Abstract
Objective
The goal of this study was to investigate whether the location of HIV integration differs in resting versus activated T cells, a feature that could contribute to the formation of latent viral reservoirs via effects on integration targeting.
Design
Primary resting or activated CD4+ T cells were infected with purified X4-tropic HIV in the presence and absence of nucleoside triphosphates and genomic locations of integrated provirus determined.
Methods
We sequenced and analyzed a total of 2661 HIV integration sites using linker-mediated PCR and 454 sequencing. Integration site data sets were then compared to each other and to computationally generated random distributions.
Results
HIV integration was favored in active transcription units in both cell types, but integration sites from activated cells were found more often in genomic regions that were dense in genes, dense in CpG islands, and enriched in G/C bases. Integration sites from activated cells were also more strongly correlated with histone methylation patterns associated with active genes.
Conclusion
These data indicate that integration site distributions show modest but significant differences between resting and activated CD4+ T cells, and that integration in resting cells occurs more often in regions that may be suboptimal for proviral gene expression.
Keywords: CD4+ T cells, gene expression, HIV integration, latency
Introduction
HIV infection is able to persist in the presence of antiretroviral therapy (ART) due in part to persistence of latentviral reservoirs found in resting CD4+ T cells [1–4]. It is unclear how proviruses establish latent infection, but invitro models suggest two possibilities. One possibility is that HIV infects activated cells, which then revert to a resting state [2,5] and another possibility is that HIV directly infects and integrates into resting CD4+ T cells [6–8].
Previous studies have suggested that the location of proviral integration in the host cell genome may influence viral gene activity, possibly contributing to latency [9]. In the extensively studied Jurkat cell model, inducible (i.e. latent) proviruses were found to be enriched in alphoid repeats, which are characteristic of centromeric heterochromatin; in gene deserts, which correlate with low host gene expression; and in very highly expressed host cell genes, consistent with gene repression by transcriptional interference (see also [10,11]). These studies were limited by the use of transformed cell models. However, studies of latently infected cells in individuals on long-term successful ART are severely limited by the background of replication-incompetent proviruses that accumulate in circulating cells; thus, so far, it has not been possible to study integration site distributions directly in latently infected cells due to masking by much larger numbers of integrated sequences from genetically inactive proviruses [12].
Here we investigate possible mechanisms of latency in resting and activated CD4+ T cells by asking whether the distributions of de-novo integration sites might differ between the two cell types, a feature that could bias proviruses in resting cells toward entering the latent state. We used DNA bar coding and pyrosequencing [13] to recover 1474 sites from resting CD4+ T cells and 1187 sites from activated cells infected in cell culture. We found that integration sites were enriched within active transcription units in both cell types. However, quantitatively modest but significant and reproducible differences could be detected between the resting and activated data sets, in which the bias toward integration in actively transcribed regions and associated features was reduced in resting cells. Although the magnitudes of these changes were small, the differences in integration site distributions in the resting cell pool were in the direction that would be predicted to result in less efficient viral gene expression following integration.
Methods
Purification, activation, and maintenance of CD4+ T cells
CD4+ T cells were purified by the University of Pennsylvania Immunology Core from a mononuclear leukapheresis product using the RosetteSep Human CD4+ T Cell Enrichment kit (Stem Cell Technologies, Bethesda, Maryland, USA) following the recommendations of the manufacturer. To obtain pure resting CD4+ T cells, rosette-purified CD4+ T cells were then stained with saturating concentrations of phycoerythrin-labeled antibodies that recognize the T cell activation markers CD25, CD69, and human leukocyte antigen (HLA-DR; BD Biosciences, San Jose, California, USA). Following staining against activation markers, the cells were labeled with anti-phycoerythrin magnetic beads and applied to a magnetic column (Miltenyi Biotec, Auburn, California, USA) to separate activated from resting CD4+ T cells. Cell purity and level of activation was monitored by flow cytometry using a FACSCalibur instrument (BD Biosciences) and the data were analyzed using FlowJo software (Treestar, Ashland, Oregon, USA). To obtain activated CD4+ T cells, rosette-purified cells were cultured in the presence of CD3/CD28 beads (Invitrogen, Madison, Wisconsin, USA) at approximately 1 bead/cell for 72 h at 378C. CD4+ T cells were cultured at 378C in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (Gibco/Invitrogen, Carlsbad, California, USA).
Infection of CD4+ T cells
Resting or activated CD4+ T cells were inoculated by spinoculation with the X4-tropic HIV molecular clone pNL4-3 [14] (obtained from the University of Pennsylvania Center for AIDS Research Virology Core) as previously described [15]. When testing the effect of deoxynucleosides on integration, the cells were treated with a deoxynucleoside mixture at 50 mmol/l (Sigma, StLouis, Missouri, USA; with equal content of each deoxynucleoside) during spinoculation. Following spinoculation, the cells were washed twice to remove unbound virus. The cells were then resuspended in medium (RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin) containing 1.25 mmol/l of saquinavir (Roche Pharmaceuticals, Nutley, New Jersey, USA) to prevent viral spread and 50 mmol/l of deoxynucleosides (wherever indicated). The inoculated cells were then incubated at 378C for 36 h (activated cells) or 60 h (resting cells).
