation19 came to a similar conclusion and predicted that the premature folding of the α2β3α3β4 tetrad towards the C-terminus of CheY was responsible for the kinetic trap. A sequence-local cluster of isoleucine, leucine and valine side-chains in precisely the same C-terminal segments identified in the Gō-model simulations was hypothesized to provide the core of stability in the off-pathway folding intermediate.18 A previous Gō-model simulation of CheY reported the premature folding of the five α-helices, but did not describe a role for the β-strands.20 The dissipation of structure in the α2β3α3β4 tetrad in the kinetically-trapped species allowed the formation of the productive TSE involving the same N-subdomain containing the β1α1β2α2β3 elements of secondary structure identified in the mutational analysis of CheY (Fig. 1a).21 The C-subdomain containing the α3β4α4β5α5 elements of secondary structure is unstructured in this TSE (Fig. 1a).
The variations in sequence for these three proteins also allow an exploration of the relationship between the structural characteristics of their folding reactions and their functional properties. Phosphorylation of an aspartic acid residue, D57, in the loop connecting β3 and α3 of CheY causes the α4β5 surface to undergo a conformational rearrangement that enable binding of CheY to its downstream target, the flagellar motor protein, FliM.22–24 The flexibility of the α4β5 surface of the protein has been attributed to the lack of a strong N-capping residue in α4 and an alanine-lined cavity21,25 (Fig. 1d) between this helix and the rest of the protein.26,27 Analogous to the conformational dynamics of CheY, phosphorylation of D54 in NT-NtrC induces a structural rearrangement in β5 and α4 of the receiver domain that transmits the signal to the C-terminal DNA-binding domain.11,28 NMR relaxation measurements and atomistic molecular dynamics simulations have identified flexibility in α4 in the inactive state,29,30 and results of a recent combined NMR and X-ray analysis,31 support a population-shift activation mechanism as has also been suggested for CheY.22,24,32,33 By contrast to CheY and NT-NtrC, the most significant conformational rearrangement in Spo0F during phosphorylation of D54 occurs in α1. This rearrangement enables Spo0F to interact directly with its immediate downstream partner in the phosphor-relay signaling pathway, Spo0B.12,34–36 Unlike CheY and NT-NtrC, the cavity between helix α4 and the β-sheet of Spo0F is filled with bulky side-chains, some of which participate in a large hydrophobic cluster (Fig. 1c, 1d and 1e).
The complementary insights into the energetic and structural aspects of the folding free energy surfaces for NT-NtrC and Spo0F, provided by a combined experimental and computational analysis, reveal a significant role for the sequences in modulating their folding reactions. Prematurely-folded intermediates, stabilized by local-in-sequence clusters of aliphatic side-chains37 appear to be a common feature in the folding of CheY-like proteins.
Results
Experimental Analysis
Equilibrium folding reactions
Equilibrium unfolding free-energy surface
Far-UV CD and fluorescence spectroscopy (FL) were employed to monitor the loss of secondary and tertiary structure of NT-NtrC and Spo0F in the presence of the chemical denaturant urea (Supplemental Fig. 1). The urea-induced equilibrium unfolding transitions, monitored by CD at 222 nm and by FL emission at 315 nm for NT-NtrC and 305 nm for Spo0F, show single sigmoidal transitions for both proteins (Fig. 2a and 2d). The normalized CD and FL equilibrium transition curves are coincident within error (Supplemental Fig. 1c and 1f), and the reversibility of the urea denaturation reaction was demonstrated by the coincidence of the unfolding and refolding CD transitions (Fig. 2a and 2d). Assuming a two-state equilibrium model for the unfolding reaction, a global analysis of the CD and FL spectral changes with urea concentration yielded values for the Gibbs free energy of unfolding from the native, N, to the unfolded, U, state in the absence of urea, ΔG° (H2O), the dependence of ΔG° on the denaturant concentration, the m-value, and the mid-point of the transition, Cm, as shown in Table 1. For comparison, the ΔG° (H2O), the m-value and the Cm for a two-state fit of the urea-induced equilibrium unfolding reaction for CheY are also shown.18
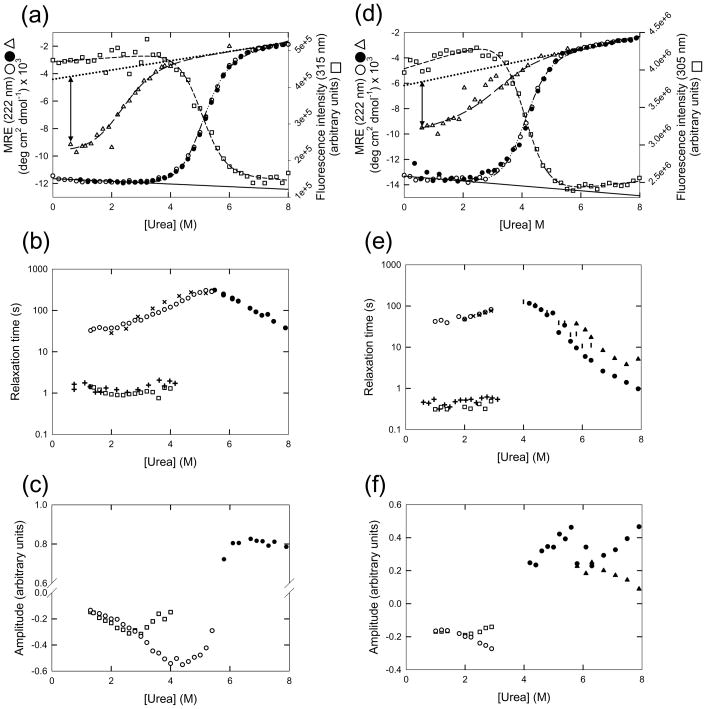
Figure 2.
Equilibrium and kinetic experimental analyses of NT-NtrC and Spo0F. (a) Equilibrium unfolding and refolding of NT-NtrC. The equilibrium denaturation is completely reversible as is seen by the coincidence of the unfolding (○) and refolding (●) CD signal at 222 nm plotted as a function of denaturant concentration. The fit to a two-state model is shown (broken and dotted line). The baselines for the native state (continuous line) and the unfolded state (dotted line) predicted from the two-state model are also shown. The burst-phase amplitude measured by stopped-flow CD refolding of NT-NtrC from 6 M urea is plotted as a function of final urea concentration (△) and fit to a two-state model (thick broken line). The magnitude of the burst-phase amplitude under strongly refolding conditions (0.6 M urea) is represented by the double-headed arrow. The FL intensity at 315 nm is plotted as a function of urea concentration (□) and fit to a two-state model (thin dashed line). (b) Chevron analysis of NT-NtrC. The recovery of the native signal upon refolding from high denaturant concentration occurs by bi-exponential kinetics. The relaxation times determined by CD [(×), slow phase; (+), fast phase] and by FL [(○), slow phase; (□), fast phase] are shown. The amplitudes associated with the fast refolding phase by both CD and FL at denaturant concentrations > 4 M urea and the amplitudes associated with the slow refolding phase by CD at denaturant concentrations < 2 M urea were too small to obtain accurate relaxation times and were thus excluded from the chevron analysis. A single phase is observed in unfolding kinetics; the relaxation times determined by FL (●) are also shown. (c) Amplitudes associated with the relaxation times determined by FL. The symbols used are the same as in (b). (d) Equilibrium unfolding and refolding of Spo0F. The symbols used are the same as in (a). The reversibility of the equilibrium denaturation is seen by the coincidence of the unfolding (○) and refolding (●) CD signal at 222 nm plotted as a function of denaturant concentration. The burst-phase amplitude (△) is measured by stopped-flow CD refolding of Spo0F from 6 M urea. The FL intensity (□) at 305 nm is plotted as a function of urea concentration. (e) Chevron analysis of Spo0F. The refolding relaxation times determined by CD [(×), slow phase; (+), fast phase] and by FL [(○), slow phase; (□), fast phase], and the unfolding relaxation times determined by CD (|) and by FL [(●), fast phase; (▲), slow phase] are shown. (f) Amplitudes associated with the relaxation times determined by FL. The symbols used are the same as in (e). Buffer conditions: 10 mM potassium phosphate at pH 7.0 and 25 °C.
Table 1.
Apparent thermodynamic properties of CheY-like proteins and their respective sub-millisecond intermediatesa,b
| Protein | NT-NtrC | Spo0F | CheY16 | |||
|---|---|---|---|---|---|---|
| State | Native | Intermediate | Native | Intermediate | Native | Intermediate |
| ΔG° (H2O) kcal mol−1 | 7.52 ± 0.14 | 2.36 ± 0.40 | 5.99 ± 0.12 | 2.98 ± 0.23 | 5.37 ± 0.21 | 2.30 ± 0.40 |
| m-value kcal mol−1 M−1 | 1.49 ± 0.03 | 1.04 ± 0.09 | 1.47 ± 0.03 | 0.93 ± 0.07 | 1.59 ± 0.06 | 0.92 ± 0.34 |
| Cm M | 5.05 ± 0.2 | 2.27 ± 0.58 | 4.08 ± 0.16 | 3.20 ± 0.49 | 3.33 ± 0.26 | 2.5 ± 1.30 |
The native state stability and m-values are obtained by a global analysis of refolding and unfolding urea denaturation curves at multiple wavelengths monitored by both FL and CD spectroscopy. The stability of the intermediate and its dependence on urea concentration is determined by fitting the amplitude of the burst-phase reaction, monitored by CD at 222 nm, to a two-state model. The errors reported are standard errors from the global fits.
These thermodynamic parameters are regarded as apparent because the two-state fits ignore the contribution by the cis/trans isomerization of the native cis-prolyl peptide bond K104-P105 in the unfolded state.
Kinetic folding reactions
Burst-phase reaction
The formation of secondary structure during the refolding of NT-NtrC and Spo0F induced by stopped-flow mixing methods was monitored by the changes in ellipticity at 222 nm. Over 50% (66% for NT-NtrC and 52% for Spo0F) of the native ellipticity at 222 nm was recovered in the dead-time of the stopped-flow instrument (~5 ms) for both proteins (Fig. 2a and 2d), compared to the 95% signal recovered in the same time frame for CheY.18,38 The apparent thermodynamic properties of the burst-phase species were estimated by measuring the amplitude of the burst-phase CD reaction as a function of the final denaturant concentration in refolding. The sigmoidal loss in the ellipticity at 222 nm for both NT-NtrC and Spo0F at increasing final urea concentrations (Fig. 2a and 2d) is consistent with the cooperative disruption of secondary structure in a stable partially-folded state, IBP. Fitting the urea-dependence of the burst-phase amplitude to a two-state model provided an estimate of the stability for the IBP species in the two proteins (Table 1) along with estimated stability for the burst-phase intermediate in CheY.18 Compared to the native state, the decreased stabilities and m-values for the intermediates suggest that the interiors of these early partially-folded states are less well packed than their native counterparts. For CheY18 and, to a lesser extent, for NT-NtrC and Spo0F, a substantial amount of secondary structure appears early in the folding process.
