Abstract
The androgen receptor (AR) is critical for disseminated prostate cancer proliferation and survival. AR activity is targeted either through prevention of ligand synthesis or through the use of antagonists that bind the COOH-terminal ligand-binding domain. Although initially effective, treatment fails due to restored AR activity in the presence of therapeutics. Thus, new means must be developed to target AR activity. The SWI/SNF chromatin remodeling complex is critical for AR transcriptional activity, and the BAF57 SWI/SNF subunit facilitates direct interaction with the receptor. Although selected SWI/SNF subunit expression is reduced in prostate cancer, we show that BAF57 is retained in human disease and is elevated in a subset of tumors. Functional analyses showed that BAF57 contributes uniquely to androgen-mediated stimulation of transcription without compromising the effectiveness of AR antagonists. Subsequent studies revealed that BAF57 is recruited to the AR DNA-binding domain/hinge region, which occurs concomitant with receptor activation. These data provided the basis for a novel inhibitor derived from BAF57 [BAF57 inhibitory peptide (BIPep)], which blocked AR residence on chromatin and resultant AR-dependent gene activation. Importantly, BIPep expression was sufficient to inhibit androgen-dependent prostate cancer cell proliferation in AR-positive cells. In summary, these data identify blockade of AR-BAF57 interaction as a novel means to target agonist-induced AR function in prostate cancer, and provide the first evidence that abrogation of SWI/SNF function can be developed as a point of therapeutic intervention in prostate cancer.
Introduction
The androgen receptor (AR) is a ligand-activated transcription factor required for prostate cancer development and progression (1). AR is activated through androgen [testosterone or dihydrotestosterone (DHT)] binding to the receptor COOH-terminal ligand-binding domain (LBD; ref. 2). Thereafter, AR is released from heat shock proteins, forms a homodimer, and translocates to the nucleus, where the receptor uses a zinc finger DNA-binding domain (DBD) and COOH-terminal extension (CTE; within the hinge region of AR) to bind androgen-responsive elements (ARE) located within the promoter/enhancer regions of AR target genes [e.g., prostate-specific antigen (PSA); refs. 3, 4]. AR occupancy initiates the recruitment of cofactors (such as p160 coactivators and the SWI/SNF chromatin remodeling complex) that assist in inducing transcriptional activation and are necessary for generating an open chromatin environment (5). This open chromatin state permits subsequent binding of the basal transcription machinery to the promoter region and initiates transcription of target genes through the action of RNA polymerase II (6–8).
Given the stringent dependence of prostate cancer cells on AR action, first-line treatment for disseminated tumors is directed toward ablation of AR function, as achieved either through ligand (androgen) depletion or through the use of AR antagonists that compete for LBD binding and initiate formation of transcriptional repressor complexes (9, 10). Although initially effective, tumor cells override these inhibitory mechanisms and restore AR activity through alterations that include mutation of the LBD, over-expression of AR cofactors, AR amplification, or signal transduction–mediated (ligand independent) AR reactivation (1). As this stage of disease is incurable, it is evident that new means must be developed to inhibit AR function, independent of the receptor COOH terminus.
The SWI/SNF chromatin remodeling complex is recruited by AR in response to androgen and is thought to assist the receptor in promoting transcriptional transactivation through its ability to mobilize nucleosomes (11–14). SWI/SNF complexes alter nucleosome structure through the action of core ATPases (Brg1 or Brm), with specificity of SWI/SNF function rendered via the remaining 8 to 10 subunits, termed BRG1-associated factors (BAF). It has been previously reported that AR predominantly uses a subfamily of SWI/SNF complexes that depend on the Brm ATPase, but direct association of the receptor with SWI/SNF is likely fostered by direct interaction with the BAF57 or BAF155 subunits (13, 15). Because the BAF57 subunit is not required for SWI/SNF ATPase activity (16), but is thought to direct specificity of the complex, it was hypothesized that this SWI/SNF subunit could be targeted for disruption in prostate cancer and therefore provide novel means of therapeutic intervention.
In our present study, the relevance of BAF57 in AR signaling and as a novel therapeutic target in prostate cancer was determined. First, it was revealed that BAF57 expression is maintained in all disease states and elevated in a subset of tumors, suggesting that BAF57 likely persists in supporting AR activity throughout disease progression. Subsequent functional studies indicated that BAF57 is required for the response to AR agonists but is dispensable for the response to therapeutic AR antagonists, defining a unique role for the protein in the protumorigenic functions of AR. Biochemical analyses indicated that BAF57 is recruited early after androgen stimulation to the AR DBD/hinge region. Mapping of the protein-protein interface facilitated development of a BAF57 fragment that is sufficient for association with this AR region. Expression of this fragment, termed BAF57 inhibitory peptide (BIPep), was shown to destabilize AR-chromatin association and block resultant gene activation from clinically relevant target genes. Most importantly, BIPep expression blocked androgen-dependent cell proliferation in AR-positive (but not AR negative) prostate cancer cells. Together, these data are the first to identify SWI/SNF subunits as therapeutic targets in prostate cancer, and provide a new means to thwart AR activity, independent of the receptor COOH-terminal domain.
