Abstract
The innate immune response is critical for host defence against respiratory coronaviruses (CoVs). This study demonstrated that an ongoing respiratory virus infection compromises innate immune responses and affects the pathogenesis of a respiratory CoV co-infection. An innate immunosuppressive respiratory virus infection was established by infecting weaned pigs with porcine reproductive and respiratory syndrome virus (PRRSV); 10 days later, the pigs were exposed to porcine respiratory coronavirus (PRCV). The PRRSV/PRCV dual-infected pigs had reduced weight gains, a higher incidence of fever and more severe pneumonia compared with either single infection. Significant suppression of innate immune responses [reduced alpha interferon (IFN-α) levels in the lungs and reduced blood natural killer cell cytotoxicity] by the ongoing PRRSV infection was observed in dual-infected pigs, which coincided with exacerbated pneumonia during early PRCV infection. The subsequent PRCV infection led to enhanced PRRSV replication in the lungs and a trend towards increased serum T-helper type 1 (Th1) (IFN-γ) but decreased Th2 [interleukin (IL)-4] responses, further exacerbating PRRSV pneumonia. Following PRCV infection, more severe PRRSV-related pulmonary alveolar macrophage (PAM) apoptosis occurred, as determined by an in situ terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling assay, suggesting increased PRRSV replication in PAMs. Collectively, these observations suggest interactive effects between PRCV and PRRSV via early innate (IFN-α) and later adaptive Th1 (IFN-γ) and Th2 (IL-4) immune responses. These findings imply that an existing immunomodulating respiratory viral co-infection may be a contributing factor to more severe pneumonia in respiratory CoV disease. This study provides new insights into host–pathogen interactions related to co-infection by CoVs and other respiratory viruses.
INTRODUCTION
The innate immune response is critical for host defence against respiratory coronaviruses (CoVs) (Charley et al., 2006; Frieman et al., 2008; Thiel & Weber, 2008). Most CoVs are sensitive to the antiviral effects of virus-induced alpha/beta interferon (IFN-α/β). Specifically, the group 1 CoVs in the family Coronaviridae, order Nidovirales, porcine respiratory coronavirus (PRCV) and transmissible gastroenteritis virus (TGEV), are potent IFN-α inducers (Charley et al., 2006; Van Reeth et al., 1999; Zhang et al., 2008). The recently identified human CoV, severe acute respiratory syndrome coronavirus (SARS)-CoV, which belongs to subgroup 2b of the group 2 CoVs (Kuiken et al., 2003; Lau et al., 2005; Saif, 2004), also elicits type I IFNs, but may also evade their antiviral activity (Frieman et al., 2008; Thiel & Weber, 2008). The innate immune responses of the host against respiratory viruses involve alveolar macrophages, pulmonary epithelial cells, natural killer (NK) cells, dendritic cells and IFN-α/β responses in the lung. These responses influence the initial virus infection and also regulate adaptive immune responses. We hypothesized that innate immunity, as compromised by a prior respiratory viral infection, might alter the pathogenesis of a respiratory CoV.
Co-infections by respiratory group 1 CoVs, such as PRCV and human CoV-229E, with other respiratory viruses have frequently been identified in swine and humans, respectively (Canducci et al., 2008; Kamogawa et al., 1996). In addition, in group 2 SARS-CoV disease cases, respiratory viral co-infections, including human metapneumovirus, cytomegalovirus and reovirus, are common in patients (Chan et al., 2003; He et al., 2006; Hwang et al., 2005; Kuiken et al., 2003; Louie et al., 2004). However, it is unknown how these co-infections influence CoV disease outcome, and whether the co-infections actually enhance CoV shedding in co-infected hosts (‘super-spreaders’). There is a dearth of information on the mechanisms of interactions between respiratory virus co-infections and their effect on disease outcome in outbred hosts (cf. inbred mouse models), mimicking the heterogeneity of the human population.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a member of the family Arteriviridae and, like CoVs, belongs to the order Nidovirales. It induces reproductive disorders such as abortion in pregnant sows, as well as severe respiratory tract disease in young pigs (Mateu & Diaz, 2008; Rossow, 1998). The virus attenuates innate immune responses, evades the antiviral cytokine (IFN-α) response, and blocks IFN-α production in the cytoplasm of infected alveolar macrophages (Mateu & Diaz, 2008). Subsequently, adaptive immune responses are compromised, leading to weak cell-mediated immune responses, the delayed appearance of neutralizing antibody, often prolonged viraemia and persistent infection of pigs (Mateu & Diaz, 2008). The immunosuppression induced may prolong PRRSV pathogenesis and enhance the severity of other respiratory viral co-infections (Mateu & Diaz, 2008; Rossow, 1998; Van Reeth et al., 1996). In addition, PRRSV predisposes pigs to co-infection by other respiratory viruses, possibly due to compromised immunity and the destruction of pulmonary tissues or cells following PRRSV infection; however, the mechanisms involved remain obscure (Mateu & Diaz, 2008; Rossow, 1998). Therefore, we used PRRSV as a common respiratory virus of swine to induce compromised innate immunity in the lung microenvironment and to reflect co-infections by other common respiratory viruses observed in humans as well as in swine.
Porcine respiratory coronavirus, a spike gene deletion mutant of TGEV that causes acute epidemic diarrhoea in neonatal piglets, is classified as a group 1 CoV (Saif, 2004). PRCV causes mainly upper and lower respiratory tract disease, infects type 1 and 2 pneumocytes, and induces acute pulmonary cell damage (Cox et al., 1990; Halbur et al., 1993; Jabrane et al., 1994; Jung et al., 2007). Even up to 10 days after PRCV infection of lung tissues, the virus simultaneously induces inflammatory (cell necrotizing) and proliferative (alveolar septa thickening) pneumonia, i.e. chronic–active bronchointerstitial pneumonia (Jung et al., 2007). PRCV also stimulates production of IFN-α in the lungs and thus promotes local innate immune responses, similar to those seen in SARS in humans (Charley et al., 2006; Frieman et al., 2008; Thiel & Weber, 2008; Van Reeth et al., 1999; Zhang et al., 2008).
In this study, we examined the effect of a pre-existing and ongoing PRRSV infection (a downregulator of innate immunity) on PRCV (which, in contrast to PRRSV, is an upregulator of innate immunity, i.e. IFN-α) infection and disease outcome, and also clarified the influence of PRCV co-infection on the persisting or ongoing PRRSV disease. Ultimately, we investigated whether a CoV co-infection with other respiratory viruses enhances pneumonia and disease compared with the single infection.