Slow refolding reactions
Subsequent to the burst-phase reaction, the remainder of the CD and FL signal for the refolding of NT-NtrC is recovered by bi-exponential kinetics whose relaxation times are shown in a chevron plot in Fig. 2b. The two phases are nearly equal in amplitude (Fig. 2c), and both relaxation times are independent of denaturant concentration under strongly folding conditions. Because both refolding phases at low urea concentration are accelerated modestly in the presence of a prolyl isomerase, cyclophilin (Supplemental Fig. 2a), the cis/trans isomerization of an Xaa-Pro peptide bond must limit folding to some extent. This behavior has been observed previously for the single slow refolding phase in CheY18 and reflects the presence of a cis prolyl peptide bond (K104-P105) in the native conformation. When these response regulators are unfolded, the trans isomer becomes dominant and must convert to the cis isomer via a rate-limiting reaction that is coupled to folding. The weak cyclophilin dependence of this rate implies limited accessibility to the prolyl bond, which suggests that the isomerization reaction occurs in a partially folded state.
Similar kinetic results are seen for Spo0F (Fig. 2e and 2f). Refolding in the presence of cyclophilin modestly decreased the relaxation time for the slow phase but had no discernible effect on the fast folding phase under strongly refolding conditions (Supplemental Fig. 2b).
Unfolding reactions
The unfolding reaction of NT-NtrC monitored by fluorescence and CD is well-described by a single exponential phase, whose relaxation time decreases exponentially above 5.8 M urea (Fig. 2b). The amplitude of the unfolding phase accounts for the entire ellipticity change expected from the equilibrium unfolding profile, eliminating the possibility of rapid unfolding reactions (Fig. 2c).
The unfolding reaction of Spo0F is more complex (Fig. 2e). Above 5.5 M urea concentration, unfolding occurs by bi-exponential kinetics; the urea dependence of the major, fast phase is collinear with the single unfolding phase observed below 5.5 M urea. Only the faster phase is observed when unfolding of Spo0F is monitored by CD. The CD amplitude associated with this unfolding phase is within error of that expected from equilibrium measurements (data not shown); the absence of the slower phase detected by fluorescence may reflect the lower signal to noise ratio in the CD experiment.
Global analysis
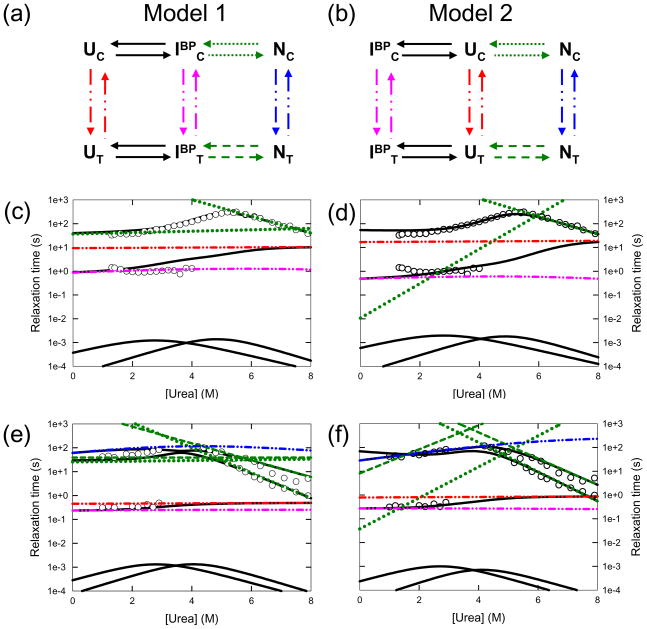
The largely comparable equilibrium and kinetic responses of NT-NtrC and Spo0F with those of CheY led to a test of the hypothesis that NT-NtrC and Spo0F also fold via prolyl isomer-dictated parallel channels with either early on- (Model 1 - Fig. 3a) or off-pathway (Model 2 - Fig. 3b) intermediates, as has been done previously for CheY.18 In these models, NC and NT correspond to the native conformation with cis and trans isomers at the native cis prolyl peptide bond, UC and UT correspond to the unfolded states with the respective prolyl isomers and IBPC and IBPT correspond to the burst-phase intermediate with cis and trans prolyl isomers.
Figure 3.
Global analysis of NT-NtrC and Spo0F (a) Model 1: The on-pathway model. The folding mechanism occurs via parallel channels based on the isomerization state of the K104–P105 peptide bond. The burst-phase species, IBP is placed on-pathway, between the unfolded and native states along either channel. (b) Model 2: The off-pathway model. The burst-phase species, IBP is placed off-pathway from the unfolded states in both channels. (c) Predicted chevron for NT-NtrC from the on-pathway model. The predicted observable relaxation times are shown as continuous black lines; the microscopic rate constants determined by the model are shown as dotted green lines for the cis channel. The isomerization relaxation times in each state are shown as broken and dotted lines, blue for the native states, red for the unfolded states, and magenta for the burst-phase intermediates. The chevron analysis from Fig. 2b is shown as open circles for comparison. (d) Predicted chevron for NT-NtrC from the off-pathway model. The legends are the same as in (c). (e) Predicted chevron for Spo0F from the on-pathway model. The legends are the same as in (c) and the microscopic rate constants for the trans channel are shown as broken green lines. The chevron analyses from Fig. 2e are shown as open circles for comparison. (f) Predicted chevron for Spo0F from the off-pathway model. The legends are the same as in (e).
A comprehensive set of unfolding and refolding traces were fit globally to these kinetic models with initial estimates for the parameters based on (1) experimentally determined equilibrium properties (Table 1), (2) the microscopic rate constants, k, and their urea dependences, m‡, obtained from the chevron plot of the dependences of the relaxation times (k = 1/τ) on the final denaturant concentrations and an in-house algorithm, Chevron Fitter,18 and (3) the experimentally determined distribution of prolyl isomers in the unfolded state for each protein obtained from penta-peptide models (Supplemental Fig. 3a and 3b). The sequence identity adjacent to the cis prolyl residue in NT-NtrC, Spo0F and CheY, Lys-Pro-Phe, resulted in trans:cis distributions that are equivalent within error, 90:10. The equilibrium analysis provided the stability and denaturant dependence of the major species observed, NC relative to UT, and the starting and final amplitudes for the kinetic traces. Although the rate constants for the burst-phase refolding reaction cannot be determined by stopped-flow mixing, the equilibrium constants for the IBPC/UC and the IBPT/UT reactions were assumed to be equal and were obtained by fitting the urea dependence of the CD burst-phase amplitude to a two-state model (Fig. 2a and 2d). The folding rate constants for the burst-phase intermediates were assumed to be >104 s−1 to account for their appearance within 5 ms. Because the rates of formation of the intermediates are at least >105 faster than the observed rate constant for the appearance of the native conformation, these two processes do not kinetically couple with each other. However, simulations show that the significant stabilities and the nonzero m-values of the intermediates (Figs. 2a and 2d) in rapid pre-equilibrium with the unfolded states result in a significant impact on the observed relaxation time (Supplemental Fig. 4). As a first approximation, the urea dependences of the refolding and unfolding rate constants for the IBPT and IBPC species, m‡rBPT and m‡uBPT and m‡rBPC and m‡uBPC, were each assigned to be half of the m-value for the equilibrium unfolding reaction, 1.04 kcal mol−1 M−1 for NT-NtrC and 0.93 kcal mol−1 M−1 for Spo0F. The procedure is described in more detail in a previous paper.18
A total of 32 FL kinetic traces for NT-NtrC and 27 for Spo0F obtained under a variety of unfolding and refolding conditions were then fit to the two models, and the parameters were optimized using the Levenberg–Marquardt algorithm. The microscopic rates, kinetic m-values, native and unfolded signals and the Z values (relative signal contribution from each species normalized to the difference between the signals for the native and unfolded species) were modeled globally. The kinetic m-values were constrained such that refolding m-values are ≥ 0 and unfolding m-values are ≤ 0. The signal offsets were allowed to vary for each kinetic trace. The protein concentration for each kinetic trace was allowed to vary, albeit with strongly constraints (within 3% of the measured value) to account for possible errors in the measurement of the protein concentration (Supplemental Fig. 5).
The quality of the fits to on- and off-pathway models was assessed by comparison of their reduced chi-square values and by visual comparisons of the predicted chevrons and the amplitudes for the globally-minimized parameters for both proteins. The equilibrium populations of the intermediates for both NT-NtrC (Supplemental Fig. 6a and Fig. 6b) and Spo0F (Supplemental Fig. 6c and Fig. 6d) in both models are sufficiently low at all urea concentrations (< 6%) as to remain undetectable, and the predicted equilibrium denaturation profiles are consistent with the experimentally observed two-state behavior (Fig. 2a and 2d). Although either model provides credible fits of the kinetic traces, the reduced chi-square value for the off-pathway model is 10% lower than that obtained from the fit for the on-pathway model for both NT-NtrC and Spo0F (number of degrees of freedom ~ 3,000, p-values < 0.01).
Representative refolding and unfolding traces for NT-NtrC and Spo0F along with their fits using the parameters from the global analysis of the off-pathway model are shown in Supplemental Fig. 7. The microscopic rate constants and their urea-dependences for the on-pathway model, Model 1, are provided in Supplemental Table 1 and Fig. 3c for NT-NtrC and Fig. 3e for Spo0F and for the off-pathway model, Model 2, in Table 2 and Fig. 3d for NT-NtrC and Fig. 3f for Spo0F. The large errors for the rates related with the NT species in Spo0F and the IBPC species in both proteins reflect the small/negligible contributions of these species to fitted kinetic traces.
Table 2.