Materials and Methods
BAF57 antibody generation
The BAF57 antibody was generated with the assistance of Bethyl Laboratories. Briefly, a 20–amino acid peptide sequence (amino acids 291–310) of BAF57 was synthesized, purified by high-performance liquid chromatography, and verified by mass spectrometry. The peptide was conjugated to KLH and rabbits were immunized. Anti-BAF57 was affinity purified from rabbit serum.
Tissue culture
BT549, LNCaP, LAPC4, 22Rv1, DU-145, and PC-3 cells were cultured as previously described (13, 17–20). For culture in steroid-free conditions, cells were cultured in phenol red–free medium supplemented with charcoal dextran–treated FBS (CDT; Atlanta Biologicals).
Immunoblots
Immunoblotting was carried out as previously described (13). Antibodies used were generated against BAF57 (described above), lamin B (Santa Cruz Biotechnology), AR (N-20; Santa Cruz Biotechnology), cyclin-dependent kinase 4 (H-22; Santa Cruz Biotechnology), and FLAG (Sigma).
Immunohistochemistry
Briefly, tissue sections were treated with the Vectastain Elite avidin-biotin complex method rabbit staining kit and developed for 2 min with the 3,3′-diaminobenzidine substrate kit according to the manufacturer’s specifications (Vector Laboratories, Inc.). Specificity and optimal dilution of the rabbit polyclonal anti-BAF57 antibody (1:2,000) was determined using tissue sections from cell culture pellets obtained from BT549 (BAF57 negative) and LNCaP (BAF57 positive) cells (protocol adapted from ref. 21). Cell culture pellets for immunohistochemistry were generated by scraping asynchronous cell cultures in PBS. Cell pellets were suspended in three drops of plasma and thrombin was added to produce a cell clot. The clot was suspended in 10 mL of 10% neutral buffered formalin for 24 h, then embedded in paraffin, and sectioned for analysis. Further validation of the anti-BAF57 antibody was determined using tissue sections from localized and lymph node metastatic prostate cancer specimens obtained from the University of Cincinnati Department of Pathology in accordance with Institutional Review Board standards. BAF57 expression was determined using a tissue microarray (TMA) slide containing 80 cores (PR801; US Biomax). Patient tumors and the TMA were evaluated, graded, and semiquantitatively scored by a pathologist (M.P.R.) according to established guidelines (22). Immunoreactivity of BAF57 was scored on intensity (0, none; +, low; ++, moderate; +++, high) and extent of tumor staining (0, none; 1, <25%; 2, >25% to <50%; 3, >50%). The final BAF57 immunohistochemical score is displayed as a composite (intensity + extent; ref. 23). Mean expression composite and SDs are shown. Statistical analyses were performed using two-tailed Student’s t test.
Reagents
Dihydrotestosterone (DHT) was obtained from Sigma. Casodex (bicalutamide) was obtained from AstraZeneca Pharmaceuticals and used at 1 μmol/L for a 48-h treatment period where indicated.
Chromatin immunoprecipitation
Time course chromatin immunoprecipitations (ChIP) were performed as previously described (13). Quantification of the data was performed by quantitative real-time PCR on an ABI Step-One apparatus using Power SYBR Green Master Mix and primers directed against the PSA enhancer region (PSA G/H), the PSA promoter (PSA A/B), or the TMPRSS2 promoter. Primer sequences have been previously described (24, 25). Input reactions and negative control (IgG) immunoprecipitations were used to assess relative recruitment, as indicated. To examine AR and BAF57 recruitment in the presence and absence of BIPep (BAF57 1-145), LNCaP cells were seeded at 1 × 106 on poly-L-lysine–coated 10-cm dishes in improved MEM (IMEM), 5% ΔFBS. Cells were transfected as previously described (13) using Lipofectin transfection reagent (Invitrogen) with a 1:4 ratio of H2B-GFP to pcDNA3 vector or pcDNA3.1-1-145-3Xflag (BiPep). After 18 h, medium was changed to IMEM containing 5% CDT. Forty-eight hours after transfection, cells were stimulated with 10 nmol/L DHT for 1 h and harvested for ChIP analyses. Transfection efficiencies exceeded 70% to 80%, as judged by the percent of cells scoring positive for GFP (data not shown).