METHODS
Viruses and cells.
The North American SD23983 strain of PRRSV was passaged five times in MARC-145 cells. Titres of the strain were expressed as 50 % tissue culture infectious dose (TCID50) ml−1 on MARC-145 cells. The ISU-1 strain of PRCV was plaque purified twice and passaged 14 times in swine testicle (ST) cells. Virus titres were determined by plaque assay as p.f.u. ml−1, as described previously (Jung et al., 2007).
Animals.
Specific-pathogen-free, 20–25-day-old, Large White×Duroc crossbred weaned pigs (n=178) were obtained from a swine herd of the Ohio Agricultural Research and Development Center (The Ohio State University, Wooster, OH, USA). The animals were serologically negative for PRRSV, PRCV, TGEV and porcine circovirus type 2, and were used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Ohio State University.
Experimental animal infection.
Pigs were randomly assigned to one of four groups: PRRSV/PRCV (n=48), PRRSV alone (n=39), PRCV alone (n=48), and mock (n=43). To establish subsequent PRCV infection in ongoing PRRSV infection of pigs, they were first inoculated intranasally with 3×104 TCID50 and intramuscularly with 2×104 TCID50 PRRSV, or were mock inoculated, and 10 days later (when PRRSV pneumonia was most severe and virus nasal shedding and viraemia were undetectable), they were subsequently inoculated intranasally with 4×106 p.f.u. and intratracheally via a bronchoscope with 6×106 p.f.u. PRCV, or were mock inoculated. All inoculation procedures were performed using aseptic techniques. After PRRSV inoculation, we monitored clinical signs and body weight gain every other day, as described previously (Jung et al., 2007). Before inoculation [post-inoculation day (PID) −2 ] and at early (PIDs 2 and 4), middle (PIDs 8 and 10) and late (PIDs 14 and 21) stages of PRCV infection, five to eight pigs per group were euthanized by electrocution as recommended by the 2000 American Veterinary Medical Association (AVMA) panel report (AVMA Panel on Euthanasia, 2001).
Gross and histological analysis.
Necropsies were performed on all of the pigs and tissues/organs [lung, heart, tonsil, kidney, spleen, pancreas, liver, tracheobronchial lymph nodes (TBLNs), inguinal LNs and intestines] were examined grossly and histologically. Gross and histological pulmonary lesions were given an estimated score based on the percentage of macroscopic lesions (consolidation) in all lobes and on the distribution and severity of histopathology, respectively, as described previously (Jung et al., 2007). Sections of lung tissue were obtained randomly from the cranial, middle and caudal lung lobes. To evaluate the effect of co-infection with PRRSV and PRCV on lymphadenopathy, the left TBLNs from each pig were weighed, and lymph node to body weight (LN : BW) ratios were calculated as described previously (Jung et al., 2007).
Determination of viral lung titres by quantitative (q)RT-PCR.
Lung homogenates from each pig were tested by real-time qRT-PCR for PRRSV and PRCV RNA quantification as described previously (Jung et al., 2007; Molina et al., 2008). For PRRSV, all samples were tested at the Animal Disease Research and Diagnostic Laboratory (Veterinary Science Department, South Dakota State University, SD, USA). For PRCV, viral RNA was extracted using an RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. cDNA was generated using random primers (Promega) and avian myeloblastosis virus reverse transcriptase (Promega). Real-time qRT-PCR was performed using a LightCycler DNA Master SYBR Green I kit (Roche Diagnostics) in a SmartCycler real-time machine (Cepheid). A standard curve was developed using tenfold serial dilutions of the stock PRCV. The standard curve was used to convert Ct values for each specimen to p.f.u. ml−1 equivalents.
Immunohistochemistry.
Paraffin-embedded lung tissues were prepared and evaluated by immunohistochemistry (IHC) for PRRSV and PRCV antigen detection as described previously (Jung et al., 2007). For PRRSV, SDOW-17 monoclonal antibody against the PRRSV nucleocapsid protein was provided by Y. Fang (Veterinary Science Department, South Dakota State University, USA) and used for PRRSV antigen detection. For PRCV, three monoclonal antibodies (all diluted 1 : 200) against the nucleocapsid proteins (25H7 and 14E3) and the spike protein (25C9) of TGEV, which cross-react with PRCV, were used (Jung et al., 2007). PRRSV or PRCV antigen-positive scores were computed by estimating the number of IHC-positive cells in the lung section per microscopic area, at ×200 magnification based on the following criteria: 0, no positive cells; 1, a few positive cells (<10); 2, a moderate number of positive cells (11–25); 3, a high number of positive cells (>26).
NK cell function assay.
The cytotoxic function of NK cells (effectors) isolated from peripheral blood mononuclear cells (PBMCs) of virus- or mock-infected pigs was evaluated by a lactate dehydrogenase (LDH) release assay as described previously (Korzeniewski & Callewaert, 1983). K-562 (human myeloblastoid cell line) or Yac-1 (mouse T lymphoma cell line) cells were used as target cells. Effectors and targets at different ratios were co-cultured and the supernatants were harvested after 24 h. The amount of LDH released was measured using LDH substrate and the absorbance evaluated at 490 nm. For each treatment group, the mean percentage NK cytotoxicity (%) at an effector : target ratio of 100 : 1 on different PIDs was determined.
Cytokine analysis.
Innate cytokines (IFN-α) in lung lysates and T-helper type 1 (Th1) (IFN-γ) and Th2 [interleukin (IL)-4] cytokines in serum were evaluated by antigen-capture ELISA as described previously (Azevedo et al., 2006).
In situ terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) assay.
Paraffin-embedded lung tissues were prepared as described above and evaluated by an in situ TUNEL assay kit (Roche Applied Science) for apoptosis according to the manufacturer's instructions. The severity of apoptosis was estimated based on the distribution and number of in situ TUNEL-positive cells in the lung per microscopic area, at ×50 magnification: −, no positive cells; +, a few positive cells; ++, a moderate number of positive cells; +++, a high number of positive cells.
PRCV nasal shedding titre analysis.
Nasal swabs were collected every other day from each animal throughout the experiment (PRRSV PIDs 0–31). The swabs were placed in 4 ml minimal essential medium supplemented with antibiotic/antimycotic. The samples were tested using a cell culture immunofluorescence test as described previously (Jung et al., 2007). Fourfold serial dilutions of the nasal swab supernatants were inoculated onto ST cells grown in 96-well tissue culture plates and incubated for 18 h at 37 °C in 5 % CO2. The cells were fixed with 80 % acetone, stained with hyperimmune porcine anti-TGEV serum conjugated to fluorescein isothiocyanate, analysed by fluorescence microscopy and expressed as fluorescent focus units (f.f.u.) ml−1.