Microscopic rate constants and their associated urea dependences determined by a global fit of kinetic and equilibrium folding data to the off-pathway model (Model 2).
| Microscopic step | NT-NtrC | Spo0F | ||
|---|---|---|---|---|
| k (s−1) | m-value (kcal mol−1 M−1) | k (s−1) | m-value (kcal mol−1 M−1) | |
| UC → UT | 5.34×10−2 ± 8.10×10−3 | 0 | 1.15 ± 8.65 | 0 |
| UT → UC | 5.95×10−3 ± 1.02×10−3 | 0 | 1.26×10−1 ± 1.03 | 0 |
| UC → IBPCa | > 1.95×104 | ~ 0.52 | > 3.17×104 | ~ 0.52 |
| IBPC → UCa | > 4.22 | ~ −0.51 | > 2.69×101 | ~ −0.52 |
| UT → IBPTa | > 1.65×103 | ~ 0.40 | > 4.23×103 | ~ 0.47 |
| IBPT → UTa | > 40.9 | ~ −0.39 | > 5.25×101 | ~ −0.48 |
| IBPC → IBPTb | 1.52×10−1 ± 1.16×10−2 | −0.16 ± 1.47×10−2 | 1.42 ± 5.52 | −0.04 ± 1.09 |
| IBPT → IBPCb | 1.95 ± 2.79×10−1 | 0.07 ± 3.89×10−2 | 2.27 ± 9.78 | 0.03 ± 1.46 |
| UC → NC | 93.4 ± 1.83 | 0.99 ± 1.54×10−3 | 2.68×101 ± 8.82 | 0.89 ± 3.87×10−2 |
| NC → UC | 2.84×10−5 ± 9.94×10−7 | −0.51 ± 1.60×10−3 | 2.14×10−5 ± 9.88×10−6 | −0.84 ± 3.91×10−2 |
| UT → NTbc | ND | ND | 1.24×10−1 ± 4.68×10−1 | 0.69 ± 7.08×10−1 |
| NT → UTbc | ND | ND | 1.48×10−5 ± 3.86×10−5 | −0.75 ± 4.75×10−1 |
| NC → NTbc | ND | ND | 2.02×10−3 ± 2.08×10−2 | −0.03 ± 1.01 |
| NT → NCbc | ND | ND | 3.33×10−2 ± 3.47×10−1 | 0.24 ± 1.07 |
Data for NT-NtrC were obtained from a global analysis of 22 refolding and 10 unfolding kinetic traces and for Spo0F from a global analysis of 10 refolding and 17 unfolding kinetic traces. The errors reported are standard errors from the global fits of the data obtained by standard propagation methods.
The lower limits of the rate constants for the burst-phase reactions are reported. The m-values for the burst-phase species are equally distributed to the forward and reverse reactions.
The uncertainty in the rates associated with the IBPC and NT species reflect the small/negligible contributions of these species to the fitted kinetic traces.
The NT species is not detected (ND) in NT-NtrC.
Although the folding of the intermediate is too fast to be directly detected by stopped-flow methods, the parameters derived for the on- and off-pathway models, in combination with the reduced chi-square statistic, enable one to choose the more likely model. Specifically, for the on-pathway model, the refolding m-value for the IBP → N reaction was constrained to be greater than or equal to zero as expected for a progressive folding reaction. If this constraint is relaxed, the m-value for the IBP → N reaction becomes less than zero and, effectively, Model 1 reverts to Model 2, with an off-pathway intermediate (Supplemental Fig. 8). Thus, the optimal fitting of the data is achieved with the off-pathway intermediates.
A triangular model, wherein the native states NC and NT have direct access to both the corresponding unfolded states and the corresponding intermediate states, was also tested (data not shown). For NT-NtrC, the model reverts back to a five-state off-pathway model (Model 2, with the NT state being inaccessible). However, the fit with Spo0F is equally good as the off-pathway model. Model 2 is favored for Spo0F because it accounts for the observed responses as well as the triangular model, but uses fewer parameters.
The lower chi-square values for the off-pathway models provide support for off-pathway intermediates but are not conclusive in eliminating the on-pathway model. Additional support for the off-pathway model in NT-NtrC and Spo0F is provided by the inspection of urea dependence of the microscopic rate constants. In the on-pathway model for NT-NtrC (Fig. 3c), the nearly urea independent refolding rate constant of the IBPC to NC reaction implies that the productive TSE does not bury a significant amount of surface area relative to that observed in the intermediate. The same prediction is made by the on-pathway model for the refolding of Spo0F (Fig. 3e). While the direct refolding of compact non-native intermediates to the native state via a TSE that requires internal repacking has been observed under extreme conditions of high salt39 or high denaturant,40 the folding kinetics of many proteins typically reveal that the TSE is more similar to the native state in terms of surface area buried.6 In accordance with this expectation, the typical urea dependencies for the UC → NC refolding reactions in the off-pathway models for both NT-NtrC and Spo0F demonstrate the burial of a significant fraction of exposed surface area in the productive TSE of both proteins (Fig. 3d and 3f). Thus, by both statistical and folding behavior criteria, the global analyses of the experimental data favor, but do not definitively prove an off-pathway folding mechanism for NT-NtrC and Spo0F.
Simulation Analysis
Thermodynamic as well as kinetic Gō-model simulations of folding were carried out for NT-NtrC and Spo0F using methodology previously developed for the homologous CheY folding reaction.19 To aid comparison of the influence of sequence variation on the folding of the topologically-equivalent proteins a flavored variant of the traditional Gō-model was employed in which heterogeneity of the native contact energies is added to incorporate sequence effects (see Materials and Methods).
The sequence of events was mapped by examining the dependence of the free energy on several structural properties. For the equilibrium thermodynamic calculations, multi-canonical umbrella sampling was used to ensure the entire accessible landscape was sampled, including unfolded, native and high-energy intermediate species. Although more exact methods for characterizing the folding TSE are available,41,42 the goal of the present work was to elucidate the folding mechanism by determining the most probable order of events in the formation of structure. Multi-canonical equilibrium simulations have proven to be useful in the study of complex folding behavior when multiple reaction coordinates are necessary to describe the essential features of folding mechanisms.43–45 To lend support to the mechanism defined by the equilibrium simulations, 100 independent, unbiased kinetic folding simulations were also performed starting from a random coil unfolded structure under conditions promoting the native state. The relative sequence of folding events observed in the ensemble kinetic simulations was in good agreement with the most probable pathway revealed from thermodynamic landscape calculations.
Thermodynamic simulations
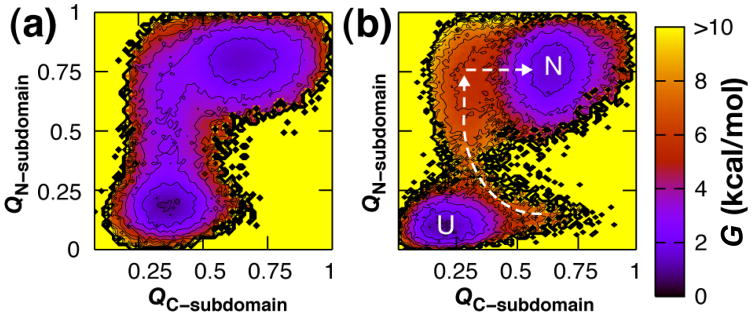
The free energy landscape was characterized at the folding transition temperature so that both the native and unfolded basins could be clearly defined. The fraction of native contacts formed, denoted Q, is a useful progress variable for monitoring the formation of secondary and tertiary structure. The free energy was computed as a function of the fraction of native contacts formed in the N- and C-subdomains (Fig. 1a) at the transition temperature (Fig. 4). The resulting Gō-simulation landscapes for NT-NtrC and Spo0F are similar to CheY21 in that the N-subdomain is partially structured in the productive folding transition state whereas the C-subdomain is not. The C-subdomain does not access its folded basin until the N-subdomain has folded, and the C-subdomain relies on contacts at the subdomain-interface for its stability. This behavior suggests that the N-subdomain serves as the folding nucleus for the C-subdomain. Additionally, the C-subdomain for NT-NtrC and Spo0F exhibited dynamic instability as evidenced by the large width of their native basins where both structured and largely unstructured states are sampled.
Figure 4.
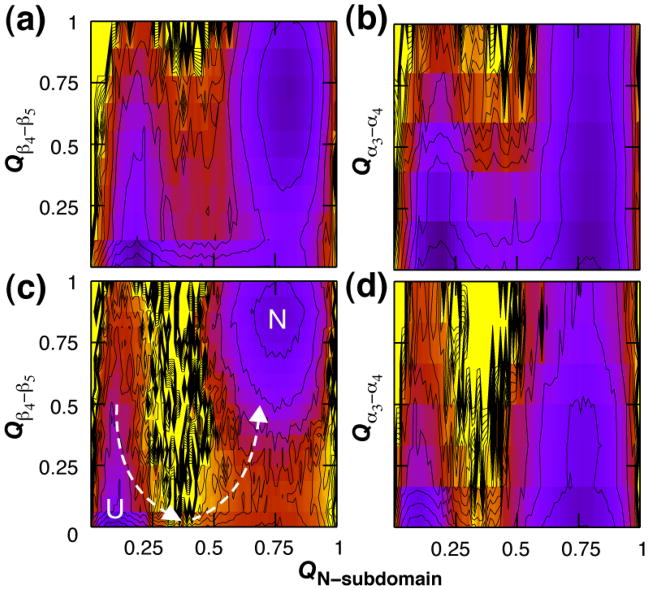
Gō-model results for the thermodynamic characterization of the N-terminally nucleated folding landscapes for (a) NT-NtrC (b) and Spo0F. The free energy, G, is shown as a function of the fraction of native contacts formed within the N-subdomain (QN-subdomain) and the fraction of native contacts formed within the C-subdomain (QC-subdomain). Off-pathway frustration is evident for Spo0F for which the prematurely structured C-subdomain must unfold in order for the N-subdomain to fold and drive the progression to the native state. Contours are drawn every kcal mol−1; values exceeding 10 kcal mol−1 and regions not sampled are shown in yellow. The free energy is computed at the folding transition temperature such that the folded and unfolded states are equally populated.
The instability of the C-subdomain can be attributed to a lower density of van der Waals contacts. The Gō-model assigned an average of 1.2/1.1 (NT-NtrC) and 1.6/1.3 (Spo0F) native contacts per residue in the N-/C-subdomains, respectively. The lower average number of contacts for NT-NtrC compared to Spo0F reflects the limitations of the solution NMR structure for NT-NtrC compared to the crystal structures for Spo0F and CheY.46,47 The smaller number of native contact potentials to promote folding for NT-NtrC results in a lower energy barrier between the unfolded and folded basins than in the case of Spo0F (Fig. 4) or CheY.19 Recent simulations of protein G also demonstrated the dependence of the folding barrier on the experimental structure from which the Gō-model is derived.47
Insight into the sequence of folding events for NT-NtrC and Spo0F can be gained by examining the formation of three topologically equivalent βαβ modules in the flavodoxin fold, β1α1β2, β3α3β4 and β4α4β5. The dependence of the free energy on the fraction of contacts formed in each of these three modules reveals the relative order of their formation. In NT-NtrC, elongation of the central β-sheet proceeds from the N-terminus, as has been observed for CheY.19 The most probable folding pathway involves the progressive formation of the β1α1β2 module, the β3α3β4 module and the β4α4β5 module (Supplemental Fig. 9). In Spo0F, folding spreads in both directions from the central β3α3β4 module, first towards the β1α1β2 module and then towards the β4α4β5 module (Supplemental Fig. 10). This finding suggests that the folding nucleus initially identified by Lopez-Hernandez et al. as β1α1β2α2β3 in CheY21 be extended in the case of Spo0F to include helix α3 and strand β4.