Measuring HIV integration
HIV integration was measured at multiple time points postinoculation by quantitative Alu-PCR as previously described [8,16,17]. To determine the number of integration events per cell, cell numbers were estimated by quantitative PCR using primers that detect β-globin [15].
Integration site analysis
Recovery of integration sites was performed as described [18]. DNA was analyzed from the sample collected 36 h after inoculation from the activated T cells and from the sample collected 60 h after inoculation from the resting T cells. Two micrograms of genomic DNA was digested overnight with MseI, ligated to linkers overnight at 16°C, and digested a second time with SacI. Nested PCR was then performed using primers and conditions described in [18,19]. DNA barcodes were included in the second round PCR primers in order to track sample origin [20]. Amplification products were gel purified and sequenced by massively parallel pyrophosphate sequencing. Only sequences that showed unique best alignments to the human genome by BLAT (BLAST-like alignment tool, hg18, version 36.1, >98% match score) and began within three base pairs of the long terminal repeat (LTR) end were used in downstream analyses. All sequences will be deposited in publicly accessible databases (NCBI) upon acceptance of this article for publication.
Comparisons to genomic features and histone modifications were carried out as described [21,22]. Details of statistical analyses can be found in study by Berry et al. [22]. Analyses of gene expression utilized data from SupT1 cells, with expression measured using the Affymetrix HU133 plus 2.0 gene chip array. Transcriptional profiles from resting and activated T cells showed relatively modest differences, and analysis of integration site placement against either type of data showed no major differences. Spinoculation did not have a measurable effect on chromatin structure, as revealed by a comparison of the activated cell data sets with other data sets for HIV vectors in activated T cells, in which the frequency of integration near DNAseI-hypersensitive sites showed no significant differences after correction for multiple comparisons. Consensus sequence analysis at the point of integration was performed using WebLogo (http://weblogo.berkeley.edu/logo.cgi). LEDGF expression levels of resting and activated T cells [23] were compared using an unpaired t-test.
Interactive supplementary data
An interactive heat map, summarizing statistical tests of integration frequency relative to genomic features, can be found at http://bushmanlab.pbwiki.com/f/RestVsActGenomicHeatmap.zip. The interactive supplementary data can be viewed using a standard web browser. Please download and unzip the supplementary data file and follow the instructions in the ReadMe file to load into a web browser.
To view statistical comparisons of experimental data to matched random controls, click on the text to the right of the screen ‘Compare to Area = 0.05’. To view comparisons between data sets (columns), click on the column headings (e.g. ‘Act + NTP’). To view comparisons of genomic features to each other (rows), click on the labels to the left of the heat map (e.g. ‘gc10000’). The P value, determined by a logistic regression method that respects the pairing in the data (clogit), is overlaid on each heatmap tile (*P < 0.05; **P < 0.01; ***P < 0.001).
Results
Infections and integration site sequencing
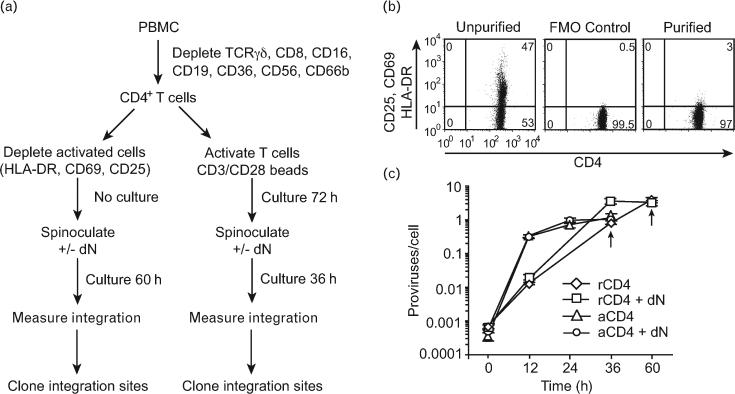
CD4+ T cells were isolated from healthy volunteers. Cell subsets were prepared as described in Fig. 1a. Briefly, peripheral blood mononuclear cells (PBMCs) were depleted with antibodies against T cell receptor (TCR)-γ/δ, CD8, CD16, CD19, CD36, CD56, and CD66b to yield CD4+ T cells. Cells expressing the activation markers for HLA-DR, CD69, and CD25 were depleted using phycoerythrin-labeled antibodies against these markers and antiphycoerythrin magnetic beads. In what follows, cells purified by this method are referred to as ‘resting cells’. This method for purifying resting CD4+ T cells yields more than 97% activation marker negative CD4+ T cells. Furthermore, a few (<3%) contaminating cells express very low levels of the activation markers CD25, CD69, and HLA-DR (Fig. 1b). To prepare the ‘activated cell’ subset, CD3/CD28 beads were added to the culture for 3 days. Resting cells purified as described above do not proliferate detectably over the time period of the study as assessed by DNA/RNA analysis and BrdU incorporation [24], whereas the activated cells enter the cell cycle and divide as shown by carboxyfluorescein succinimidyl ester (CFSE) staining [25–27]. Previous studies have also shown that activation induces the resting CD4+ T cells to express high levels of activation markers [7].