Virus-specific antibody titre analysis in serum and bronchoalveolar lavage (BAL).
Serum virus neutralizing (VN) antibody titres against PRCV were evaluated by a plaque-reduction test as described previously (VanCott et al., 1993). Antibody titres were expressed as the reciprocal of the specimen dilution that resulted in an 80 % reduction in the number of plaques. PRCV-specific IgA antibody titres in BAL fluids were tested by a semi-quantitative ELISA method as described previously (Du et al., 2008), with slight modifications. PRRSV serum antibodies were identified by a commercial PRRSV ELISA (Idexx Laboratories) and evaluated based on progression of the sample : positive (S : P) ratios.
Statistical analysis.
All values were expressed as means±sem. Data from the four treatment groups (PRRSV/PRCV, PRRSV only, PRCV only and mock) were analysed and compared by a Kruskal–Wallis test (non-parametric) using sas software (SAS Institute Inc.). A value of P<0.05 was considered statistically significant. Fischer's exact test was used to evaluate the proportion of pigs with fever and decreased body weight gain between the PRRSV/PRCV-infected and PRRSV only-infected pigs.
RESULTS
Ongoing PRRSV and subsequent PRCV co-infection exacerbates clinical illness compared with PRRSV or PRCV alone
Rectal temperatures of pigs ≥39.5 °C were considered to be febrile responses. PRRSV induced recurrent and persisting fever and various clinical signs, such as sneezing, coughing, polypnoea and anorexia, throughout the study period, whereas PRCV caused only mild sneezing and coughing without fever. After the subsequent PRCV infection, a significantly higher incidence of fever (P<0.05) was observed in the PRRSV/PRCV-infected pigs (93/139 pigs with fever; 66.9 %) than in the PRRSV only-infected pigs (60/116 pigs with fever; 51.7 %). We also observed that PRRSV alone, but not PRCV alone, markedly decreased the rates of body weight gain in infected pigs compared with mock-infected pigs. Moreover, at PRCV PIDs 2–21, a significantly higher proportion of pigs with decreased body weight gains was observed in the PRRSV/PRCV-infected pigs than in the PRRSV alone pigs (84.9 vs 66.9 % of pigs with decreased body weight gains, respectively; P<0.05).
Ongoing PRRSV and subsequent PRCV co-infection exacerbate lymphadenopathy and pulmonary disease compared with pigs infected with PRRSV or PRCV alone
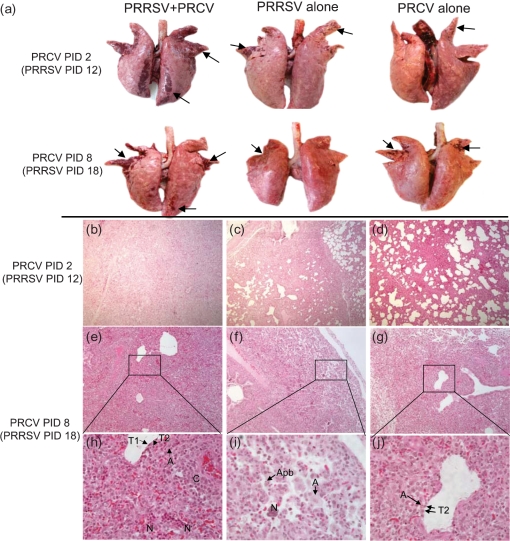
Both PRRSV and PRCV infections induced lymphadenopathy, and the left TBLNs were enlarged on PRCV PIDs 2–21 compared with mock-infected pigs. The LN : BW ratios indicated that the left TBLNs of PRRSV/PRCV pigs were more hyperplasic at PRCV PIDs 4, 8, 14 and 21 than those of the single PRRSV infection, and at PIDs 2–21 (significant at PID 14; P<0.05) than the single PRCV infection, respectively (data not shown). The PRRSV and/or PRCV-induced macroscopic and microscopic lesions were mainly limited to the lungs, and were commonly observed as a reddish dark consolidation of the lung (Fig. 1a) and bronchointerstitial pneumonia with frequent peribronchiolar and perivascular cuffing, respectively (Fig. 1b–j). The PRRSV histological lesions consisted of cellular infiltration with moderate to large numbers of alveolar macrophages that frequently had apoptotic bodies in their nuclei (Fig. 1i). No apoptotic bodies were evident in PRCV-infected epithelial cells and alveolar macrophages (Fig. 1j). By gross and histological lung lesion scores, the PRRSV/PRCV-infected pigs had a significantly more severe pneumonia than either single infection group throughout the experiment (Figs 1a–j and 2a, b), indicative of the pathological interactions between PRRSV and PRCV.
Fig. 1.
Ongoing PRRSV and subsequent PRCV co-infection exacerbate pneumonia compared with pigs infected with PRRSV or PRCV alone. (a) Gross evidence of pneumonia at PRCV PIDs 2 and 8. (b–j) Lung sections from: (b) a PRRSV/PRCV-infected pig at PRCV PID 2, showing severe bronchointerstitial pneumonia; (c) a PRRSV-infected pig at PRCV PID 2, showing moderate bronchointerstitial pneumonia; (d) a PRCV only-infected pig at PRCV PID 2, showing moderate interstitial pneumonia; (e, h) a PRRSV/PRCV-infected pig at PRCV PID 8, showing severe bronchointerstitial pneumonia characterized by accumulation of necrotic cells (N) in alveolar spaces and type 2 pneumocyte (T2) hyperplasia and alveolar macrophage infiltration (A) in alveolar septa, accompanying lymphohistiocytic perivascular cuffing (C); sometimes, type 1 pneumocytes (T1) formed the alveolar epithelium; (f, i) a PRRSV-infected pig at PRCV PID 8, showing moderate to severe bronchointerstitial pneumonia characterized by accumulation of necrotic cells (N) and massive infiltration of alveolar macrophages (A) that had apoptotic bodies (Apb) in their nuclei; (g, j) a PRCV only-infected pig at PRCV PID 8, showing severe interstitial pneumonia in which type 2 pneumoytes have infiltrated the alveolar septa. Magnification, ×100 (b–g); ×400 (h–j).