Topological frustration in Spo0F
The α2β3α3β4 region in CheY was previously shown to cause significant frustration in folding simulations by partially forming prior to the folding of the N-subdomain.19 This topologically-frustrated structure was observed to unfold before productive folding in the N-subdomain could occur and proceed to the native state. Topological frustration was not observed in the C-subdomain of NT-NtrC; premature structure in β4 and β5, and the α3 and α4 helices do not preclude productive folding in the N-subdomain of NT-NtrC (Fig. 4a, Fig. 5a and Fig. 5b). In Spo0F, the β3α3β4 module is no longer a site of frustration as the region serves to initiate productive folding. Frustration in the thermodynamic landscape is evident, however, within its C-subdomain, α3β4α4β5 (Fig. 4b, Fig. 5c and Fig. 5d).
Figure 5.
Frustration in the C-subdomain of NT-NtrC and Spo0F. The free energy is shown as a function of the fraction of native contacts formed within the N-subdomain and between β4 and β5 (a, c), and within the N-subdomain and between α3 and α4 (b, d). Premature structure in the C-subdomain of NT-NtrC (a, b) does not preclude N-subdomain folding. For Spo0F (c, d), N-subdomain folding is seen to accompany an initial unfolding of C-subdomain contacts, as evidenced by the high energy barrier bisecting the path to N at QN-subdomain = 0.4. The energy scale is described in the caption to Figure 5.
The dissolution of prematurely formed native contacts, termed backtracking, ascribed to topological frustration has been previously reported in the literature.20,48–50 Computational and experimental studies of TIM barrel proteins by Finke and colleagues have implicated backtracking as a general mechanism for assisting the protein in reaching the native state.51,52 Premature structure formation followed by backtracking is a likely scenario in the maturation of tertiary structure in multicomponent proteins, in which subdomains must compete for structural contacts.
Kinetic simulations
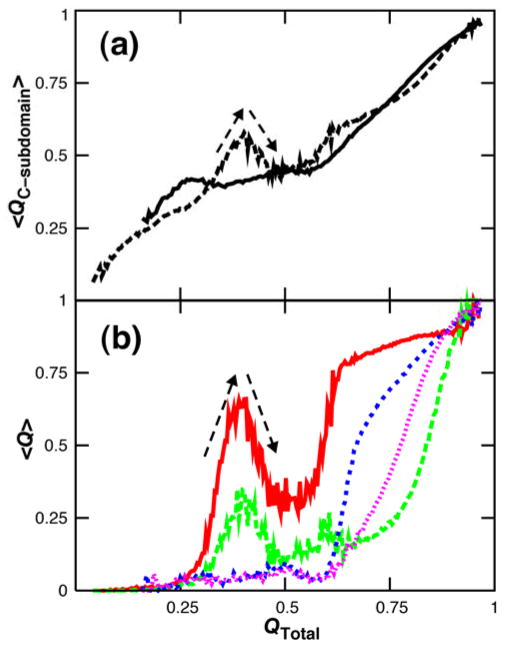
Ensemble kinetic simulation data are in accord with the thermodynamic results. The fraction of contacts formed at each time point was computed for the 100 kinetic folding simulations and ensemble-averaged. Unfolding of prematurely-folded structure in the C-subdomain is evident in the overall ensemble-averaged kinetic time course for Spo0F but not for NT-NtrC (Fig. 6). The negative slopes at QTotal = 0.4 indicate local disruption of contacts between β4 and β5 and between α3 and α4 in Spo0F while N-terminal folding is still in progress.
Figure 6.
Influence of frustration on kinetics. The fraction of native contacts formed at each time point was computed for 100 independent kinetic folding simulations and ensemble-averaged. (a) The mean fraction of C-subdomain contacts formed is shown as a function of the fraction of native contacts formed in the entire protein for NT-NtrC, solid line, and Spo0F, broken line. (b) The mean fraction of contacts formed in different regions of a protein is shown as a function of the fraction of native contacts formed in the entire protein. For Spo0F, contacts between β4 and β5 are in red and those between α3 and α4 in green. The corresponding regions for NT-NtrC are in blue and pink respectively. The large negative slopes at QTotal = 0.4 indicate local unfolding, or backtracking, of C-subdomain contacts in Spo0F.
Discussion
As hypothesized for the (βα)5 motif, both chain topology and amino acid sequence modulate its folding properties. The equilibrium unfolding mechanisms of NT-NtrC and Spo0F are best described by a two-state model, demonstrating that only the unfolded and native states of these proteins are measurably populated at equilibrium. However, a kinetic analysis reveals a more complicated picture of the folding free energy landscapes for NT-NtrC and Spo0F, which are in many ways similar to the landscape observed for CheY.18
Experimental analysis
Global analysis of equilibrium unfolding transitions, the stabilities of the burst-phase intermediates and the kinetic FL traces derived from a series of unfolding and refolding reactions under a variety of denaturant concentrations for NT-NtrC and Spo0F were best described by parallel channel models with off-pathway intermediates. The previous conclusion that a similar mechanism is operative for CheY,18 underscores the role of topology in defining the basic features of the folding energy landscape of these three βα-repeat proteins.
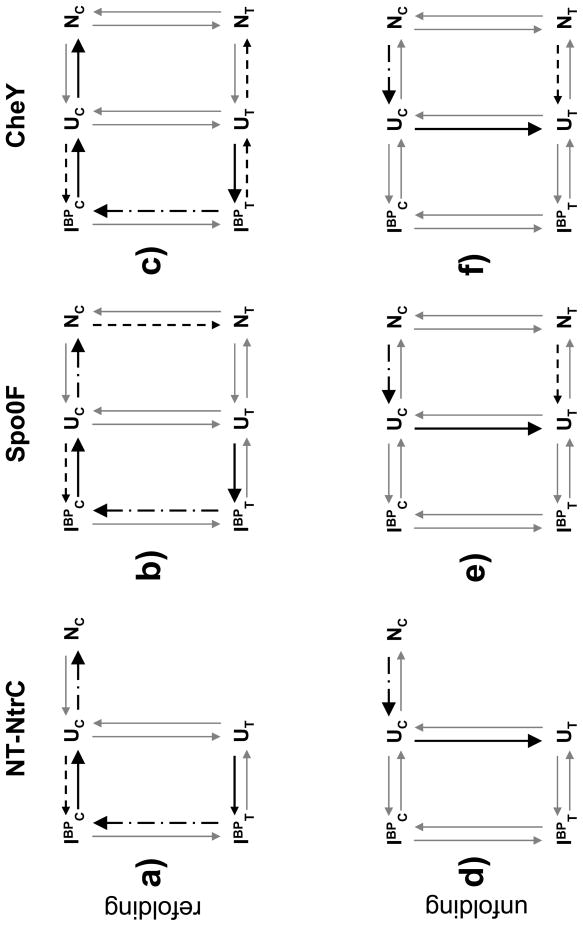
However, significant differences between the populations of kinetic species and the rate-limiting reactions were observed during the refolding of the three proteins. Under strongly refolding conditions for NT-NtrC (Fig. 7a), both the major and minor unfolded populations, UT and UC, rapidly collapse to the corresponding burst-phase, off-pathway intermediates, IBPT and IBPC, respectively (Fig. 2a and Table 1). The subsequent fast refolding reaction corresponds to the isomerization of the prolyl peptide bond accompanying the conversion of IBPT to IBPC (Fig. 3d and Table 2). The slow refolding reaction gives rise to the acquisition of native structure and is dependent on at least partial unfolding of the IBPC species, the rate-limiting step in the conversion of UC to NC. However, at higher urea concentrations where the intermediate is destabilized, the direct refolding of UC to NC becomes rate-limiting as evidenced by the roll-over of the slow refolding phase (Fig. 2b). The acceleration of the slow phase by cyclophilin can be explained by its dependence on the flow of material from the preceding isomerization reaction. Unlike in CheY,18 (Fig. 7c), the slow equilibration to an alternate native state NT during refolding is not observed in NT-NtrC. The refolding reaction in Spo0F, under strongly refolding conditions (Fig. 7b), is similar to that observed in NT-NtrC, with the exception that the native state, NC, slowly isomerizes to an alternate conformation, NT, that is also seen in CheY.18 CheY differs from both NT-NtrC and Spo0F in that the slow refolding phase corresponds to the prolyl isomerization reaction and the fast refolding reaction to the refolding of UC to NC (Fig. 7c).18
Figure 7.
Mechanism for (a–c) refolding of NT-NtrC, Spo0F and CheY, respectively, under strongly refolding conditions and (d–f) unfolding under strongly unfolding conditions predicted by the off-pathway model. The progress of the reaction is shown as thick arrows, while the reactions not accessible under the respective conditions are represented by thin gray arrows. The rate-limiting reactions are shown as broken and dotted lines, and the minor channels are shown as broken lines. (a) Refolding of NT-NtrC. The dominant unfolded state with the K104-P105 bond in the trans isomer, UT, collapses within the burst-phase of stopped-flow instrumentation (~ 5 ms) to an off-pathway intermediate, IBPT. Isomerization of the prolyl bond gives rise to the fast refolding phase followed by the slow phase corresponding to at least partial unfolding of the intermediate to access the productive TSE between UC and NC. A small contribution to the burst-phase reaction from the minor unfolded population, UC is also shown. (b) Refolding of Spo0F. The progression of events is identical to that of NT-NtrC, with the exception that the native state slowly isomerizes to an alternate native state, NT, which is populated to ~ 5% at equilibrium. (c) Refolding of CheY. The isomerization reaction of the IBPT intermediate in CheY is significantly slower than that observed in the other two proteins. This reaction gives rise to the only observable refolding phase that masks all subsequent reactions. A small fraction of the intermediate can also fold to the NT state, which is populated to ~ 15% at equilibrium. (d) Unfolding of NT-NtrC. Under strongly unfolding conditions, the native state unfolds globally by a single unfolding phase. The acquisition of the equilibrium population of the UT state is optically silent. (e) Unfolding of Spo0F is similar to that of NT-NtrC. An additional small amplitude unfolding phase is explained by the independent unfolding of a small population of the NT state, similar to that observed during the unfolding of CheY (f).
Under strongly unfolding conditions, NC, the only measurably populated state in NT-NtrC (Fig. 7d) and the dominant native state in Spo0F (Fig. 7e) and CheY (Fig. 7f),18 rapidly unfolds to UC. The subsequent slow equilibration to the dominant unfolded state, UT, is spectrally silent in all three proteins. A small amplitude unfolding phase corresponding to the unfolding of the minor NT population is also observed in Spo0F (Figs. 2f and 7e) and CheY (Fig. 7f).