Plasmids
Most constructs have been previously described (13, 26). GST-AR 553-635 and GST-AR 634-668 were gifts from S. Balk (Harvard Medical School, Boston, MA; ref. 27). GST-AR 1-173 and GST-AR 1-660 were received from E. Wilson (University of North Carolina, Chapel Hill, NC; ref. 28) and GST-AR 505-676 was obtained from Z. Sun (Harvard Medical School, Boston, MA; ref, 29). pcDNA3.1(−)BAF57-flag, pcDNA3.1(−)BAF57ΔPR-flag, and pcDNA3.1(−)BAF57ΔHMG-flag were provided by Dr. T. Archer (NIEHS, NIH, Research Triangle Park, NC; ref. 30). pcDNA3-BAF57ΔN-flag was generated by subcloning BAF57-flag from the pBabe plasmid using BamHI and EcoRI restriction sites. pcDNA3.1-1-145-3Xflag (BIPep) was generated by PCR amplification of coding regions for the first 145 amino acids of BAF57 with the addition of a BamHI restriction site at the 3′ end. PCR amplification was followed by TA cloning (Invitrogen). This fragment was then cloned in frame into the pcDNA3.1-3Xflag vector using EcoRI and BamHI restriction sites. The construct was verified by sequencing.
GST pull-down
GST fusion protein production and immobilization was performed as previously described (13). Immobilized proteins were incubated for 16 h at 4°C with the indicated in vitro transcribed/translated [35S]Met-labeled proteins (generated using TNT-Coupled Reticulocyte Lysate systems, Promega). Binding reactions were carried out as previously described (13). Samples were separated by SDS-PAGE, stained with Coomassie Blue, and subjected to fluorography using Fluoro-Hance (Research Products International). Bound and input proteins were detected by autoradiography and quantified using a PhosphoImager (Molecular Dynamics).
Transfection and reporter analyses
BT549 cells were transfected in steroid-free medium with FuGENE6 (Roche Molecular Biochemicals) as per the manufacturer’s protocol. To assess BAF57 activation of wild-type AR (wtAR) or ARΔ629-636, transfections in six-well dishes consisted of 0.5 μg pSG5AR or pSG5ARΔ629-636, 0.5 μg pARR2-Luc, 0.25 μg pTK-Renilla-Luc, and 2.25 μg pBabe-wtBAF57-flag or vector control, as indicated. Following transfection, medium was changed and 0.1 nmol/L DHT or 0.1% ethanol vehicle was added as indicated. Cells were treated for 24 h, harvested, and lysed. Reporter analysis was carried out using the Dual Luciferase Assay Reporter System (Promega). To determine the effect of BIPep on BAF57-mediated AR activity, transient transfections were performed using 0.25 μg pSG5AR, 0.25 μg pARR2-Luc, 0.125 μg pTK-Renilla-Luc, 2.25 μg pBabe-wtBAF57-flag, and/or 0.5, 1.125, or 2.25 μg of pcDNA3.1-1-145-3Xflag (BIPep). Reporter assays reflect the means and SDs of experiments with at least nine independent data points. Statistical analyses were performed using two-tailed Student’s t test.
Reverse transcription-PCR
For semiquantitative analyses, RNA was obtained and converted to cDNA as previously described (13). Primers were used for PSA (primer pair: 5′-CTTGTAGCCTCTCGTGGCAG-3′ and 5′-GAC-CTTCATAGCATCCGTGAG-3′), TMPRSS2 (primer pair 5′-AATCGGTGTGT-TCGCCTCTAC-3′ and 5′-GCGGCTGTCACGATCC-3′), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; loading control; primer pair: 5′-CCACCCCATGGCAAATTCCATGGCA-3′ and 5′-TCTAGACGGCAGGT-CAGGTCCACC-3′). For PSA and GAPDH, PCR conditions were as follows: 94°C for 2 min; 21 cycles (PSA) or 28 cycles (GAPDH) of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s; and 72°C for 10 min. TMPRSS2 amplifications were performed using cycles of 94°C for 5 min; 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and 72°C for 5 min. PCRs were separated on a 1% agarose gel and representative data were shown from a minimum of at least two to three independent experiments. Quantification was performed by real-time PCR using the Taqman Fast Universal PCR Master Mix. PSA and GAPDH probe and primer sets were obtained from Applied Biosystems (HS00266705 and HS00426859). TMPRSS2 was quantified using SYBR Green real-time analyses using the following primer pair: 5′-GTG-ATGGTATTCACGGACTGG-3′ and 5′-CAGCCCCATTGTTTTCTTGTA-3′.