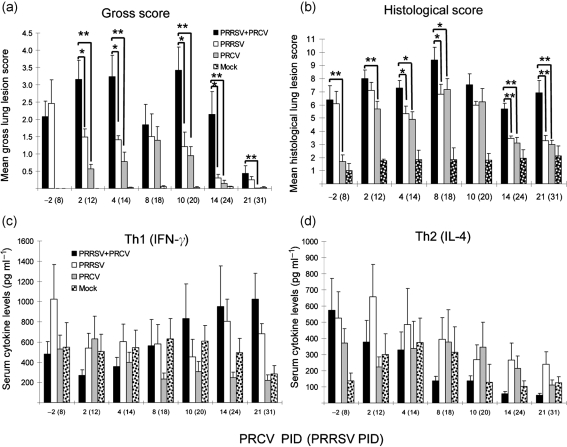
Fig. 2.
High Th1 (IFN-γ) and low Th2 (IL-4) serum cytokine responses coincide with the prolonged pneumonia observed in dual-infected pigs. The pigs (n=6–8 at PRCV PIDs −2 to 14 and n=5 at PID 21 for PRRSV/PRCV and PRCV alone; n=5–6 at each PID for PRRSV alone; n=6–7 at each PID for mock infections) were necropsied and assessed for serum cytokine ELISA. (a) Gross lung lesion score. Gross and histological lung lesions were given an estimated score based on the percentage of macroscopic consolidation in all lobes and the distribution and severity of histopathology, respectively. (b) Histological lung lesion score. (c) Th1 (IFN-γ) cytokine serum levels. (d) Th2 (IL-4) cytokine serum levels. Each bar represents the mean±sem. *P<0.05; **P<0.01 (statistically significant differences between the dually and singly infected pigs by the Kruskal–Wallis test).
High Th1 (IFN-γ) and low Th2 (IL-4) serum cytokine responses coincide with the prolonged pneumonia observed in dual-infected pigs
A trend towards increased Th1 (IFN-γ) serum cytokine responses was observed in the dual-infected pigs compared with either group of singly infected pigs after PRCV PID 10 (Fig. 2c). In contrast, a trend towards decreased Th2 (IL-4) serum cytokine response was evident in the dual-infected pigs during this period (Fig. 2d). Although there were no significant differences in mean Th1/Th2 serum cytokine levels among treatment groups during the study period due to data variability between each independent trial, the same trend in Th1/Th2 serum responses was consistent and reproducible during each independent trial. The trend of high Th1 (IFN-γ) and low Th2 (IL-4) serum cytokine responses coincided with the later (PRCV PIDs 10 and 21) severe lung lesions in the dual-infected pigs. More detailed cytokine responses including analysis of additional cytokines have been reported by G. J. Renukaradhya, K. P. Aleekseev, K. Jurg and L. J. Saif (unpublished data).
Suppression of innate immune responses (reduced IFN-α levels in lung and blood NK cell cytotoxicity) by ongoing PRRSV infection is observed in dual-infected pigs, coinciding with exacerbated pneumonia during early PRCV infection (PRCV PIDs 2 and 4)
Lung IFN-α levels remained low after PRRSV infection. The PRCV only-infected pigs had increased IFN-α levels in their lungs at PRCV PIDs 2 and 4. However, at PRCV PIDs 2 and 4, the dual-infected pigs had reduced IFN-α levels compared with the single PRCV infection; these were significantly lower at PRCV PID 4 (P<0.05) (Fig. 3a). For PRCV PID 8 and beyond, lung IFN-α levels of virus-infected pigs did not differ significantly from those of mock-infected pigs. The NK cells of PRRSV singly or dual-infected pigs had reduced cytotoxicity compared with PRCV- or mock-infected pigs at PRCV PIDs −2 to 14 (PRRSV PIDs 8–24), and significantly reduced lytic activity (undetectable) at PRCV PIDs 2 and 8 (P<0.05) compared with mock-infected pigs (Fig. 3b). Suppression of innate immune responses by PRRSV seen in the dual-infected pigs compared with PRCV only-infected pigs at PRCV PID 4 coincided with the exacerbated pneumonia (Figs 2a, b, 3a, b).
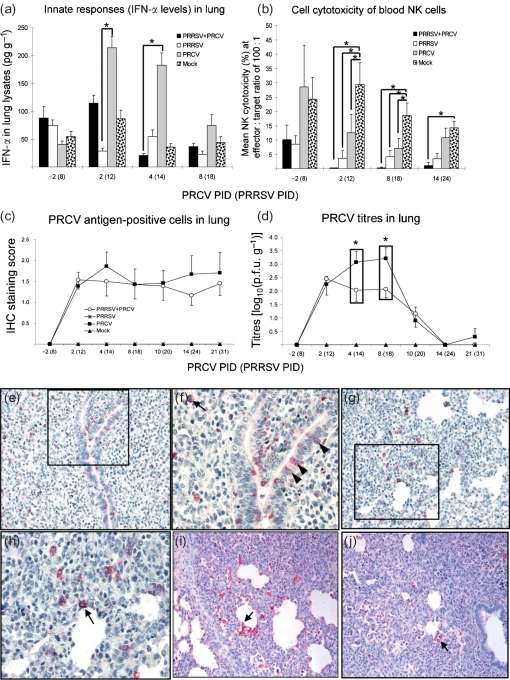
Fig. 3.
Suppression of innate immune responses (reduced IFN-α levels in lung and reduced blood NK cell cytotoxicity) by ongoing PRRSV infection coincides with exacerbated pneumonia during early PRCV infection. (a) IFN-α in the lungs. Lung lysates were prepared from pigs at each PID with the numbers of pigs indicated in the legend for Fig. 2 and tested for IFN-α levels by ELISA. (b) NK cell cytotoxicity (%) was measured using PBMCs (effectors) harvested from pigs at each PID against target cells (K-562 or Yac-1). Effectors and targets at the indicated ratio (100 : 1) were co-cultured and the supernatants harvested after 24 h. The amount of LDH released was measured by using LDH substrate and measuring absorbance at 490 nm. Each bar represents the mean percentage of NK-specific lysis of targets from two or three pigs±sem. (c, d) PRCV replication in the lungs. Paraffin-embedded lung tissues were evaluated by IHC for PRCV antigen detection (c). Lung homogenates were also tested by qRT-PCR for viral RNA quantification (d). Each data point represents the mean±sem. *P<0.05 (statistically significant difference between the dual-infected and the singly or mock-infected pigs by the Kruskal–Wallis test). (e, f) Lung section from a PRCV only-infected pig at PRCV PID 4, showing the large number of PRCV-positive cells in the lung by IHC. PRCV antigens were identified in bronchiolar epithelial cells (f; arrowheads) and type 2 pneumocytes (arrow). (g, h) Lung section from a PRRSV/PRCV-infected pig at PRCV PID 4, showing moderate numbers of PRCV-positive cells (h; arrow) in the alveolar septa by IHC. (i) Lung section from a PRRSV/PRCV-infected pig at PRCV PID 8, showing large numbers of PRRSV-positive cells in the alveolar septa and spaces by IHC. PRRSV antigens were identified in alveolar macrophages (arrow). (j) Lung section from a PRRSV-infected pig at PRCV PID 8, showing small numbers of PRRSV-positive cells (arrow) by IHC. Magnification: ×200 (e, g, i, j); ×400 (f, h).