The progressive increase in the stability of the NC state for CheY, 5.4 kcal mol−1, to Spo0F, 6.0 kcal mol−1, to NT-NtrC, 7.5 kcal mol−1, provides a rationale for the inverse correlation with the fractional population of the NT state, 20%, < 10% and 0%. The higher stability of the NC state also appears to be reflected in the higher stability of the IBPC state in NT-NtrC and Spo0F relative to the same species in CheY. While the enhanced stability accelerates the isomerization reaction ~ 100-fold in these two proteins,53,54 (2.10 s−1 in NT-NtrC, and 3.69 s−1 in Spo0F) (Table 2), relative to CheY, (0.08 s−1),18 it impedes access to the TSE that is primarily structured in the N-subdomain and distal to the site of prolyl bond isomerization. Access to the TSE for the IBPC to NC reaction (k(UC→NC) × k(IBPC→UC) ÷ k(UC→IBPC)) (Table 2) is slowest for NT-NtrC (2.02×10−2 s−1), intermediate for Spo0F (2.27×10−2 s−1) and fastest for CheY (6.40×10−1 s−1).18 This anti-Hammond behavior,55 of inverse correlation of refolding rate with stability (Table 1) of the native state possibly relates to the packing densities of the folding nuclei (see below).
Gō-model simulations
The simulations of NT-NtrC and Spo0F reveal the development of a productive folding nucleus in their N-subdomains. However the sequence of events and the extent of topological frustration differ from that seen in CheY.19 In NT-NtrC the reaction proceeds from N-subdomain to the C-subdomain, while in Spo0F topological frustration in the C-subdomain, precedes the appearance of the folding nucleus at the interface of the N- and C-subdomains. In CheY, C-subdomain frustration is readily apparent by both experiment18 and simulation.19 However, the site of nucleation is distinctly restricted to the N-subdomain for the subsequent productive folding reaction.
As noted above, CheY32 and NT-NtrC30 contain an alanine-rich cavity between α4 and β4β5, which is flexible in the inactive state before undergoing rearrangement upon phosphorylation. Sequence alignment and structural visualization of the three proteins reveals that the same region in Spo0F is filled in with several bulkier side-chains (Fig. 1c, 1d and 1e). As a consequence of these sequence variations, CheY, NT-NtrC and Spo0F have 0.83, 0.88 and 1.06 native contacts per residue in this cavity. Comparing the overall amino acid abundances for the three proteins, they have rather similar numbers of each of the twenty amino acids with the exception that Spo0F has half as many alanines (7 in Spo0F vs. 16 in CheY and 15 in NT-NtrC) and twice as many isoleucines (15 in Spo0F vs. 6 in CheY and 8 in NT-NtrC) as NT-NtrC and CheY (Supplemental Table 2). Several of the alanine replacements by bulkier side-chains occur in the cavity between α4 and β4β5 (Fig. 1d and 1e).
Topological frustration in Spo0F can be understood in terms of a high density of native contacts in β4α4β5. This property drives the early development of structure in this region as evidenced by the high QC-subdomain contacts, > 0.6, while the QN-subdomain contacts are low, < 0.4 (Fig. 4b and 5c). The absence of C-subdomain contacts when QN-subdomain increases to 0.4 (Fig. 5c) suggests that disruption of preformed structure in the C-subdomain of Spo0F is required to access the productive folding TSE. Indeed, the disruption of C-subdomain contacts is seen to coincide with the appearance of N-subdomain folding in kinetic simulations. The N-subdomain, with its greater density of native contacts, eventually out-competes the C-subdomain and induces its local unfolding. The shift in the location of folding initiation in Spo0F to β3α3β4 can also be understood in terms of the packing density. Spo0F contains approximately 82% more contacts between helices α2 and α3 and 39% more contacts between strands β3 and β4 than CheY and NT-NtrC. The lower density of contacts in the β1α1β2 module is consistent with the flexibility of α1 observed in NMR relaxation measurements of the unphosphorylated state of Spo0F.34 Finally, a kinetic trap in the C-subdomain of NT-NtrC was not observed in the simulation results, possibly due to the lower packing density of the C-subdomain of NT-NtrC when compared to the packing densities of the corresponding regions in CheY and Spo0F. Thus, the density of native contacts in the C-subdomain appears to be a good predictor of early misfolding reactions in the (βα)5 motif.
Kinetic traps in βα-repeat proteins
βα-repeat proteins belonging to the TIM barrel family, α subunit of tryptophan synthase (αTS) from E. coli,37 indole-3-glycerol phosphate synthase (IGPS) Sulfolobus solfataricus (sIGPS)56 and a hypothetical protein IOLI from Bacillus subtilis,16 and the flavodoxin fold proteins, E. coli CheY,18 apo-flavodoxin from Anabaena sp.40 and from Azotobacter vinelandii,57 experience early kinetic traps during folding. The local-in-sequence/local-in-space structure characteristic of these motifs provides ready access to intermediates that cannot directly access the native conformation.
Sequence local clusters of branched aliphatic side-chain (BASiC) residues, isoleucine, leucine and valine (ILV), have previously been implicated in the formation of off-pathway intermediates in CheY18 (Fig. 8a) and in TIM barrel proteins.16,37,56 The correlation between the location, size and connectivity of clusters of ILV side-chains and the predicted location of prematurely-formed structure in both (βα)5 and (βα)818,19,37 proteins is consistent with an important role for ILV clusters in the appearance of kinetic traps. These branched aliphatic side-chains, along with alanine and glycine, are the only side-chains that do not spontaneously transfer from the vapor phase to water.58 Therefore, these clusters are especially resistant to fluctuations that would allow the penetration of water into the cluster or to the underlying peptide linkages that support the hydrogen-bonding networks in β-strands and α-helices. The synergy between the tertiary and secondary structures, mediated by the exclusion of water from the peptide linkages,59 would provide a molecular explanation for the two-state cooperativity observed for many proteins. Locally-connected ILV clusters of sufficient size appear to be able to drive off-pathway unproductive folding reactions while both local and non-local clusters could serve to stabilize the native conformations of their resident proteins. Because NT-NtrC and Spo0F also have ILV clusters fusing both of the helical layers to the intervening β sheet, it was of interest to explore their relationship to the folding mechanisms of both proteins.
Figure 8.
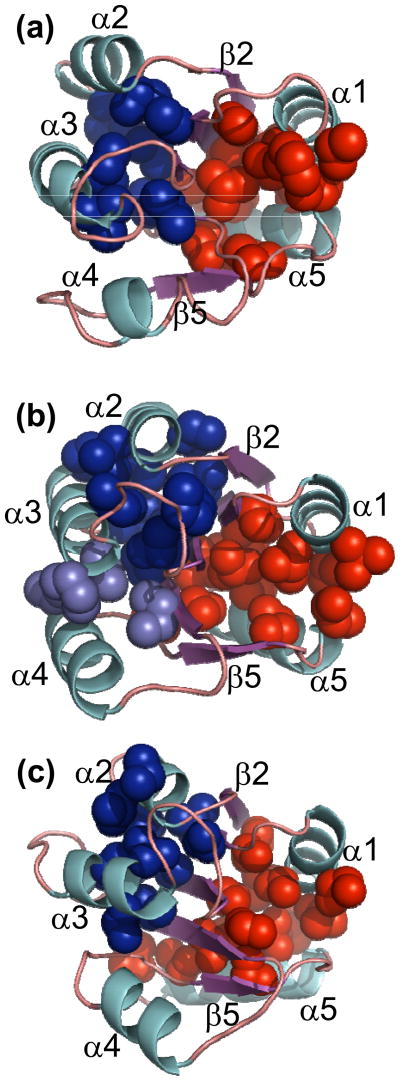
Clusters of branched aliphatic side-chain residues in (a) NT-NtrC (1DC7.pdb11), (b) Spo0F (1SRR.pdb12) and (c) CheY (3CHY.pdb10). Cartoon representation of the NMR solution structure of NT-NtrC and the crystal structures of Spo0F and CheY are shown. α-helices are colored cyan, β-strands are magenta and loops are in light orange. ILV residues that bury greater than 10 Å2 by contacting other ILV residues are highlighted, and the VDW surfaces of the heavy atoms of these residues are shown as spheres. Two major clusters of ILV residues are observed in all three proteins, one on either side of the central β-sheet. The cluster on the side facing helices α2, α3 and α4 is designated Cluster 1 and is colored blue, while the cluster on the side facing helices α1 and α5 is designated Cluster 2 and is colored red. An additional group of four ILV residues (light blue) that appears contiguous with Cluster 1 is observed in Spo0F and is considered to be a part of Cluster 1. Cluster 1 in all three proteins comprises residues that are closer in sequence that those in Cluster 2.
For NT-NtrC, a large cluster of 16 ILV side-chains from β1, β3, β4, β5, α1 and α5 and a total buried surface area (BSA) of 1219 Å2 is observed on one face of the β-sheet (Fig. 8b). A smaller cluster of eight side-chains from β1, β3, β4, α2 and α3, is also observed on the opposing face of the β-sheet. While the latter cluster only buries a total of 407 Å2 and does not reach the cut-off previously chosen to define a stable cluster, 10 side-chains and a BSA of 500 Å2,18 it was retained in the analysis for comparison with the corresponding cluster in CheY.18 Retaining the nomenclature adopted for CheY, the cluster on the α2, α3 and α4 side of the β-sheet in NT-NtrC is designated as Cluster 1 and the cluster on the α1 and α5 face of the β-sheet is designated as Cluster 2.
In Spo0F (Fig. 8c), Cluster 1 is comprised of 12 ILV side-chains from β1, β3 and β4 and helices α2 and α3, and it buries 655 Å2. Cluster 2 is comprised of 13 ILV side-chains from β1, β3 and β4 and helices α1 and α5, and it buries 731 Å2. These two clusters resemble CheY in their size and the elements of secondary structure involved. However, an additional group of four residues that is adjacent to Cluster 1 is also seen in Spo0F (Fig. 8c). While none of the individual residue-residue contacts between members of this smaller group and those in Cluster 1 bury more than the 10 Å2, previously defined for participation in a cluster,18 the total surface area buried between the two clusters is greater than 50 Å2. Moreover, the continuity of secondary structure elements between Cluster 1 (β1, α2, β3, α3 and β4) and the small cluster represented by these four residues (α3, β4 and α4) suggests that both should be considered in Cluster 1 (Fig. 8c).