Bromodeoxyuridine incorporation
LNCaP or PC-3 cells were seeded on poly-L-lysine–coated coverslips in steroid-free conditions and transfected as previously described (13). Cells were cotransfected with vectors encoding H2B-GFP (as a marker of transfection efficiency) and one of the following expression plasmids: pcDNA3 (vector control), pBabe-BAF57ΔN-flag, pCMV-p16ink4a, dnAR (pSG5ARΔ46-408), or pcDNA3.1-1-145-3Xflag (BIPep). After transfection, bromodeoxyuridine (BrdUrd; GE Healthcare) was added for a 16-h incubation period. Indirect immunofluorescence was performed as previously described (13) to detect BrdUrd. H2B-GFP–positive cells (marking transfection) were screened for BrdUrd-positive staining. Results are plotted as percent BrdUrd inhibition versus vector control, as indicated. Data shown are reflective of at least three experiments, wherein each experiment consisted of at least two to three independent replicates.
Results
BAF57 expression is maintained in prostate cancer specimens and elevated in a subset of tumors
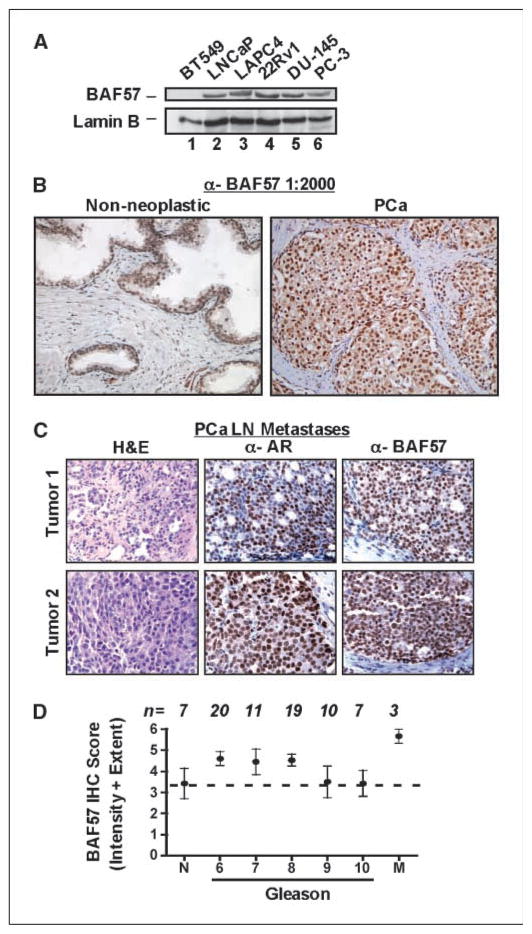
To assess the effect of BAF57 on AR signaling in prostate cancer, BAF57 expression patterns were examined in prostate cancer specimens. Antisera directed against BAF57 were first generated, as described in Materials and Methods. Specificity was assessed in multiple systems; no immunoreactivity was detected in BT549 cells by immunoblot (Fig. 1A, lane 1), which are devoid of this SWI/SNF subunit (13, 31). By contrast, BAF57 was apparent in LNCaP (prostatic adenocarcinoma) cells (lane 2) consistent with previous reports (13). Screening of additional prostate cancer cell lines (lanes 3–6) showed that BAF57 was detected in all tested model systems. Although these data were suggestive that BAF57 expression is retained in prostate cancer, immunohistochemical studies were undertaken to quantify expression in primary tumors. Specificity for BAF57 immunoreactivity in paraffin-embedded specimens was validated using fixed and embedded BT549 and LNCaP cells (Supplementary Fig. S1A). Subsequently, expression of BAF57 in nonneoplastic and primary prostate cancer specimens was examined. No positive staining was observed in the absence of primary antibody (Supplementary Fig. S1B). However, significant nuclear expression of BAF57 was evident in the presence of antibody for the nonneoplastic sample (Fig. 1B, left), with the majority of protein localized within the epithelial layer of the prostate glands and minimal expression observed in the stromal layer of the tissue. High nuclear BAF57 expression was also observed in the prostate cancer sample (Fig. 1B, right). Furthermore, BAF57 expression was high in metastatic samples of prostate cancer (Fig. 1C). AR staining was used to validate the specimen as prostate cancer (middle); H&E control sections are also shown (left). Thus, BAF57 expression can be clearly observed in both localized and metastatic prostate cancer.
Figure 1.