Fig. 1. Diagram of the experiment and purification of CD4+ T cells.
(a) Experimental design to investigate HIV integration distributions in resting and activated CD4+ T cells. (b) Resting CD4+ T cells were purified by negative selection using antibodies against CD25, CD69, and human leukocyte antigen (HLA-DR) to deplete cells expressing these activation markers. The purity of the resting CD4+ T cells was monitored by flow cytometry before and after purification. A fluorescence-minus-one control (FMO; cells labeled for CD4 but not for activation markers) was used as negative control to set the gate that measures activation in purified resting CD4+ T cells. The quadrants were set such that 0.5% of cells were present in the upper right quadrant when the cells were not stained with activation markers. (The activation markers used for staining were CD25, CD69, and HLA-DR). (c) Kinetics of viral infection measured using quantitative Alu-PCR. ‘+ dN’ indicates addition of deoxynucleosides to the cell culture. Arrows indicate the time point at which cells were harvested for genomic DNA extraction. PBMC, peripheral blood mononuclear cell.
Cells were infected by spinoculation as described [15] using pNL4-3 derived virus particles packaged with the native HIVenvelope for cell entry. As reported previously, resting cells do not divide after spinoculation as measured by BrdU incorporation, nor does the treatment induce expression of activation markers, and viability of resting cells after spinoculation is typically 100% and the yield of cells 3 days after infection is typically 50% [24]. Because low pools of deoxynucleoside triphosphate substrates in resting T cells are associated with inefficient reverse transcription [7], we also infected some aliquots of cells in the presence of added deoxynucleosides to boost efficiency (though this did not turn out to be necessary to recover sufficient integration sites). Quantitative PCR [16,17] showed approximately one to three proviruses per cell for all DNA pools (Fig. 1c). Consistent with earlier studies [6,7,24], the kinetics of HIV integration in activated cells was faster than in resting cells.
Integration sites were isolated as described previously [18,19,21]. Briefly, genomic DNA was purified, digested with the restriction enzyme MseI, and linkers ligated to the digested ends. Proviral-host DNA junctions were amplified by PCR in a first round using primers annealing to the linker and to the U5 region of the LTR. A second round of nested PCR introduced DNA barcodes and binding sites for the 454/Roche sequencing primers. Samples were pooled and sequenced using 454/Roche pyrosequencing [28].
A total of 2661 unique sites were recovered and mapped to the human genome (Table 1). For comparison, matched random control sets were generated computationally by randomly choosing three genomic sites lying the same distance from an MseI cut site as each of the integration sites. This method for generating matched random controls accounts for biases in the recovery of integration sites based on their proximity to MseI sites and allows for more accurate statistical analysis [18,21,22,29–31].
Table 1.
Integration site data sets used in the study.
| Average values per 1 Mb surrounding integration sitec |
|||||||
|---|---|---|---|---|---|---|---|
| Virus | Target cell | Set name | Total sequence readsa | Unique integration sitesb | % G/C content/Mb | No. Transc. units/Mb | No. CpG islands/Mb |
| HIV | Activated CD4+ T cell | Activated | 1183 | 524 | 46.7*** | 9.5*** | 60.9*** |
| HIV | Resting CD4+ T cell | Resting | 1955 | 947 | 44.5*** | 6.7*** | 40.0*** |
| HIV | Activated CD4+ T cell | Activated+dN | 1500 | 663 | 47.3*** | 9.5** | 67.0*** |
| HIV | Resting CD4+ T cell | Resting+dN | 1076 | 527 | 44.4*** | 6.9** | 38.0*** |
Transc., transcription.
The number of sequences recovered by pyrosequencing that contained the proper barcode and long terminal repeat (LTR) primer.
The number of total sequence reads (a) that had a single best match to the human genome of >98% identity, where the terminal viral sequence (5′-CA-3′) is within 3 bp of the high quality match and where all duplicate integration sites were condensed into a single entry.
The average values of ‘% G/C content/Mb’, ‘No. Transc. units/Mb’ and ‘No. CpG islands/Mb’ correspond to data sets used to generate heatmap tiles in Fig. 4 for ‘GC content, 1 Mb’, ‘Expression density, Unigene, gene density, 1 Mb’ and ‘CpG Islands, Density, 1 Mb’, respectively.
*P < 0.05.
P < 0.01.
P < 0.001.