In assessing the roles of these clusters in the folding reactions of NT-NtrC and Spo0F, it has proven to be useful to include information on the sequence disposition of the side-chains in the clusters. While the BSA provides a measure of the hydrophobicity and the van der Waals energy contributions to stability, the connectivity reflects the conformational entropy penalty required to form the mutual ILV contacts. The absolute contact order (ACO) algorithm, developed previously for CheY,18 is based upon earlier algorithms devised by Baker and his colleagues,3,4 and is similar in spirit to other approaches,5 and provides a useful metric for comparing the clusters in NT-NtrC, Spo0F and CheY (Table 3). The striking difference in the connectivity of Clusters 1 and Clusters 2 in all three proteins suggests that these two clusters may play different roles in the folding of these proteins. The low ACO values for Cluster 1 in all three proteins (Table 3) may drive the early formation of non-productive intermediates. The premature formation of these clusters may preclude direct folding to the native state because the side-chains in Cluster 2, on the opposing face, require the formation of non-local interactions. Although the unfavorable kinetic competition with Cluster 1 formation may preclude access to the productive TSE, the larger sizes of Cluster 2 may ultimately drive the folding reaction to the native conformation.
Table 3.
Cluster analysis of CheY-like proteins
| Protein | NT-NtrC | Spo0F | CheY16 | |||
|---|---|---|---|---|---|---|
| Cluster | Cluster 1 | Cluster 2 | Cluster 1 | Cluster 2 | Cluster 1 | Cluster 2 |
| Secondary structure elements | β1, β3, β4, α2 & α3 | β1, β3, β4, β5, α1 & α5 | β1, β2, β3, β4, α2, α3 & α4 | β1, β3, β4, β5, α1 & α5 | β1, β3, β4, α2 & α3 | β1, β2, β3, β4, β5, α1 & α5 |
| # of residues (> 50 Å2)a | 8 (4) | 15 (11) | 16 (9) | 13 (8) | 10 (7) | 15 (8) |
| BSA Total (BSA/residue)b | 407 (50.8) | 1050 (70.0) | 870 (54.4) | 731 (56.3) | 608 (60.8) | 838 (55.9) |
| ACO (BSA/contact)c | 23.33 (18.3) | 45.26 (27.2) | 23.88 (34.8) | 47.56 (29.0) | 21.64 (27.6) | 36.09 (26.2) |
Clusters represent networks of Isoleucine, Leucine and Valine side-chains that are in contact with each other and bury a surface area of 10 Å2 or more per contact.
The number of residues that bury more than 50 Å2 of their side-chain atoms by contacts within the cluster are shown in parentheses.
The average surface area of each residue buried within the cluster is shown in parentheses.
The average surface area buried by each contact between two residues is shown in parentheses.
While the sources of frustration are readily apparent in coarse-grained simulations of folding for CheY and Spo0F, the simulations do not report this behavior for NT-NtrC. It is interesting to speculate that the smaller size of Cluster 1 in NT-NtrC does not provide a sufficient contribution to the simplified potential function to be apparent in the Gō-model simulations.
Topology vs. sequence and folding energy landscapes
As has been observed previously for other motifs, 8,9,60–62 both topology and sequence contribute to defining the folding free energy landscapes for the three CheY-like proteins examined in the present study. Similar two-state thermodynamic behavior and off-pathway folding intermediates have also been observed for a pair of proteins with the closely-related flavodoxin fold.40,57 The misfolding reaction may be an inherent property of the βα-repeat motif that is a defining feature of the native conformation and, therefore, cannot be eliminated through evolution.
Similarities in the folding free energy landscape of structurally homologous proteins is expected to arise from constraints of their shared topology, which in turn may be defined by a set of conserved, structurally important residues. This core set of residues may serve not only to maintain the native state topology, but also to direct the rapid folding of the polypeptide chain to the native state. Analysis of the β-sandwich motif63 revealed a strong preference for VLIF residues at the interface between a quartet of interlocking β-strands contributed by both β-sheet layers. Mutational analysis of one member of this family, azurin, demonstrated that a sub-set of the equivalent side-chains also are involved in the TSE leading to the native state.64 A subsequent mutational analysis of another β-sandwich protein, a fibronectin domain, showed that the position of the folding nucleus can vary slightly, depending on variations in the sequence.61 In both examples, clusters of ILV residues serve to define the β-sandwich motif. A statistical analysis of several different motifs, including CheY,65 also found preferred conservation of ILV side-chains in their folding nuclei. In this case, the conservation was at the level of the group of branched beta residues, not at the level of the individual amino acids. The conservation of the group of side-chains is quite logical, given the propensity of the ILV side-chains to form clusters of a significant size that can stabilize the underlying hydrogen bond network by the preferential exclusion of water.
These and many other results show that a great deal of variation can be tolerated in sequence space without altering the topology. The sequence variations enable the development of entirely novel functional properties, a case in point being the plethora of reactions catalyzed by the TIM barrel, (βα)8, motif.66 The present study shows, however, that sequence-induced variations on topology-defined folding landscapes can result in substantial redistributions of the flow of protein through partially-folded states during the folding reaction or along folding trajectories. Thus, the variations in sequence that support functional divergence can also modulate folding mechanisms that are primarily defined by the topology.
Materials and methods
Protein expression and purification
The expression plasmid pJES820 with the gene encoding NT-NtrC was obtained from Dr. David Wemmer at UC Berkeley, and the plasmid pET20 with the gene encoding Spo0F was obtained from Dr. James A. Hoch at the Scripps Research Institute. The DNA sequence was confirmed at the UC Davis sequencing facility. The Escherichia coli strain BL21 Codonplus®(DE3)RIL was used for expression of NT-NtrC and BL21(DE3)PlysS® was used for expression of Spo0F. Both proteins were isolated from inclusion bodies by dissolving the insoluble fraction of the cell lysate in 8 M urea and refolding into 10 mM potassium phosphate buffer at pH 7.0 and 4 °C. The refolded protein was concentrated, applied to a Q Sepharose column and eluted using a salt gradient from 0 to 400 mM NaCl for NT-NtrC and 0 to 200 mM NaCl for Spo0F. Further purification was done using a Sephadex® G-75 gel filtration column in 10 mM potassium phosphate at pH 7.0. The purity was confirmed (> 98%) using nano-spray mass-spectrometry at the Proteomics Facility at the University of Massachusetts Medical School. An extinction coefficient of 14060 M−1cm−1 at 280 nm and 7000 M−1cm−1 at 275 nm was used for NT-NtrC67 and Spo0F,68 respectively, to determine the protein concentration.
Stability analysis
Samples of 10 μM NT-NtrC in 10 mM potassium phosphate at pH 7.0 were equilibrated overnight in 0 M to 8 M urea at concentration increments of 0.2 M urea. The far-UV CD spectra of each sample at 25 °C, using a 1cm cuvette in a Peltier-style thermostatted sample compartment, were recorded on a JASCO model J810 CD spectrophotometer. The CD spectra were recorded between 215 nm and 260 nm, with a band width of 2.5 nm, and a step size of 0.5 nm, integrated for 1 s and averaged over three traces. The measurements were repeated twice, and the reversibility of the reaction was confirmed by coincidence of the equilibrium transition curve obtained by starting from the unfolded state in 8 M urea. The steady-state FL emission spectra of 8 μM NT-NtrC under similar conditions to the CD equilibrium titration were recorded between 295 nm and 500 nm at a 1 nm interval, after excitation at 290 nm using a T-format Horiba Fluorolog fluorimeter. After correcting the spectra for contributions from the buffer, the transition curves at 222 nm for CD and 315 nm for FL emission were plotted as a function of urea concentration and fitted to a two-state model, N ⇆ U, where N is the native form of the protein and U is its denatured form. The free energy change associated with unfolding in the absence of denaturant was determined by assuming a linear dependence of the apparent free-energy change on the denaturant concentration.69,70
| (1) |
where ΔG°(H2O) is the standard unfolding free energy change in the absence of urea, ΔG°[Urea] is the standard unfolding free energy change at any urea concentration, [Urea], and m is its dependence on the concentration of urea.69,70 A nonlinear regression analysis module of the software Savuka16 was used to fit the data to this model. Fitting to a three-state model did not improve the fit significantly. The two-state fit was confirmed by globally fitting the FL and CD data across all wavelengths using singular value decomposition (SVD) vectors (for description, see Ionescu et al.71 and Gualfetti et al.72 and references therein). Only two significant vectors were observed.
Similar unfolding and refolding equilibrium titrations between 0 M and 8 M urea were performed for 5 μM samples of Spo0F using CD and 10 μM samples using steady state FL emission after excitation at 280 nm. The data were fitted using the method described above.
Kinetics
Fluorescence
The change in FL emission associated with refolding or unfolding of NT-NtrC was monitored using an Applied Photophysics SX 17MV instrument (dead time 2 ms). The excitation wavelength was 290 nm, and emission was monitored using a 320 cut-off filter. The relaxation times and the associated amplitudes were calculated by fitting the kinetic data to the equation
| (2) |
where A(∞) is the observed signal at infinite time, A(t) is the observed signal at time t, Ai is the signal and τi is the relaxation time associated with phase (i) and n is the number of exponentials. The kinetic data were fit to a series of exponentials using an in-house non-linear least squares fitting program, Savuka.16 The logarithm of the relaxation time was plotted as a function of final denaturant concentration in the form of a chevron analysis.73
Refolding
NT-NtrC was equilibrated overnight in 7.4 M urea and 10 mM potassium phosphate at pH 7.0 and at 25 °C and refolded by rapid mixing into refolding buffer with varying final concentrations of denaturant (1 M urea to 4 M urea), and 10 μM final protein concentration. The same experiments were performed with 10 μM Spo0F equilibrated in 6 M urea and 10 mM potassium phosphate at pH 7.0 and at 25 °C
Unfolding
NT-NtrC was unfolded by rapid mixing into high concentration of urea buffered with 10 mM potassium phosphate at pH 7.0 and 25 °C, to final urea concentrations ranging from 2.5 M to 8.0 M, and 10 μM protein. The same experiments were performed with 10 μM Spo0F in 10 mM potassium phosphate at pH 7.0 and at 25 °C.
Circular dichroism
Refolding was also monitored by the far-UV CD ellipticity at 222 nm using an AVIV model 202 stopped-flow CD spectrophotometer (dead time 5 ms) and a JASCO model J810 CD spectrophotometer (manual mixing dead time ~10 s). The conditions for the experiments were as described above, and the data were fitted by the same method used for FL emission.
Stability of burst-phase intermediate
The refolding kinetics upon 10-fold dilution of 100 μM protein unfolded with 6 M urea into refolding buffer were monitored by CD at 222 nm, buffer-corrected and extrapolated to 0 s to determine the signal associated with the burst-phase intermediate. The amplitude of the signal was then plotted against the final denaturant concentration. The sigmoidal unfolding curve was then fitted to a two-state model, IBP ⇆ U, where IBP is the burst-phase intermediate and U is the denatured form. The free energy change associated with the unfolding of the intermediate in 0 M urea and its dependence on urea concentration was determined by the method described above.