BAF57 is highly expressed in human prostate cancer specimens. A, immunoblotting was performed for BAF57 and lamin B (loading control) in the LNCaP, LAPC4, 22Rv1, DU-145, and PC-3 prostate cancer cells. BT549 cells were used as the negative control. B, nonneoplastic (left) and primary (right) human prostate cancer (PCa) specimens were analyzed by immunohistochemistry for BAF57 using the generated antisera (BAF57, 1:2,000 dilution). C, independent lymph node (LN) metastases of prostate cancer were stained by standard H&E (left) and immunostained for AR (middle) or BAF57 (right). D, immunohistochemical (IHC) staining was performed for BAF57 on a TMA slide containing ~80 cores. n, number for each category. BAF57 immunohistochemical score is displayed as a composite of intensity + extent of tumor staining as indicated in Materials and Methods. X axis, Gleason scores. M, lymph node metastatic samples.
To quantify BAF57 expression in prostate specimens, a TMA containing both tumor tissue and nonneoplastic controls was stained and scored (Fig. 1D). Nonneoplastic prostate tissue scored positive for BAF57 staining, as expected. As shown, tumors of all Gleason grades were positive for BAF57, with Gleason 6 to 8 samples showing a trend toward increased BAF57 expression. These data did reach statistical significance by comparison of nonneoplastic tissue with metastatic samples (P < 0.05), which showed the highest levels of BAF57 expression. These data suggest that BAF57 expression is maintained or enhanced in prostate cancer. Congruent with this supposition, microarray profiling of BAF57 mRNA expression in metastatic prostate cancer (32) revealed a similar trend of elevated BAF57 mRNA expression in tumor tissue (Supplementary Fig. S1C). Together, these data indicate that BAF57 expression is retained in prostate cancer and elevated in a subset of tumors; therefore, BAF57 may provide a therapeutic target by which to deplete AR activity. To test this hypothesis, it was necessary to first dissect the mechanisms by which BAF57 regulates AR.
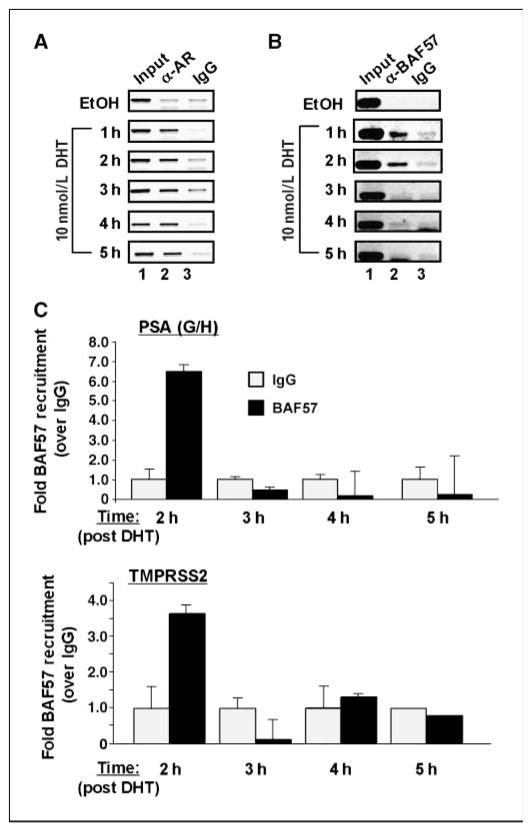
BAF57 is temporally recruited to endogenous AREs
It has been previously shown that BAF57 function is important for the response to androgen (13), but whether this requirement reflected an early or late event in AR activation is a critical determinant in dissecting BAF57 function. As shown by ChIP assay, AR was recruited to the PSA enhancer after androgen stimulation and occupied this locus for over a 5-h period (Fig. 2A), consistent with previous reports (33). By contrast, BAF57 was recruited early after DHT stimulation and cycled off the PSA enhancer after 3 h (Fig. 2B and C). Similar kinetic trends were observed on a second endogenous ARE present within the regulatory region of the AR-induced gene TMPRSS2 (Fig. 2C, bottom; ref. 34). These data indicate that BAF57 action at the ARE occurs early after agonist stimulation, consistent with a model wherein SWI/SNF activity is required for transcriptional initiation. By contrast, BAF57 (and SWI/SNF function in general) is not required for the response to AR antagonists (Supplementary Fig. S2). Although BAF57 was detected at the PSA (but not the TMPRSS2) locus after exposure to the AR antagonist bicalutamide (Supplementary Fig. S2B), functional studies showed that disruption of BAF57 activity did not alter the transcriptional or proliferative suppression after bicalutamide treatment (Supplementary Fig. S2C and D, respectively). Combined, these observations show that BAF57 is important for initiating the transcriptional response to agonist but is dispensable for antagonist function.
Figure 2.