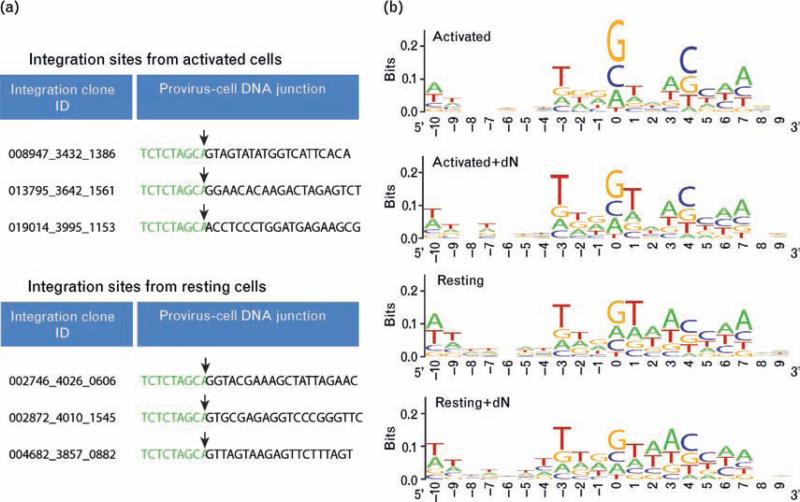
Integration site sequences were judged to be authentic if they showed a more than 98% match to a cellular sequence; showed a single best match to the human genome; and if the 5′-CA-3′ sequence of the terminal viral DNA was within three bases of the start of the high quality match to the human genome sequence. Representative junctions are shown in Fig. 2a. Note that 1474 sites from the resting cell sequences met these criteria, indicating that integrase-mediated integration takes place in the resting cell pools studied here. This is consistent with data showing that HIV DNA can be detected within the chromosomal DNA of resting CD4+ T cells inoculated with HIV by Alu-PCR [6–8,24].
Fig. 2. Primary human DNA sequences at sites of integration.
(a) Examples of sequences recovered from resting and activated cells. HIV long terminal repeat (LTR) sequence is shown in green with 25 bp of human genomic sequence in black. The viral-host DNA junction is marked by an arrow. (b) Information content at aligned integration target sites from activated and resting integration site data sets. The Y-axis indicates base conservation, with perfect conservation equaling two bits and no conservation equal to zero. The X-axis shows nucleotide positions relative to the integration site (10 bases on each side with the point of integration between positions –1 and 0).
Comparison of primary sequences at integration sites
HIV is known to favor integration at the weakly conserved palindromic sequence 5′-GT(A/T)AC-3′ at the point of integration [22,32–35]. Although this preference is weak, it can be a strong predictor of integration targeting in comparisons to randomly chosen sequences [22]. To investigate sequence preferences at the point of integration in the resting and activated data sets, we examined 20 bp of genomic sequence surrounding the point of integration for each. Consensus sequences at the point of integration did not differ whether or not infections were supplemented with deoxynucleosides or in resting versus dividing cells (Fig. 2b). These data support the idea that correct integrase-mediated integration took place in the cell populations studied here.
Quantifying genome-wide distributions of integration sites relative to mapped genomic features
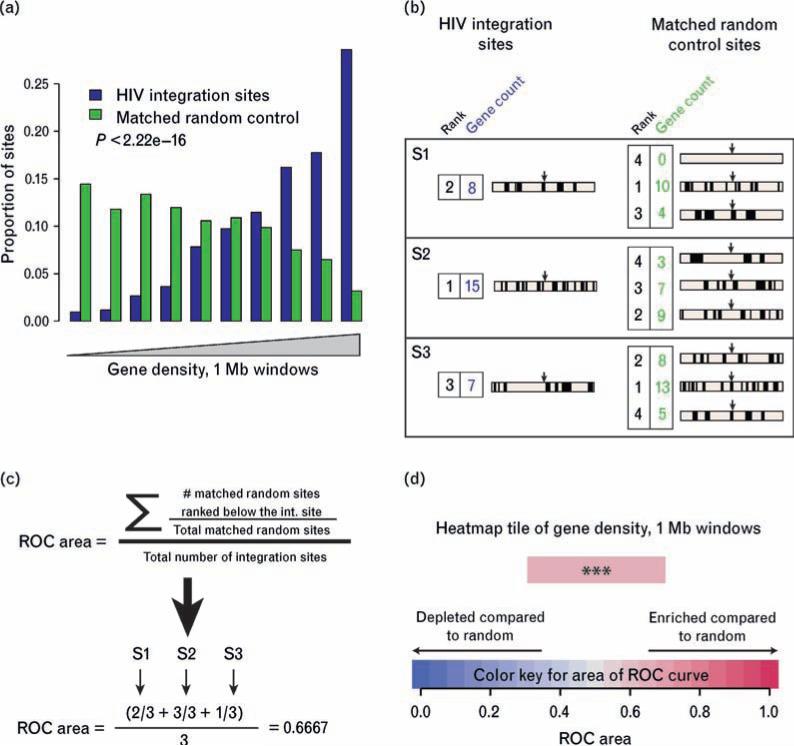
We examined the distribution of integration site patterns in resting and activated T cells relative to annotated features on the human genome sequence. For this, a heatmap format was developed to summarize many relationships using the receiver operating characteristic (ROC) area method introduced in [22]. Figure 3 summarizes the construction of an ROC curve. Figure 3a shows a conventional histogram illustrating the highly significant (P < 2.22e–16) correlation of integration frequency in activated T cells with relatively higher gene density compared with matched random controls. The experimental integration sites are more frequently found in bins of high gene density (right side of histogram) compared with the matched random control.