Global analysis
Both the CD and the FL kinetic traces from the refolding, unfolding and double jump experiments were fit globally to several different models. The Levenberg-Marquart method74 was then used to obtain the best fit to the kinetic data. Details of the methods used are described previously.18,75
Analysis of hydrophobic clusters
The contact surface area between atoms was calculated using the CSU software developed by Sobolev et al.76 The method for analysis of hydrophobic ILV clusters has been described previously.18 The application of the Absolute Contact Order4 (ACO) algorithm to the ILV clusters is also described in earlier work.18
Coarse-grained simulations
Gō-model simulations were performed with NT-NtrC and Spo0F using the coarse-grained model developed by Karanicolas and Brooks,77 previously developed for the study of CheY folding.19 Briefly, the protein backbone is represented as a string of beads connected by virtual bonds. Each bead represents a single amino acid and is located at the α-carbon position. Bond lengths are kept fixed, bond angles are subject to a harmonic restraint, and dihedral angles are subject to potentials representing sequence-dependent flexibility and conformational preferences in Ramachandran space. Nonbonded interactions are represented using a Gō-model in which only residues that are in contact in the native state (taken to be structures 1DC7.pdb11 and 1SRR.pdb12 for NT-NtrC and Spo0F, respectively) interact favorably. Backbone hydrogen bonds and side-chain pairs with non-hydrogen atoms separated by less than 4.5 Å interact via a pairwise 6-10-12 potential that consists of an energy well and a small desolvation barrier. To incorporate sequence effects, the interaction energies of side-chain native contacts are scaled according to their abundance in the Protein Data Bank as reported by Miyazawa and Jernigan.78 Residues not in contact in the native state interact via a repulsive volume exclusion term. A complete description of the model potential and its parameters can be found in Karanicolas and Brooks.77
Molecular dynamics simulations were performed in Cartesian space using CHARMM79 within the gorex.pl module of the MMTSB Tool Set.80 Langevin dynamics with a 1.36 ps−1 friction coefficient was used to maintain thermal equilibrium, and the time step was set at 22 fs. For kinetic folding simulations, 100 independent runs were each performed for 2×108 dynamics steps at a temperature highly favoring the native state, namely at 0.87 Tf, where Tf is the folding transition temperature defined by the maximum in the heat capacity curve, Cv(T). Note that absolute timescales cannot be obtained due to the coarse-grained nature of the Gō-model and the lack of explicit solvent molecules. Unfolded starting structures for the folding runs were generated by equilibration at 1.5 Tf for 107 dynamics steps starting from randomly assigned initial velocities. Conformational snapshots were recorded every 105 dynamics steps. The fraction of native contacts formed, Q, was used to monitor folding progress. Each contact was considered formed if its residue pair was within a cutoff distance chosen such that the given contact is satisfied 85% of the time in native state simulations at 0.83 Tf.
To characterize the entire accessible free energy landscape, a two-dimensional extension of replica-exchange molecular dynamics81 was performed. Each replica was assigned one of four temperatures (0.87, 0.97, 1.08 or 1.20 Tf) and one of seven harmonic biasing restraints on the radius of gyration, Rg, for a total of 28 replicas. To ensure overlap between the Rg distributions harmonic potentials were used with minima at 1.0, 1.1. 1.2, 1.3, 1.5, 1.7 and 2.0 , where is the radius of gyration of the native state, with force constants 0.5, 5.0, 5.0, 5.0, 4.0, 0.8 and 0.5 kcal/mol-Å2, respectively. Stronger restraints were required at intermediate radii to sample the high energy transition region between the unfolded and native states. Conformational exchanges between temperature windows and restraints were attempted every 40,000 dynamics steps, and the snapshots were recorded. The exchange frequency remained between ~10% and 40% throughout the 6×108-step simulation. Finally, conformations were combined from all 28 replicas for a total of 4.2×105 structures, and the multidimensional weighted histogram analysis method82,83 was used to obtain the unbiased free energy at Tf projected along various progress coordinates. The above procedure was carried out in its entirety for both NT-NtrC and Spo0F.
Supplementary Material
Acknowledgments
Simulations were performed using the modeling package available through the NIH resource (RR12255) Multiscale Modeling Tools for Structural Biology (http://mmtsb.org). The authors would like to thank Dr. David Wemmer and James A. Hoch for generously providing the plasmids with the genes encoding NT-NtrC and Spo0F, respectively. R.D. Hills Jr. thanks the La Jolla Interfaces in Science Training Program for financial support, the Center for Theoretical Biological Physics (www.ctbp.ucsd.edu) for providing a stimulating intellectual environment and D.A. Case for providing laboratory space. S.V. Kathuria thanks Osman Bilsel for guidance in the global analysis, Can Kayatekin and J.A. Zitzewitz for valuable discussions and their help with preparation of the manuscript. This work was supported by NIH grant GM48807 to C.L. Brooks III and NSF grants MCB0327504 & MCB0721312 to C.R. Matthews.
Abbreviations used
- NT-NtrC
124 residue, amino-terminal receiver domain of nitrogen regulation protein NtrC from Salmonella typhimurium
- ILV
isoleucine, leucine and valine
- TSE
transition state ensemble
- FL
fluorescence spectroscopy
- IBP
kinetic intermediate populated within the stopped-flow burst-phase (<5 ms)
- N
native state
- U
unfolded state
- BASiC
branched aliphatic side-chain
- ACO
absolute contact order
- BSA
buried surface area
- TIM
triosephosphate isomerase
- αTS
α subunit of tryptophan synthase from Escherichia coli
- IGPS
indole-3-glycerol phosphate synthase
- eIGPS
IGPS from Escherichia coli
- sIGPS
IGPS from Sulfolobus solfataricus
- IOLI
the 278 residue TIM barrel protein of unknown function encoded by the Bacillus subtilis iolI gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anfinsen CB, Scheraga HA. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- 2.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 4.Ivankov DN, Garbuzynskiy SO, Alm E, Plaxco KW, Baker D, Finkelstein AV. Contact order revisited: Influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamagata K, Kuwajima K. Surprisingly high correlation between early and late stages in non-two-state protein folding. J Mol Biol. 2006;357:1647–1654. doi: 10.1016/j.jmb.2006.01.072. [DOI] [PubMed] [Google Scholar]
- 6.Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 7.Scott KA, Batey S, Hooton KA, Clarke J. The folding of spectrin domains I: wild-type domains have the same stability but very different kinetic properties. J Mol Biol. 2004;344:195–205. doi: 10.1016/j.jmb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 8.Wensley BG, Gartner M, Choo WX, Batey S, Clarke J. Different members of a simple three-helix bundle protein family have very different folding rate constants and fold by different mechanisms. J Mol Biol. 2009;390:1074–1085. doi: 10.1016/j.jmb.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friel CT, Capaldi AP, Radford SE. Structural analysis of the rate-limiting transition states in the folding of Im7 and Im9: similarities and differences in the folding of homologous proteins. J Mol Biol. 2003;326:293–305. doi: 10.1016/s0022-2836(02)01249-4. [DOI] [PubMed] [Google Scholar]
- 10.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 11.Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 12.Madhusudan, Zapf J, Whiteley JM, Hoch JA, Xuong NH, Varughese KI. Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis. Structure. 1996;4:679–690. doi: 10.1016/s0969-2126(96)00074-3. [DOI] [PubMed] [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Wallace LA, Robert Matthews C. Highly divergent dihydrofolate reductases conserve complex folding mechanisms. J Mol Biol. 2002;315:193–211. doi: 10.1006/jmbi.2001.5230. [DOI] [PubMed] [Google Scholar]
- 15.Forsyth WR, Matthews CR. Folding mechanism of indole-3-glycerol phosphate synthase from Sulfolobus solfataricus: a test of the conservation of folding mechanisms hypothesis in (beta(alpha))(8) barrels. J Mol Biol. 2002;320:1119–1133. doi: 10.1016/s0022-2836(02)00557-0. [DOI] [PubMed] [Google Scholar]
- 16.Forsyth WR, Bilsel O, Gu Z, Matthews CR. Topology and sequence in the folding of a TIM barrel protein: global analysis highlights partitioning between transient off-pathway and stable on-pathway folding intermediates in the complex folding mechanism of a (betaalpha)8 barrel of unknown function from B. subtilis. J Mol Biol. 2007;372:236–253. doi: 10.1016/j.jmb.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Bollen YJM, van Mierlo CPM. Protein topology affects the appearance of intermediates during the folding of proteins with a flavodoxin-like fold. Biophys Chem. 2005;114:181–189. doi: 10.1016/j.bpc.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Kathuria SV, Day IJ, Wallace LA, Matthews CR. Kinetic traps in the folding of beta/alpha-repeat proteins: CheY initially misfolds before accessing the native conformation. J Mol Biol. 2008;382:467–484. doi: 10.1016/j.jmb.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 19.Hills RD, Jr, Brooks CL., III Subdomain competition, cooperativity, and topological frustration in the folding of CheY. J Mol Biol. 2008;382:485–495. doi: 10.1016/j.jmb.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Hernandez E, Serrano L. Structure of the transition state for folding of the 129 aa protein CheY resembles that of a smaller protein, CI-2. Fold Des. 1996;1:43–55. [PubMed] [Google Scholar]
- 22.Formaneck MS, Ma L, Cui Q. Reconciling the “old” and “new” views of protein allostery: A molecular simulation study of chemotaxis Y protein (CheY) Proteins. 2006;63:846–867. doi: 10.1002/prot.20893. [DOI] [PubMed] [Google Scholar]
- 23.Lee SY, Cho HS, Pelton JG, Yan DL, Henderson RK, King DS, Huang LS, Kustu S, Berry EA, Wemmer DE. Crystal structure of an activated response regulator bound to its target. Nat Struct Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 24.Stock AM, Guhaniyogi J. A new perspective on response regulator activation. J Bacteriol. 2006;188:7328–7330. doi: 10.1128/JB.01268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson ED, Grishin NV. Alternate pathways for folding in the flavodoxin fold family revealed by a nucleation-growth model. J Mol Biol. 2006;358:646–653. doi: 10.1016/j.jmb.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Sola M, Lopez-Hernandez E, Cronet P, Lacroix E, Serrano L, Coll M, Parraga A. Towards understanding a molecular switch mechanism: Thermodynamic and crystallographic studies of the signal transduction protein CheY. J Mol Biol. 2000;303:213–225. doi: 10.1006/jmbi.2000.4507. [DOI] [PubMed] [Google Scholar]
- 27.Zhu XY, Rebello J, Matsumura P, Volz K. Crystal structures of CheY mutants Y106W and T871/Y106W - CheY activation correlates with movement of residue 106. J Biol Chem. 1997;272:5000–5006. doi: 10.1074/jbc.272.8.5000. [DOI] [PubMed] [Google Scholar]