BAF57 is intermittently recruited to AREs. LNCaP cells were cultured in steroid-free medium for 3 d and then treated with either 0.1% ethanol (EtOH) or 10 nmol/L DHT for the indicated time. Cells were harvested for ChIP analyses and antibodies to AR (A), BAF57 (B), or preimmune serum (IgG) were used to examine recruitment to the PSA enhancer region. At least three independent experiments were performed and a representation is depicted. C, quantification of BAF57 recruitment after DHT stimulation was determined by real-time PCR using primers that flank the ARE regions of the PSA enhancer (PSA G/H; top) or the TMPRSS2 locus (bottom). Fold recruitment over negative control (IgG, set to “1” for each time point) is plotted, and SDs of samples assessed in triplicate are shown.
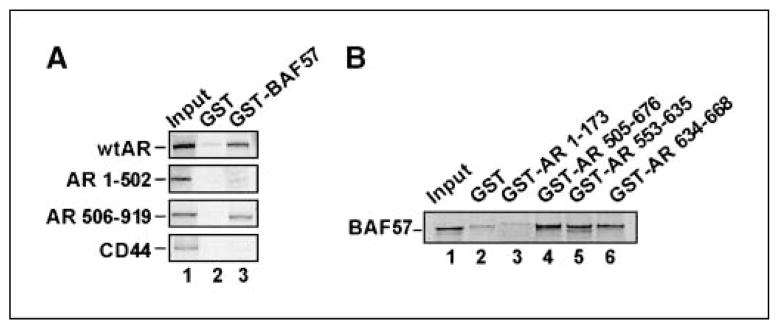
BAF57 binds to the DBD and hinge region of AR
Given the importance of BAF57 in the response to agonist, biochemical strategies were used to develop means of targeting the AR/BAF57 interface. First, to identify the means by which BAF57 impinges on AR, in vitro binding assays were performed. AR constructs and expression thereof are shown in Supplementary Fig. S3. As expected, BAF57 associated with full-length AR in vitro but not with CD44 (negative control; Fig. 3A). BAF57 showed no binding capacity to the NH2 terminus of AR (amino acids 1–502), whereas strong association with the COOH terminus (amino acids 506–919) was observed. The BAF57 association site was further localized to the central DBD and hinge regions within AR, whereas no association was observed with control NH2-terminal domains or the GST control (Fig. 3B, compare lanes 1–4). Subsequent dissection showed that the DBD and hinge region each harbor some BAF57-binding capacity (lanes 5 and 6), signifying that these combined motifs provide a means for BAF57 to exert its function on AR. Together, these studies identify the AR DBD/hinge region as the principal site of BAF57 association and indicate that this interaction uses AR motifs not currently targeted in cancer therapy.
Figure 3.
BAF57 binds AR DBD/hinge region. A, immobilized GST or GST-BAF57 was incubated with [35S]Met-labeled AR mutants or CD44 (negative control) overnight at 4°C, washed, and subjected to SDS-PAGE. Five percent input and bound protein were detected by autoradiography. B, immobilized GST or GST-AR fusions were incubated with [35S]Met-labeled BAF57 and pull-down analysis was carried out as in A.
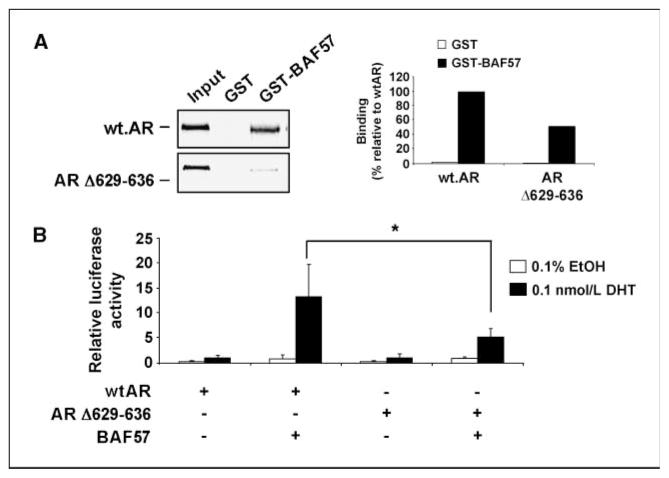
Loss of CTE within AR causes reduction in BAF57 binding and transactivation potential
The observation that BAF57 binds the hinge domain was of importance, as this region contains a CTE that facilitates AR binding to AREs (3, 4). To characterize the effect of the CTE on BAF57-mediated AR activation, a small, 8–amino acid deletion within the CTE (amino acids 629–636) of AR was used in GST pull-down analyses. A 50% reduction in BAF57 binding to ARΔ629-636 was observed compared with wtAR (Fig. 4A), indicating that this region is of importance for BAF57 binding. The functional importance of the AR CTE for BAF57-mediated activation of AR was examined through reporter analyses (Fig. 4B). As expected, minimal activation of wtAR was detected in the absence of BAF57. Identical results were observed for ARΔ629-636. Addition of BAF57 strongly induced activation of wtAR (13-fold), but not ARΔ629-636 (<5-fold), despite equal protein expression (shown in Supplementary Fig. S4B). Increasing dosage of BAF57 expression failed to enhance ARΔ629-636 in reporter assays, thus further emphasizing that this mutant is compromised for activation by BAF57 (Supplementary Fig. S4C). In short, these data show that the CTE is critical for BAF57-mediated activation of AR and that disrupting the BAF57-AR interaction may provide a means of attenuating AR activity.