By using IHC for viral antigen and qRT-PCR for RNA quantification in the lungs, reduced PRCV replication (i.e. numbers of PRCV antigen-positive cells at PRCV PIDs 4 and 10–21 and viral RNA titres at PRCV PIDs 4 and 8) was observed in the lungs of dual-infected pigs compared with PRCV only-infected pigs, with significantly reduced PRCV titres by qRT-PCR at PRCV PIDs 4 and 8 (P<0.05) (Fig. 3c–h). However, the reverse trend was notable early at PRCV PID 2 in the dual-infected pigs (Fig. 3c, d), although the differences between PRRSV/PRCV-infected and PRRSV only-infected pigs were not statistically significant, probably due to data variability between independent trials consistent with the use of outbred swine. Conversely, at this time (PRCV PID 2), PRRSV replication was significantly reduced in the dual-infected pigs compared with single PRRSV infection (P<0.05) (Fig. 4b, c). This is also likely to represent an early antagonistic interaction between PRCV and PRRSV replication in the lungs.
Fig. 4.
Subsequent PRCV infection promotes PRRSV replication in the lungs and induction of severe PRRSV-related apoptotic lesions at PRCV PIDs 4–10. (a, b) PRRSV replication in the lungs. Each data point represents the mean±sem. *P<0.05 (statistically significant differences between dual-infected pigs and pigs singly infected by either virus by the Kruskal–Wallis test. (c) Paraffin-embedded lung tissues were evaluated by an in situ TUNEL assay (black staining) for detection of apoptosis. Magnification: ×50. Cells were counterstained with methyl green. +, A few positive cells; ++, moderate numbers of positive cells; +++, many positive cells.
Subsequent PRCV infection promotes PRRSV replication in lungs, and severe PRRSV-related apoptotic lesions are induced at PRCV PIDs 4–10
We performed an in situ TUNEL assay to assess the severity and distribution of PRRSV-related apoptotic lesions. After PRCV PID 4, subsequent PRCV infection led to increased PRRSV replication in the lungs at PRCV PIDs 4–21 (significant at PRCV PIDs 8 and 14 by IHC; P<0.05; Fig. 4a, b) and more severe apoptotic lesions (Fig. 4c). During this time, PRCV replication in the lungs of co-infected pigs decreased more compared with single PRCV infection (significant at PRCV PIDs 4 and 8 by qRT-PCR; Fig. 3d). Thus, the opposite antagonistic interactions between PRCV and PRRSV replication in the lung were observed at later stages of PRCV infection (PRCV PIDs 4–14).
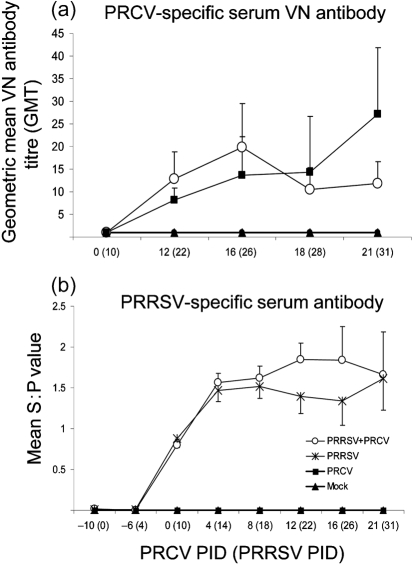
Increased PRRSV replication in the lungs coincides with increased virus-specific serum antibody titres
No significant differences in PRCV-specific serum VN antibody geometric mean titre between the dual- and singly infected pigs were observed (Fig. 5a). On the other hand, after PRCV PID 4, PRRSV-specific serum antibody titres of the dual-infected pigs tended to be higher (but not statistically different) compared with single PRRSV infection (Fig. 5b). This result coincided with the increased PRRSV replication and severe apoptotic lung lesions observed in the dual-infected pigs.
Fig. 5.
PRCV- and PRRSV-specific serum VN and ELISA antibody titres in pigs co-infected with PRRSV and PRCV. Serum was collected at each PID from the numbers of pigs indicated in the legend of Fig. 2. (a) Serum VN antibody titres against PRCV were evaluated by a plaque-reduction test. Antibody titres were expressed as the reciprocal of the specimen dilution that resulted in an 80 % reduction in the number of plaques. (b) PRRSV serum antibodies were identified by a commercial PRRSV ELISA (Idexx Laboratories, ME) and evaluated based on progression of the S : P ratios. Each data point represents the mean±sem.
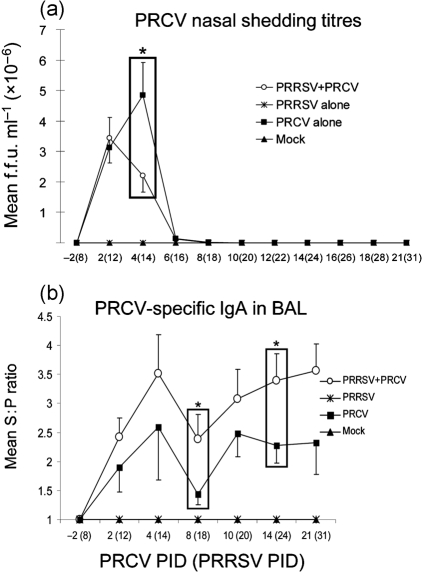
Ongoing PRRSV infection decreases PRCV nasal shedding titres of pigs subsequently co-infected with PRCV, coinciding with higher PRCV-specific IgA antibody titres in BAL at PRCV PIDs 2–21 compared with PRCV only-infected pigs
At PRCV PID 4, prior PRRSV infection coincided with significantly decreased PRCV nasal shedding titres in the dual-infected pigs (P<0.05), compared with single PRCV infection (Fig. 6a), which further corresponded to decreased PRCV replication in the lungs at this time (Fig. 3d). The PRCV-specific IgA antibody titres in BAL of the dual-infected pigs were higher than for the single PRCV infection throughout the experiment and were significantly higher at PRCV PIDs 8 and 14 (P<0.05; Fig. 6b). These data suggest that PRRSV infection might promote the production of IgA antibodies against PRCV in the respiratory tract of dual-infected pigs. The higher local (BAL) PRCV-specific IgA antibody titres might be associated with the significant reduction in PRCV nasal shedding (PRCV PID 4) and virus replication (PRCV PID 4 and 8) in the lungs of dual-infected pigs.