- 28.De Carlo S, Chen BY, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu XH, Wang YM. Molecular dynamic simulations of the N-terminal receiver domain of NtrC reveal intrinsic conformational flexibility in the inactive state. J Biomol Struct Dyn. 2006;23:509–517. doi: 10.1080/07391102.2006.10507075. [DOI] [PubMed] [Google Scholar]
- 30.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 31.Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dyer CM, Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonovic M, Volz K. A distinct meta-active conformation in the 1.1-angstrom resolution structure of wild-type apoCheY. J Biol Chem. 2001;276:28637–28640. doi: 10.1074/jbc.C100295200. [DOI] [PubMed] [Google Scholar]
- 34.Gardino AK, Volkman BF, Cho HS, Lee SY, Wemmer DE, Kern D. The NMR solution structure of BeF3--activated Spo0F reveals the conformational switch in a phosphorelay system. J Mol Biol. 2003;331:245–254. doi: 10.1016/s0022-2836(03)00733-2. [DOI] [PubMed] [Google Scholar]
- 35.Madhusudan, Zapf J, Hoch JA, Whiteley JM, Xuong NH, Varughese KI. A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: Crystal structure of Spo0F from Bacillus subtilis. Biochemistry. 1997;36:12739–12745. doi: 10.1021/bi971276v. [DOI] [PubMed] [Google Scholar]
- 36.Varughese KI, Tsigelny I, Zhao HY. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J Bacteriol. 2006;188:4970–4977. doi: 10.1128/JB.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Vadrevu R, Kathuria S, Yang X, Matthews CR. A tightly packed hydrophobic cluster directs the formation of an off-pathway sub-millisecond folding intermediate in the alpha subunit of tryptophan synthase, a TIM barrel protein. J Mol Biol. 2007;366:1624–1638. doi: 10.1016/j.jmb.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz V, Lopez EM, Jager M, Serrano L. Kinetic characterization of the chemotactic protein from Escherichia coli, CheY. Kinetic analysis of the inverse hydrophobic effect. Biochemistry. 1994;33:5858–5866. doi: 10.1021/bi00185a025. [DOI] [PubMed] [Google Scholar]
- 39.Otzen DE, Oliveberg M. Salt-induced detour through compact regions of the protein folding landscape. Proc Natl Acad Sci U S A. 1999;96:11746–11751. doi: 10.1073/pnas.96.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez-Recio J, Genzor CG, Sancho J. Apoflavodoxin folding mechanism: An alpha/beta protein with an essentially off-pathway lntermediate. Biochemistry. 2001;40:15234–15245. doi: 10.1021/bi010216t. [DOI] [PubMed] [Google Scholar]
- 41.Du R, Pande VS, Grosberg AY, Tanaka T, Shakhnovich ES. On the transition coordinate for protein folding. J Chem Phys. 1998;108:334–350. [Google Scholar]
- 42.Snow CD, Rhee YM, Pande VS. Kinetic definition of protein folding transition state ensembles and reaction coordinates. Biophys J. 2006;91:14–24. doi: 10.1529/biophysj.105.075689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juraszek J, Bolhuis PG. Rate constant and reaction coordinate of Trp-cage folding in explicit water. Biophys J. 2008;95:4246–4257. doi: 10.1529/biophysj.108.136267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho SS, Levy Y, Wolynes PG. P versus Q: Structural reaction coordinates capture protein folding on smooth landscapes. Proc Natl Acad Sci U S A. 2006;103:586–591. doi: 10.1073/pnas.0509768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karanicolas J, Brooks CL., III Improved Go-like models demonstrate the robustness of protein folding mechanisms towards non-native interactions. J Mol Biol. 2003;334:309–325. doi: 10.1016/j.jmb.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 46.Rey-Stolle MF, Enciso M, Rey A. Topology-based models and NMR structures in protein folding simulations. J Comput Chem. 2009;30:1212–1219. doi: 10.1002/jcc.21149. [DOI] [PubMed] [Google Scholar]
- 47.Prieto L, Rey A. Simulations of the protein folding process using topology-based models depend on the experimental structure. J Chem Phys. 2008;129:115101. doi: 10.1063/1.2977744. [DOI] [PubMed] [Google Scholar]
- 48.Chavez LL, Gosavi S, Jennings PA, Onuchic JN. Multiple routes lead to the native state in the energy landscape of the beta-trefoil family. Proc Natl Acad Sci U S A. 2006;103:10254–10258. doi: 10.1073/pnas.0510110103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gosavi S, Chavez LL, Jennings PA, Onuchic JN. Topological frustration and the folding of interleukin-1 beta. J Mol Biol. 2006;357:986–996. doi: 10.1016/j.jmb.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 50.Gosavi S, Whitford PC, Jennings PA, Onuchic JN. Extracting function from a beta-trefoil folding motif. Proc Natl Acad Sci U S A. 2008;105:10384–10389. doi: 10.1073/pnas.0801343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finke JM, Onuchic JN. Equilibrium and kinetic folding pathways of a TIM barrel with a funneled energy landscape. Biophys J. 2005;89:488–505. doi: 10.1529/biophysj.105.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu Z, Rao MK, Forsyth WR, Finke JM, Matthews CR. Structural analysis of kinetic folding intermediates for a TIM barrel protein, indole-3-glycerol phosphate synthase, by hydrogen exchange mass spectrometry and Go model simulation. J Mol Biol. 2007;374:528–546. doi: 10.1016/j.jmb.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jakob RP, Schmid FX. Energetic coupling between native-state prolyl isomerization and conformational protein folding. J Mol Biol. 2008;377:1560–1575. doi: 10.1016/j.jmb.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 54.Jakob RP, Schmid FX. Molecular determinants of a native-state prolyl isomerization. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 55.Fowler SB, Clarke J. Mapping the folding pathway of an immunoglobulin domain: structural detail from Phi value analysis and movement of the transition state. Structure. 2001;9:355–366. doi: 10.1016/s0969-2126(01)00596-2. [DOI] [PubMed] [Google Scholar]
- 56.Gu Z, Zitzewitz JA, Matthews CR. Mapping the structure of folding cores in TIM barrel proteins by hydrogen exchange mass spectrometry: the roles of motif and sequence for the indole-3-glycerol phosphate synthase from Sulfolobus solfataricus. J Mol Biol. 2007;368:582–594. doi: 10.1016/j.jmb.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bollen YJM, Kamphuis MB, van Mierlo CPM. The folding energy landscape of apoflavodoxin is rugged: Hydrogen exchange reveals nonproductive misfolded intermediates. Proc Natl Acad Sci U S A. 2006;103:4095–4100. doi: 10.1073/pnas.0509133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radzicka A, Wolfenden R. Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry. 1988;27:1664–1670. [Google Scholar]
- 59.Schell D, Tsai J, Scholtz JM, Pace CN. Hydrogen bonding increases packing density in the protein interior. Proteins. 2006;63:278–282. doi: 10.1002/prot.20826. [DOI] [PubMed] [Google Scholar]
- 60.Olofsson M, Hansson S, Hedberg L, Logan DT, Oliveberg M. Folding of S6 structures with divergent amino acid composition: pathway flexibility within partly overlapping foldons. J Mol Biol. 2007;365:237–248. doi: 10.1016/j.jmb.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 61.Lappalainen I, Hurley MG, Clarke J. Plasticity within the obligatory folding nucleus of an immunoglobulin-like domain. J Mol Biol. 2008;375:547–559. doi: 10.1016/j.jmb.2007.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lam AR, Borreguero JM, Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich E. Parallel foldng pathways in the SH3 domain protein. J Mol Biol. 2007;373:1348–1360. doi: 10.1016/j.jmb.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 63.Kister AE, Finkelstein AV, Gelfand IM. Common features in structures and sequences of sandwich-like proteins. Proc Natl Acad Sci U S A. 2002;99:14137–14141. doi: 10.1073/pnas.212511499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson CJ, Wittung-Stafshede P. Snapshots of a dynamic folding nucleus in zinc-substituted Pseudomonas aeruginosa azurin. Biochemistry. 2005;44:10054–10062. doi: 10.1021/bi050342n. [DOI] [PubMed] [Google Scholar]
- 65.Mirny L, Shakhnovich E. Evolutionary conservation of the folding nucleus. J Mol Biol. 2001;308:123–129. doi: 10.1006/jmbi.2001.4602. [DOI] [PubMed] [Google Scholar]
- 66.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 67.Nohaile M, Kern D, Wemmer D, Stedman K, Kustu S. Structural and functional analyses of activating amino acid substitutions in the receiver domain of NtrC: evidence for an activating surface. J Mol Biol. 1997;273:299–316. doi: 10.1006/jmbi.1997.1296. [DOI] [PubMed] [Google Scholar]
- 68.Zapf JW, Hoch JA, Whiteley JM. A phosphotransferase activity of the Bacillus subtilis sporulation protein Spo0F that employs phosphoramidate substrates. Biochemistry. 1996;35:2926–2933. doi: 10.1021/bi9519361. [DOI] [PubMed] [Google Scholar]
- 69.John AS. Solvent denaturation. Biopolymers. 1978;17:1305–1322. [Google Scholar]
- 70.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 71.Ionescu RM, Smith VF, O’Neill JC, Jr, Matthews CR. Multistate equilibrium unfolding of Escherichia coli dihydrofolate reductase: thermodynamic and spectroscopic description of the native, intermediate, and unfolded ensembles. Biochemistry. 2000;39:9540–9550. doi: 10.1021/bi000511y. [DOI] [PubMed] [Google Scholar]
- 72.Gualfetti PJ, Bilsel O, Matthews CR. The progressive development of structure and stability during the equilibrium folding of the alpha subunit of tryptophan synthase from Escherichia coli. Protein Sci. 1999;8:1623–1635. doi: 10.1110/ps.8.8.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matthews CR. Effect of point mutations on the folding of globular proteins. Methods Enzymol. 1987;154:498–511. doi: 10.1016/0076-6879(87)54092-7. [DOI] [PubMed] [Google Scholar]
- 74.Marquardt DW. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. Journal of the Society for Industrial and Applied Mathematics. 1963;11:431–441. [Google Scholar]
- 75.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Folding mechanism of the alpha-subunit of tryptophan synthase, an alpha/beta barrel protein: global analysis highlights the interconversion of multiple native, intermediate, and unfolded forms through parallel channels. Biochemistry. 1999;38:1018–1029. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
- 76.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 77.Karanicolas J, Brooks CL., III The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miyazawa S, Jernigan RL. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 79.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM - A Program For Macromolecular Energy, Minimization, And Dynamics Calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 80.Feig M, Karanicolas J, Brooks CL., III MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 81.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 82.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method For Free-Energy Calculations On Biomolecules. 1 The Method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 83.Gallicchio E, Andrec M, Felts AK, Levy RM. Temperature weighted histogram analysis method, replica exchange, and transition paths. J Phys Chem B. 2005;109:6722–6731. doi: 10.1021/jp045294f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.