Figure 4.
Loss of CTE within AR hinge diminishes binding to BAF57 and transactivation potential. A, immobilized GST or GST-BAF57 was incubated with [35S]Met-labeled wtAR or CTE mutant of AR (ARΔ629-636) and pull-down analysis was performed as in Fig. 3A. Right, binding as a percentage to wtAR. B, BT549 cells were transfected with pARR2-Luc and pTK-Renilla-Luc and pSG5AR, pSG5ARΔ629-636, or pBabe-BAF57-flag as indicated. After transfection, cells were treated with either 0.1% ethanol or 0.1 nmol/L DHT for 24 h. Cells were harvested and lysed, and luciferase activity was measured. DHT-stimulated wtAR activity is set to “1” and data are presented as relative light units. *, P < 0.05.
The NH2-terminal inhibitory peptide of BAF57 (BIPep) blocks AR activity and AR-dependent cancer cell proliferation
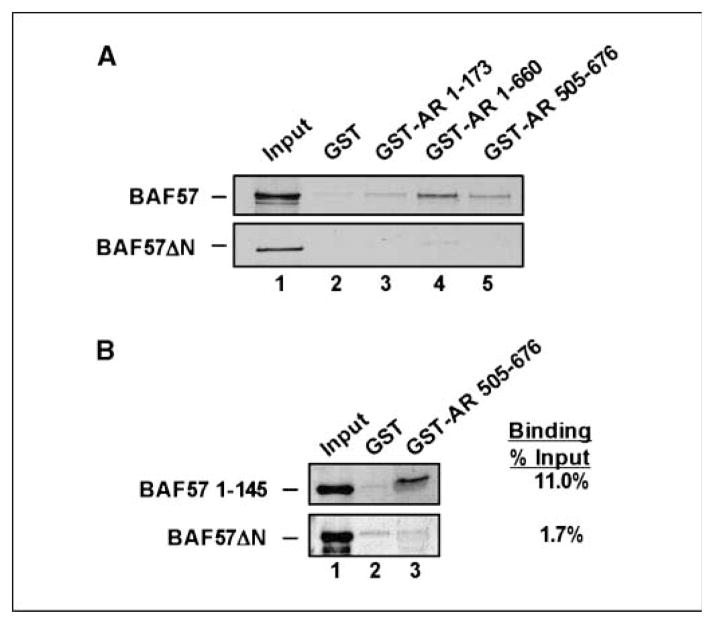
Discovery that BAF57 impinges on critical regions of AR not currently targeted in therapy highlighted the need to uncover the mechanisms to disrupt the AR-BAF57 association. BAF57 consists of several conserved domains (Supplementary Fig. S5A), although the requirement of these for SWI/SNF specificity remains largely unchallenged. To dissect the AR-binding region, GST pull-down analyses were performed, which revealed that both the BAF57 NH2-terminal proline-rich and HMG domains are required to support association with AR 505-676 (DBD/hinge region; Fig. 5A; Supplementary Fig. S5C). Furthermore, the minimal AR-binding site was shown to be sufficient for binding (Fig. 5B).
Figure 5.
BAF57 NH2 terminus is sufficient for AR association. A, immobilized GST or GST-AR fusions were incubated with [35S]Met-labeled BAF57 or BAF57ΔN and pull-down analysis was carried out as in Fig. 3A. B, immobilized GST or GST-AR 505-676 were incubated with [35S]Met-labeled BAF57 1-145 or BAF57ΔN and pull-down analysis was performed as in Fig. 3A. Right, percent binding.