Fig. 6.
PRCV nasal shedding titres (a) and PRCV-specific IgA antibodies (b) in BAL of pigs co-infected with PRRSV and PRCV. (a) Nasal swabs were collected from each pig (n=39 at PRCV PID 2, n=31 at PID 4, n=20 at PID 6, n=15 at PID 8, n=11 at PID 10 and n=5–6 at PIDs 12–21 for PRRSV/PRCV-infected and PRCV only-infected pigs) at PRRSV PIDs 0–31 and tested by a cell culture immunofluorescence assay. The numbers of PRCV only-infected cells were expressed as f.f.u. ml−1. (b) BAL fluids were prepared at each PID from the numbers of pigs indicated in the legend of Fig. 2. PRCV-specific IgA antibody titres in BAL fluids were tested by ELISA. Each data point represents the S : P ratio (mean±sem). *P<0.05 (statistically significant difference between the dual-infected and PRCV only-infected pigs by the Kruskal–Wallis test).
DISCUSSION
Our study demonstrated interactive effects between PRCV and PRRSV as influenced by early innate (IFN-α) and later Th1 (IFN-γ) and Th2 (IL-4) immune responses that involve pulmonary alveolar macrophages (PAMs), NK and T cells. Coronaviruses may predispose the host to co-infection by other viruses (Jung et al., 2008; Lanza et al., 1992; Saif, 2004; Van Reeth et al., 1996). It is unclear if or how CoV co-infections with other viruses affect the severity of CoV disease or alter shedding or infection patterns. In a previous study, simultaneous infection of pigs with PRCV and swine influenza H1N1 virus did not affect clinical disease outcome (fever or decreased body weights) compared with each virus alone (Lanza et al., 1992). However, Van Reeth et al. (1996) reported that European swine influenza H1N1 or PRRSV inoculated 2 or 3 days prior to PRCV (Belgium strain) exacerbated clinical disease in weaned pigs compared with PRCV single infection. Similarly, enhanced pathogenicity of SARS-CoV was evident in the presence of a pre-existing reovirus infection in mice (He et al., 2006). Thus, the pathogenesis of respiratory CoVs and disease outcome could be affected by the presence of a pre-existing respiratory virus infection. In our study, using a low-virulence North American PRRSV strain (SD23983), followed by co-infection with a US PRCV strain (ISU-1), exacerbated clinical disease and pneumonic lesions were evident in the PRRSV/PRCV-infected pigs. Our observations concur with the prolonged fever and decreased body weights in PRRSV/PRCV-infected pigs as reported previously (Van Reeth et al., 1996). However, the latter investigators did not find a synergistic interaction in cytokine induction in their study (Van Reeth et al., 1996). Here, we greatly expanded studies of the innate immunological and pathological responses to elucidate the interactions that contribute to the enhanced disease severity in PRRSV/PRCV-infected pigs.
Pre-existing viral infections influence the immune responses to subsequent virus infections and alter the disease outcome of the secondary viral infection (reviewed by Didierlaurent et al., 2007). Modification of immunity by a pre-existing influenza A viral infection alters disease outcome upon a subsequent respiratory viral infection. The influenza A virus co-infections resulted in either detrimental (exacerbated murine cytomegalovirus infection) or beneficial (decreased vaccinia infection) effects that influenced the subsequent respiratory viral co-infection (Chen et al., 2003). Because PRRSV infection is ubiquitous in swine herds, the virus can be isolated from pigs with PRCV co-infection (Kamogawa et al., 1996). Similarly, co-infections of human metapneumovirus and respiratory syncytial virus have been identified in many human CoV-229E patients (Canducci et al., 2008). In addition, human metapneumovirus and reovirus have been identified in many SARS patients and the former was initially proposed as the cause of SARS (Chan et al., 2003; Kuiken et al., 2003). In our study, detrimental effects of pre-existing PRRSV infection included downregulation of innate immunity (IFN-α levels in the lungs and NK cell cytotoxicity) in the initial stage of the subsequent PRCV infection, exacerbating pneumonia during early PRCV infection. It is likely that the ongoing PRRSV infection, by compromising innate immunity, may enhance early PRCV replication in the lungs. The subsequent PRCV co-infection led to further detrimental effects associated with a trend towards increased Th1 (IFN-γ) but reduced Th2 (IL-4) serum cytokines and enhanced PRRSV replication and possibly inflammatory lesions in the lung at the middle to later infection times tested. Consistent with the trend towards increased Th1 (IFN-γ) cytokine responses, the Th2 (IL-4) serum responses tended to decrease in the dual-infected pigs during the corresponding period. Similarly, increased Th1 (IFN-γ) and pro-inflammatory (IL-6, but not tumour necrosis factor-α) serum cytokine responses may have contributed to the severe lung and immunopathological damage in SARS-CoV disease (Huang et al., 2005). These findings imply that the presence of existing immunomodulating respiratory viral co-infections in CoV disease may have been a contributing factor to the more severe pneumonia and immunopathology.
The rapid production of type I IFN (IFN-α/β) innate immune responses to viral infections suppresses initial viral replication and promotes the adaptive immune response (Cameron et al., 2008; Thiel & Weber, 2008). PRCV stimulates IFN-α production by plasmacytoid dendritic cells present in the mucosal-associated lymphoid tissues of the respiratory tract (Charley et al., 2006; Van Reeth et al., 1999; Zhang et al., 2008). Significant elevation of IFN-α is evident in BAL by 4 days after PRCV infection (Van Reeth et al., 1999; Zhang et al., 2008). Likewise, PRCV increased IFN-α in the lungs at PRCV PIDs 2 and 4 in our study, but significantly reduced the cytotoxicity of NK cells isolated from the PBMCs. Nevertheless, only pneumonic lesions without any multi-organ disease were observed in PRCV only-infected pigs. Unlike PRCV, PRRSV suppresses IFN-α production by the macrophages of infected pigs (Mateu & Diaz, 2008). Interestingly, at PRCV PIDs 2 and 4, the ongoing PRRSV infection downregulated the innate immunity (IFN-α) stimulated by PRCV, as evidenced by reduced IFN-α levels in lung lysates, and further significantly suppressed the NK cell cytotoxicity in the dual-infected pigs. The reduced early innate immunity probably contributed to the exacerbated pneumonia during this period.