Fig. 3. Construction of heat maps using the receiver operating characteristic area method.
(a) Conventional histogram of data from the activated cell data set (without added deoxynucleosides) showing favored integration of HIV in gene-dense regions. All integration sites and matched random controls were annotated for gene density in the 1 Mb region surrounding the integration site. The pool of sites was then separated into 10 equal bins by relative gene density, with lower gene density to the left and higher to the right. The proportions of the experimental sites and matched random controls were then plotted for each bin, allowing visualization of the higher proportions in the experimental set at higher gene densities compared with the control set. (b) Annotating and ranking integration sites and matched random controls for construction of receiver operating characteristic (ROC) areas. Genes are represented by short vertical lines, integration sites by arrows. The gene count is shown beside each genomic interval; the rank over the pooled data set is shown colored blue for experimental integration sites or green for matched random controls. (c) Use of rank information to generate the ROC areas. For each set of one experimental site and three matched random controls, the experimental site is ranked relative to the matched random control. Positive correlation is indicated by values greater than 0.5, negative correlation as values less than 0.5. (d) Color-coding of heatmap tiles. Increasingly intense shades of red = enriched compared to random; increasing shades of blue depleted compared to random. The heatmap tile in (d) represents the same data shown in histogram form in (a). The P value, determined by a logistic regression method that respects the pairing in the data (clogit), is overlaid on the heatmap tile. (*P < 0.05; **P < 0.01; ***P < 0.001).
Figure 3(b–d) provides an example of how the comparison shown in Fig. 3a can be converted into a single colored tile of a heat map. The genomic interval surrounding each experimental or control site was extracted and the number of genes found within it quantified. In the example, 1 Mb windows were studied (Fig. 3b). Each sequence was then ranked by relative gene density in the flanking 1 Mb (Fig. 3b, numbers beside the sequences). ROC areas were then calculated by determining the number of matched random control sites with ranks lower than the integration sites, and this number was divided by the total number of matched random controls (Fig. 3c). All values for all sets of integration sites and matched random controls were then averaged, yielding the final ROC (0.667 in Fig. 3c). An ROC area greater than 0.5, as in the example, indicates positive correlation between the experimental integration site data set and the genomic feature studied. An ROC area less than 0.5 indicates negative correlation.
A single colored tile can be used to represent the resulting ROC area (Fig. 3d). Enriched associations are shown as increasing shades of red, negative associations as increasing shades of blue, and no difference from random as white (Fig. 3d). The statistical significance can be determined by regression and is represented by asterisks overlaid on the tile (*P < 0.05; **P < 0.01; ***P < 0.001) [22].
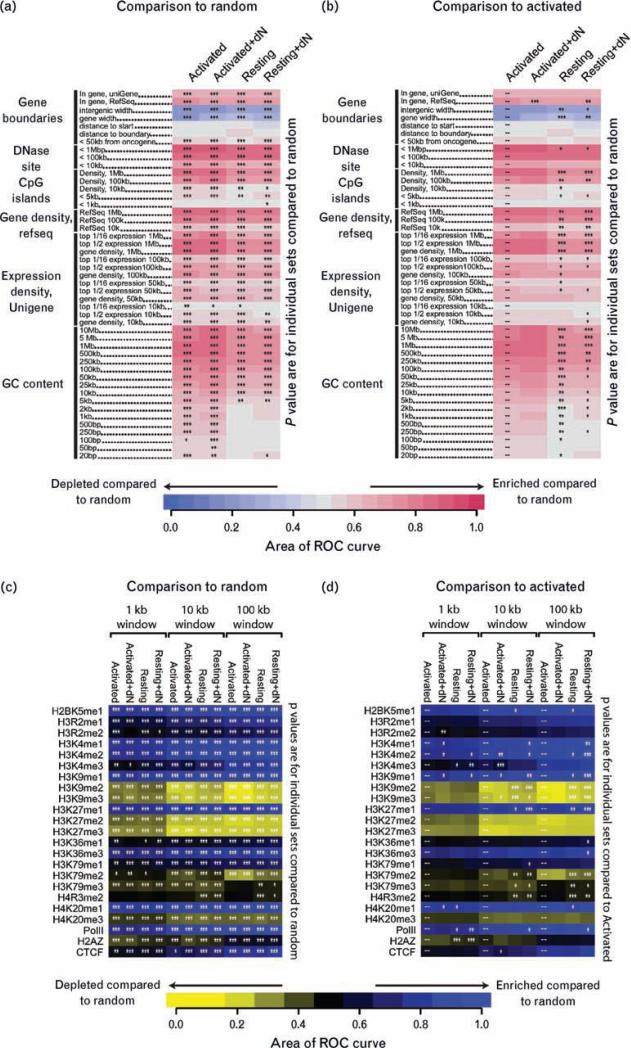
Many forms of statistical comparisons are possible among the four data sets and the dozens of genomic features queried (Fig. 4) [22]. For example, one might wish to know whether ROC areas for the experimental data sets differ from the matched random controls, or alternatively whether data sets differ from each other (e.g. resting versus activated cells). The heat maps described above have been further developed to allow users to carry out interactive statistical comparisons to investigate these questions. In interactive supplementary data 1 (found at http://bushmanlab.pbwiki.com/f/RestVsActGenomicHeatmap.zip; see the Methods section for further instructions), we introduce user-configurable statistical tests, where clicking on any row of the heat map allows statistical comparison to all other rows, clicking on any column allows statistical comparison to all columns, and clicking on the indicated button to the right of the heat map allows comparison of all experimental data to matched random controls. Thus, readers can use this automated tool to visualize the statistical significance for any comparison of interest.
Fig. 4. Genomic heat map of integration frequency relative to genomic features.
Integration site data set names are shown above the columns. Genomic features analyzed are shown to the left of the corresponding row of heat map. A colored receiver operating characteristic (ROC) area scale is shown along the bottom of the panel with increasing shades of blue indicating negative correlation relative to the genomic feature and increasing shades of red indicating positive correlation relative to the comparison set. P values showing significance of the departure from the comparison set are shown with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). (a) Comparisons of each experimental data set to the matched random controls relative to frequency of the indicated genomic feature. Asterisks summarize the statistical significance of departures from random. (b) Heat map similar to that in (a), but here the statistical test compares the activated cell data set to each of the other experimental sets. The naming of genomic features is described in the text. A version of these heat maps with user-configurable statistical tests can be found in the interactive supplementary information. (c) Heat map of integration frequency relative to epigenetic marks and chromatin-bound proteins. Associations of integration with histone methylation and chromatin-bound proteins were quantified using ROC areas [22], comparing the association of integration site data sets with the frequency in corresponding matched random controls. A colored ROC area scale is shown along the bottom of the panel with increasing shades of yellow indicating negative correlation of the experimental data set relative to matched random control and increasing shades of blue indicating positive correlation relative to the matched random control. Data sets and window sizes analyzed are shown above each column. CCCTC-binding factor (CTCF) is a DNA-binding protein proposed to be associated with chromatin boundaries. P values showing significance of the departure from the comparison set are shown with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001) Pair-wise comparisons of all sets to matched random controls relative to correlations with histone modifications or chromatin-binding proteins. Colored tiles and asterisks represent significance of departure from random. (d) Pair-wise comparisons of the activated set to each of the other T cell data sets. Asterisks summarize the significance of the departure of each experimental data set from the activated set. Colored tiles represent comparisons to matched random controls as in (c).
Comparison of HIV integration site distributions in activated and resting CD4+ T cells
Heat maps of the four HIV integration site data sets are shown in Fig. 4a and b. For both the activated and resting cell infections, deoxynucleosides were added or not to replicate cultures and integration sites analyzed separately. Addition of deoxynucleosides had only very slight effects on integration site selection. Figure 4b shows that for the two activated sets with or without deoxynucleoside addition, only one of 47 comparisons achieved significance. All comparisons between a given data set and all other sets can be carried out using the interactive tool in supplementary data 1. In what follows, the closely similar sets with or without deoxynucleosides serve as replicates documenting the reproducibility of differences between the activated and resting cell data sets.
Figure 4a shows that most comparisons to random for all four data sets were significant (asterisks on each tile). Starting at the top two rows of Fig. 4a, integration was favored within transcription units called by the uniGene and RefSeq databases, as seen in all previous studies of lentiviral integration site distributions [18,21,22,29,31,36–42]. The next two rows (intergenic width and gene width) indicate that integration was less frequent in intervals of relatively long distances between transcription units and less frequent in transcription units of relatively long lengths. Both of these features are characteristic of gene-rich regions. Tests of correlations with distances to gene 5′ and 3′ ends (distance to start, distance to gene boundary) did not achieve significance, reflecting the previous finding that HIV integration is evenly distributed over the length of transcription units [29]. Integration within 50 kb of proto-oncogene 5′ ends was increased compared with random, consistent with favored integration in gene-rich regions. Integration was enriched near sites of DNAseI cleavage and CpG islands, with the effects stronger over longer interval sizes. This length dependence reflects the fact that over long intervals DNAseI sites and CpG islands are characteristic of gene-rich regions, which is favored for HIV integration, whereas over shorter intervals these sites are enriched near gene promoters, which are disfavored for HIV integration.
Most measures of gene density were positively correlated with HIV integration frequency, and statistical tests achieved high significance. The correlation between gene density and integration frequency was tested over multiple interval sizes (10 kb to 1 Mb) and found to be significant for each. Transcriptional intensity was also compared. In this measure, gene activity in T cells was quantified using Affymetrix microarrays, and then genes ranked for relative expression levels. The expression intensity measure was quantified as for gene density; except only genes in the upper half of the expression ranks (‘top ½ expression’) or upper 16th (‘top 1/16 expression’) were scored. Most expression intensity measures were positively correlated with integration frequency.