Beyond PRCV PID 4, the increasing lung IFN-α levels following PRCV infection might potentiate PRRSV replication rather than inhibiting it in the dual-infected pigs. PRRSV replicates in well-differentiated monocytes, such as PAMs, and leads to apoptotic death of PAMs (Chang et al., 2005; Costers et al., 2008; Kim et al., 2002). For PRRSV cellular binding and internalization, sialoadhesin is one of the PRRSV cellular receptors that PAMs express on their surface (Delputte et al., 2005). Exogenous IFN-α treatment could enhance PRRSV infection of monocytes via upregulation of sialoadhesion on the monocytes (Delputte et al., 2007). The results imply that PRRSV replication in PAMs can be increased in the presence of IFN-α in the lungs, which occurred in the PRCV-infected pigs. PRRSV-infected pigs develop interstitial pneumonia characterized by infiltration of PAMs, usually derived from blood monocytes, into alveolar septa (Rossow, 1998). Therefore, PRCV-induced IFN-α in the lungs of PRRSV/PRCV-infected pigs might upregulate sialoadhesion on the surface of the infiltrating PAMs, which could further contribute to the severe interstitial pneumonia. Indeed, our study showed that PRRSV replication and apoptosis of PAMs were increased in dual-infected pigs at PRCV PIDs 4–10 compared with pigs infected with PRRSV alone.
PRRSV infection in pigs is immunosuppressive and compromises innate and adaptive immunity, leading to persistent infection of pigs and prolonged PRRSV pathogenesis (Mateu & Diaz, 2008). PRRSV also enhances co-infections by other pathogens, but serum antibodies against the secondary pathogens are not compromised (Wills et al., 2000); therefore, the effect of PRRSV on adaptive immunity remains unclear. Strikingly, in our study, PRRSV/PRCV co-infected pigs had higher BAL IgA antibodies to PRCV, suggesting that PRRSV co-infection enhanced mucosal IgA antibody responses to PRCV. The increased local (BAL) IgA antibodies to PRCV might account for the reduced PRCV nasal shedding (PRCV PID 4) and virus replication (PRCV PIDs 4 and 8) in the lungs of the dual-infected pigs. PRRSV induces B-cell hyperplasia in lymphoid tissues and increases total serum IgM, IgG and IgA by 10–80 times compared with control pigs (Lemke et al., 2004). These observations support the possibility that PRRSV could stimulate mucosal IgA antibodies against secondary respiratory viruses, possibly due to PRRSV polyclonal stimulation of B and plasma cells present in the respiratory mucosa.
In summary, an ongoing PRRSV infection compromised innate immune responses to a subsequent PRCV infection, exacerbating pneumonia during early PRCV infection. The subsequent PRCV infection led to enhanced PRRSV replication in the lungs and a trend towards increased Th1 (IFN-γ) serum cytokines, which exacerbated the severity of PRRSV infection. These findings also imply that the presence of ongoing immunomodulating respiratory viral co-infections in CoV disease may be a contributing factor to more severe pneumonia. Our study also provides valuable insights into host–pathogen interactions related to co-infections of CoVs with other respiratory viruses.
Acknowledgments
We thank Peggy Lewis for technical assistance. We also thank Dr Juliette Hanson, Todd Root, Aaron Higgins, April Eyster, Morgan Chapman, Justin Dickey and Thales DeNardo for assistance with animal care. Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. This work was supported by a grant to L. J. S. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, grant R01 AI060739.
References
- AVMA Panel on Euthanasia (2001). 2000 Report of the AVMA Panel on Euthanasia. J Am Vet Med Assoc 218, 669–696. [DOI] [PubMed] [Google Scholar]
- Azevedo, M. S., Yuan, L., Pouly, S., Gonzales, A. M., Jeong, K. I., Nguyen, T. V. & Saif, L. J. (2006). Cytokine responses in gnotobiotic pigs after infection with virulent or attenuated human rotavirus. J Virol 80, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, M. J., Bermejo-Martin, J. F., Danesh, A., Muller, M. P. & Kelvin, D. J. (2008). Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res 133, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canducci, F., Debiaggi, M., Sampaolo, M., Marinozzi, M. C., Berre, S., Terulla, C., Gargantini, G., Cambieri, P., Romero, E. & Clementi, M. (2008). Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J Med Virol 80, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, P. K., Tam, J. S., Lam, C.-W., Chan, E., Wu, A., Li, C.-K., Buckley, T. A., Ng, K.-C., Joynt, G. M. & other authors (2003). Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis 9, 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.-W., Jeng, C.-R., Liu, J.-J., Lin, T.-L., Chang, C.-C., Chia, M.-Y., Tsai, Y.-C. & Pang, V. F. (2005). Reduction of porcine reproductive and respiratory syndrome virus (PRRSV) infection in swine alveolar macrophages by porcine circovirus 2 (PCV2)-induced interferon-alpha. Vet Microbiol 108, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley, B., Riffault, S. & Van Reeth, K. (2006). Porcine innate and adaptative immune responses to influenza and coronavirus infections. Ann N Y Acad Sci 1081, 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. D., Fraire, A. E., Joris, I., Welsh, R. M. & Selin, L. K. (2003). Specific history of heterologous virus infections determines anti-viral immunity and immunopathology in the lung. Am J Pathol 163, 1341–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costers, S., Lefebvre, D. J., Delputte, P. L. & Nauwynck, H. J. (2008). Porcine reproductive and respiratory syndrome virus modulates apoptosis during replication in alveolar macrophages. Arch Virol 153, 1453–1465. [DOI] [PubMed] [Google Scholar]
- Cox, E., Hooyberghs, J. & Pensaert, M. B. (1990). Sites of replication of a porcine respiratory coronavirus related to transmissible gastroenteritis virus. Res Vet Sci 48, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delputte, P. L., Costers, S. & Nauwynck, H. J. (2005). Analysis of porcine reproductive and respiratory syndrome virus attachment and internalization: distinctive roles for heparan sulphate and sialoadhesin. J Gen Virol 86, 1441–1445. [DOI] [PubMed] [Google Scholar]