In the human genome, G/C-rich regions are correlated with regions of high gene density, high expression intensity, high densities of CpG islands, and high densities of DNAseI sites. Correlations with integration site densities and G/C richness were analyzed over interval sizes ranging from 20 bp to 10 Mb. Over longer chromosomal intervals, HIV integration was positively correlated with G/C richness, paralleling the preference for integration in gene-rich regions. Over short intervals, the strength of the trend is reduced near to or equal to random. Previous work has shown that HIV integration is favored in DNAwrapped on nucleosomes [18,43–45], and nucleosome wrapping is favored by periodic A/T-rich motifs in DNA [46], probably explaining the diminished G/C sequence preference over short intervals (<2 kb).
The resting and activated cell sets differed over many of the forms of annotation analyzed, but in most cases the differences were modest in magnitude. Table 1 shows the quantitative differences for a few values (%GC content, number of transcription units, and number of CpG islands, all measured over 1 Mb intervals). Compared with resting cells, in the 1 Mb intervals surrounding integration sites in activated cells, G/C content was 2–3% higher, about two more genes were found, and about 25 more CpG islands were found. The asterisks on Fig. 4b summarize the statistical significance of pair-wise comparisons of the activated cell data set to the other three data sets. Statistical comparisons of any data set (columns in Fig. 4) to all others are available in the interactive supplementary data.
In all cases in which significant differences were seen, the integration site distributions in the resting cell data sets more resembled the matched random controls than did the activated cell data sets. There were some slight differences in the frequencies of integration within transcription units between the resting and activated cell data sets (Fig. 4b), but onlyone of fourcomparisons between resting and activated achieved statistical significance, so we conclude that integration in transcription units was about equally favored in the resting and activated cells. However, for resting cells, integration frequency within gene-rich regions was reduced compared with activated, and integration near associated features such as DNAseI sites, CpG islands, and regions of high G/C-rich content was diminished as well. Effects were most pronounced over longer intervals. The strongest effects involved the G/C-rich regions and CpG islands.
Thus, differences between the activated and resting cell data sets could be discerned, though they were all quantitative in nature, involving less extreme departures from random for the resting cell data sets.
HIV integration frequency in activated versus resting T cells compared with histone modifications and chromatin-bound proteins
We also investigated the integration site distribution for the resting and activated cells relative to 20 types of histone modifications and selected chromatin-bound proteins (Fig. 4c and d). For this we used data from a study of resting T cells in which chromatin immunoprecipitation and Solexa sequencing were used to map between 1 and 16 million sequence tags for each of these histone modifications and proteins [47]. As with the genomic heat maps, comparisons were done over multiple window sizes to maximize detection of differences between sets. Detailed information on these epigenetic marks can be found in [47,48].
The activated and resting cell data sets showed similar patterns, positively correlating with histone modifications and bound proteins found in actively transcribed regions (e.g. H2BK5me1; H3K4me1,2,3; H3K9me1; Pol II, etc.) and negatively associating with marks common to heterochromatin and gene repression (e.g. H3K9me2 and me3; H3K27me3, etc.).
Differences between resting and activated sets were modest though statistically significant, particularly when larger genomic window sizes (100 kb) were used in the comparisons. Comparisons over larger genomic lengths may achieve significance more easily because each bin contains more total tags from the ChIP-seq experiment. Comparing the resting and activated cell data sets, less negative correlation was seen in the resting cell data sets with H3K9me2 and 3 (which localize to silent chromatin) and H3K79me2 (which has no obvious localization preference) and more negative association of H3K79me3 (found at promoters) and H4R3me2 (which has no obvious localization preference). Thus, the differences in associations with histone modifications seen for resting and activated cells are generally consistent with the differences in integration targeting observed for other types of genomic features.
Discussion
Recent studies [24,49,50] suggest that HIV is able to infect resting CD4+ T cells, raising the question of whether integration in resting cells may contribute to formation of the latent reservoir. Previous studies have supported the idea that integration in specific chromosomal regions can result in suboptimal HIV gene expression, and this in turn correlated with formation of inducible (i.e. latent) proviruses, at least in cell culture models of latency [9]. Thus, we sought to investigate whether HIV proviruses formed by integration in resting cells were found more commonly in genomic regions suboptimal for gene expression.
Overall the distributions of integration sites in resting and dividing CD+ T cells were similar, with integration favored in active transcription units in all data sets, but quantitative differences could be detected. The replicate experiments (i.e. with or without added deoxynucleo-sides) were closely similar, supporting the idea that the differences observed between resting and activated cells were not due to experimental error. We found that activated cells sustained more integration in regions that were gene-dense, CpG island-dense, and GC-dense than did resting cells. Significant differences in integration frequency near some types of histone modifications were also detected.
The HIV integrase-binding protein PSIP1/LEDGF/p75 is known to help direct HIV integration into active transcription units [21,41,51–53], raising the question of possible involvement of LEDGF protein here. A previous report showed that the level of LEDGF expression in different cell types correlated with the proportion of HIV integration occurring in transcription units [21], so we compared LEDGF expression in published transcriptional profiling data for activated and resting T cells [23]. We found that LEDGF expression was actually slightly higher in resting T cells compared with activated T cells, inconsistent with a role for LEDGF here. In addition, when LEDGF activity is reduced, the fraction of integration sites near CpG islands actually increases [21,41], whereas we found that resting cells showed less integration near CpG islands than activated cells. Thus, variations in the level of LEDGF expression probably do not explain the differences between resting and activated cells seen here.
In summary, differences between integration targeting in resting and activated T cells are detectable and statistically significant, though quantitatively modest. In previous studies [7,24], infection of resting cells prepared as described here resulted in substantial levels of integration of viral DNA but relatively low levels of Gag protein production, providing a model for latency. Here we report that integrated proviruses are found more often in relatively less gene-dense regions in resting cells than in activated cells. Proviruses in such gene deserts may be more prone to forming latent proviruses, as in functional studies integration in gene deserts correlated with an inducible or latent phenotype [30]. Weinberger et al. [54] have shown that when levels of HIV Tat protein are low and fluctuating, drops in Tat expression switch HIV gene expression into a stable off state because Tat protein positively activates its own synthesis. The differences reported here are consistent with a model in which switching off of HIV transcription might occur more frequently in resting cells than in activated cells because of increased integration in gene deserts in resting cells. Thus, according to this idea, infection of resting cells would more often lead to latent infection due to distinctive features of integration target site selection.
Acknowledgements
We are grateful to members of the O'Doherty and Bushman laboratories for help and suggestions. This work was supported by NIH grant AI52845, the University of Pennsylvania Center for AIDS Research, and the Penn Genome Frontiers Institute with a grant with the Pennsylvania Department of Health to F.D.B. and by NIH grants AI058862-06 and AI058862-04S1 to U.O. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
References
- 1.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina C, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 5.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 6.Vatakis DN, Bristol G, Wilkinson TA, Chow SA, Zack JA. Immediate activation fails to rescue efficient human immunodeficiency virus replication in quiescent CD4+ T cells. J Virol. 2007;81:3574–3582. doi: 10.1128/JVI.02569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plesa G, Dai J, Baytop C, Riley JL, June CH, O'Doherty U. Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. J Virol. 2007;81:13938–13942. doi: 10.1128/JVI.01745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenasi T, Contreras X, Peterlin BM. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe. 2008;4:123–133. doi: 10.1016/j.chom.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y, Lin YB, An W, Xu J, Yang HC, O'Connell K, et al. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe. 2008;4:134–146. doi: 10.1016/j.chom.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu S, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler S, Hansen M, Bushman FD. A quantitative assay for HIV cDNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 18.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall H, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, Schretzenmair R, et al. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J Virol. 2005;79:14179–14188. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamad AR, Srikrishnan A, Mirmonsef P, Broeren CP, June CH, Pardoll D, et al. Lack of coreceptor allows survival of chronically stimulated double-negative alpha/beta T cells: implications for autoimmunity. J Exp Med. 2001;193:1113–1121. doi: 10.1084/jem.193.10.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas AK, Maus MV, Shalaby WS, June CH, Riley JL. A cell-based artificial antigen-presenting cell coated with anti-CD3 and CD28 antibodies enables rapid expansion and long-term growth of CD4 T lymphocytes. Clin Immunol. 2002;105:259–272. doi: 10.1006/clim.2002.5277. [DOI] [PubMed] [Google Scholar]
- 27.Suhoski MM, Golovina TN, Aqui NA, Tai VC, Varela-Rohena A, Milone MC, et al. Engineering artificial antigen-presenting cells to express a diverse array of co-stimulatory molecules. Mol Ther. 2007;15:981–988. doi: 10.1038/mt.sj.6300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewinski M, Bisgrove D, Shinn P, Chen H, Verdin E, Berry CC, et al. Genome-wide analysis of chromosomal features repressing HIV transcription. J Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens SW, Griffith JD. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6462. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carteau S, Hoffmann C, Bushman FD. Chromosome structure and HIV-1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci U S A. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 38.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 39.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, et al. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Pryciak P, Muller HP, Varmus HE. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc Natl Acad Sci U S A. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruss D, Reeves R, Bushman FD, Wolffe AP. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J Biol Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 45.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci U S A. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reilly C, Wietgrefe S, Sedgewick G, Haase A. Determination of simian immunodeficiency virus production by infected activated and resting cells. AIDS. 2007;21:163–168. doi: 10.1097/QAD.0b013e328012565b. [DOI] [PubMed] [Google Scholar]
- 50.O'Doherty U. Mechanisms of human immunodeficiency virus-1 latency. Transfusion. 2005;45:88S–91S. doi: 10.1111/j.1537-2995.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 51.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Ciuffi A, Diamond T, Hwang Y, Marshall H, Bushman FD. Fusions of LEDGF/p75 to lambda repressor promote HIV DNA integration near lambda operators in vitro. Hum Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- 53.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 54.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]