- Delputte, P. L., Van Breedam, W., Barbe, F., Van Reeth, K. & Nauwynck, H. J. (2007). IFN-α treatment enhances porcine arterivirus infection of monocytes via upregulation of the porcine arterivirus receptor sialoadhesin. J Interferon Cytokine Res 27, 757–766. [DOI] [PubMed] [Google Scholar]
- Didierlaurent, A., Goulding, J. & Hussell, T. (2007). The impact of successive infections on the lung microenvironment. Immunology 122, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L., Zhao, G., Lin, Y., Sui, H., Chan, C., Ma, S., He, Y., Jiang, S., Wu, C. & other authors (2008). Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J Immunol 180, 948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman, M., Heise, M. & Baric, R. (2008). SARS coronavirus and innate immunity. Virus Res 133, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbur, P. G., Paul, P. S., Vaughn, E. M. & Andrews, J. J. (1993). Experimental reproduction of pneumonia in gnotobiotic pigs with porcine respiratory coronavirus isolate AR310. J Vet Diagn Invest 5, 184–188. [DOI] [PubMed] [Google Scholar]
- He, C., Yang, Q., Lei, M., Pang, W., Yang, J., Zhu, H. & Duan, Q. (2006). Diffuse alveolar lesion in BALB/c mice induced with human reovirus BYD1 strain and its potential relation with SARS. Exp Anim 55, 439–447. [DOI] [PubMed] [Google Scholar]
- Huang, K.-J., Su, I.-J., Theron, M., Wu, Y.-C., Lai, S.-K., Liu, C.-C. & Lei, H.-Y. (2005). An interferon-γ-related cytokine storm in SARS patients. J Med Virol 75, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, D. M., Chamberlain, D. W., Poutanen, S. M., Low, D. E., Asa, S. L. & Butany, J. (2005). Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol 18, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabrane, A., Girard, C. & Elazhary, Y. (1994). Pathogenicity of porcine respiratory coronavirus isolated in Quebec. Can Vet J 35, 86–92. [PMC free article] [PubMed] [Google Scholar]
- Jung, K., Alekseev, K. P., Zhang, X., Cheon, D. S., Vlasova, A. N. & Saif, L. J. (2007). Altered pathogenesis of porcine respiratory coronavirus in pigs due to immunosuppressive effects of dexamethasone: implications for corticosteroid use in treatment of severe acute respiratory syndrome coronavirus. J Virol 81, 13681–13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K., Kang, B-K., Lee, C.-S. & Song, D.-S. (2008). Impact of porcine group A rotavirus co-infection on porcine epidemic diarrhea virus pathogenicity in piglets. Res Vet Sci 84, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamogawa, O., Tomita, Y., Kaneko, M., Yamada, S., Kubo, M. & Shimizu, M. (1996). Isolation of porcine respiratory coronavirus from pigs affected with porcine reproductive and respiratory syndrome. J Vet Med Sci 58, 385–388. [DOI] [PubMed] [Google Scholar]
- Kim, T. S., Benfield, D. A. & Rowland, R. R. (2002). Porcine reproductive and respiratory syndrome virus-induced cell death exhibits features consistent with a nontypical form of apoptosis. Virus Res 85, 133–140. [DOI] [PubMed] [Google Scholar]
- Korzeniewski, C. & Callewaert, D. M. (1983). An enzyme-release assay for natural cytotoxicity. J Immunol Methods 64, 313–320. [DOI] [PubMed] [Google Scholar]
- Kuiken, T., Fouchier, R. A., Schutten, M., Rimmelzwaan, G. F., van Amerongen, G., van Riel, D., Laman, J. D., de Jong, T., van Doornum, G. & other authors (2003). Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza, I., Brown, I. H. & Paton, D. J. (1992). Pathogenicity of concurrent infection of pigs with porcine respiratory coronavirus and swine influenza virus. Res Vet Sci 53, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S. K., Woo, P. C., Li, K. S., Huang, Y., Tsoi, H.-W., Wong, B. H., Wong, S. S., Leung, S.-Y., Chan, K.-H. & Yuen, K.-Y. (2005). Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 102, 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke, C. D., Haynes, J. S., Spaete, R., Adolphson, D., Vorwald, A., Lager, K. & Butler, J. E. (2004). Lymphoid hyperplasia resulting in immune dysregulation is caused by porcine reproductive and respiratory syndrome virus infection in neonatal pigs. J Immunol 172, 1916–1925. [DOI] [PubMed] [Google Scholar]
- Louie, J. K., Hacker, J. K., Mark, J., Gavali, S. S., Yagi, S., Espinosa, A., Schnurr, D. P., Cossen, C. K., Isaacson, E. R. & other authors (2004). SARS and common viral infections. Emerg Infect Dis 10, 1143–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu, E. & Diaz, I. (2008). The challenge of PRRS immunology. Vet J 177, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, R. M., Nelson, E. A., Christopher-Hennings, J., Hesse, R., Rowland, R. R. & Zimmerman, J. J. (2008). Evaluation of the risk of PRRSV transmission via ingestion of muscle from persistently infected pigs. Transbound Emerg Dis 56, 1–8. [DOI] [PubMed] [Google Scholar]
- Rossow, K. D. (1998). Porcine reproductive and respiratory syndrome. Vet Pathol 35, 1–20. [DOI] [PubMed] [Google Scholar]
- Saif, L. J. (2004). Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev Sci Tech 23, 643–660. [DOI] [PubMed] [Google Scholar]
- Thiel, V. & Weber, F. (2008). Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev 19, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCott, J. L., Brim, T. A., Simkins, R. A. & Saif, L. J. (1993). Isotype-specific antibody-secreting cells to transmissible gastroenteritis virus and porcine respiratory coronavirus in gut- and bronchus-associated lymphoid tissues of suckling pigs. J Immunol 150, 3990–4000. [PubMed] [Google Scholar]
- Van Reeth, K., Nauwynck, H. & Pensaert, M. (1996). Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol 48, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth, K., Labarque, G., Nauwynck, H. & Pensaert, M. (1999). Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res Vet Sci 67, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills, R. W., Gray, J. T., Fedorka-Cray, P. J., Yoon, K. J., Ladely, S. & Zimmerman, J. J. (2000). Synergism between porcine reproductive and respiratory syndrome virus (PRRSV) and Salmonella choleraesuis in swine. Vet Microbiol 71, 177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Alekseev, K., Jung, K., Vlasova, A., Hadya, N. & Saif, L. J. (2008). Cytokine responses in porcine respiratory coronavirus-infected pigs treated with corticosteroids as a model for severe acute respiratory syndrome. J Virol 82, 4420–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]