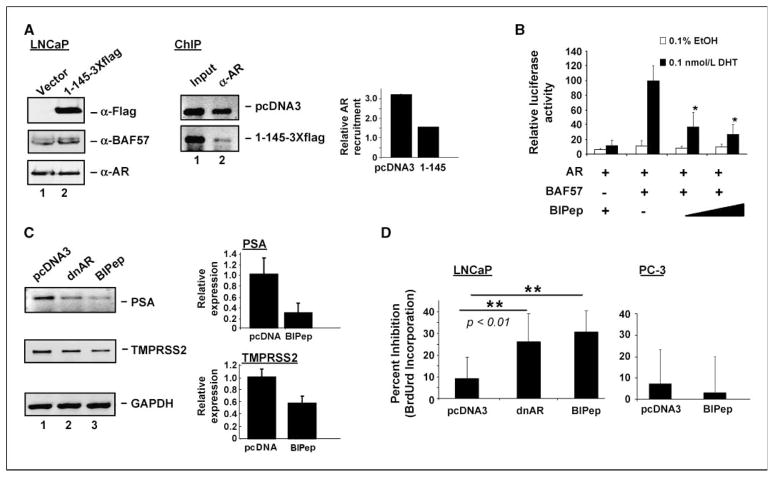
To decipher the importance of the BAF57/AR interface, a competitive inhibitor was developed. Specifically, a fragment encoding amino acids 1 to 145 of BAF57 was epitope tagged and transiently expressed in prostate cancer cells (Fig. 6A, left). Expression of the peptide did not influence endogenous AR or BAF57 levels. However, ectopic expression of BAF57 1-145 significantly reduced the recruitment of AR (Fig. 6A, middle and right) and BAF57 (data not shown) to the PSA enhancer element. Similar results were observed on examination of the AR recruitment to the TMPRSS2 locus (data not shown). Given the ability of the peptide to inhibit the AR-chromatin association, the fragment was renamed BIPep.
Figure 6.
BIPep reduces AR residence at AREs, limits AR activity, and suppresses AR-dependent prostate cancer cell proliferation. A, LNCaP cells were transfected with H2B-GFP and either pcDNA3 or pcDNA3-BAF57 1-145-3Xflag. After transfection, medium was changed to steroid-free conditions for 48 h. Cells were stimulated with 10 nmol/L DHT for 1 to 2 h and subjected to immunoblot analysis for flag, BAF57, or AR (left) or ChIP analysis for AR recruitment to the PSA enhancer region (middle). Ten percent input and AR recruitment are shown for the 1-h time point. Right, as shown by quantification of relative AR recruitment using real-time PCR, these findings were recapitulated at the 2-h time point. B, BT549 cells were transfected with pARR2-Luc, pTK-Renilla-Luc, pSG5AR, pBabe-BAF57-flag, and pcDNA3-BAF57 BIPep as indicated. Treatment and reporter analysis were carried out as in Fig. 4B. DHT-stimulated AR plus BAF57 is set to 100 and data are presented as relative light units. *, P < 0.05. C, LNCaP cells were transfected with H2B-GFP and plasmid encoding pcDNA3 vector, pSG5ARΔ46-408 (dnAR), or pcDNA3-BAF57 BIPep 3Xflag. Reverse transcription-PCR was performed for PSA, TMPRSS2, or GAPDH. Left, representative data. Quantification of relative PSA or TMPRSS2 expression (relative to GAPDH, with expression levels for vector-transfected cells set to “1”) is shown. D, LNCaP cells were transfected with H2B-GFP and pcDNA3, pSG5ARΔ46-408 (dnAR), or pcDNA3-BAF57 BIPep 3Xflag. Left, BrdUrd analysis was performed; right, parallel experiments were performed for PC-3 cells. Results from at least three independent experiments (each experiment with at least two to three independent biological replicates) are plotted as percent inhibition of BrdUrd incorporation over vector control. **, P < 0.01.
Given the effect of BIPep on AR recruitment, the influence of BAF57 1-145 overexpression on AR activity was determined through reporter analyses (Fig. 6B). As expected, AR showed no response to ligand in BT549 cells, which lack endogenous BAF57; conversely, introduction of BAF57 restored AR function in the presence of ligand (set to 100%). Under these conditions, introduction of BIPep elicited a dose-dependent reduction of AR activity (37% and 27%, respectively), providing functional evidence that targeting BAF57 can inhibit AR-dependent gene transcription. To further confirm this result with endogenous protein in prostate cancer cells, expression of two clinically relevant, prostate-specific AR target genes (PSA and TMPRSS2) was examined (Fig. 6C). As shown, robust induction of PSA and TMPRSS2 mRNA in the presence of androgen was inhibited by BIPep, similar to that observed with dominant-negative AR. These data show that BIPep significantly attenuates AR transcriptional activity on ectopic and endogenous AR targets. Given these effects of AR-BAF57 disruption on AR activity, the effect of BIPep on prostate cancer cell proliferation was investigated (Fig. 6D). As expected, dnAR showed significant inhibition of LNCaP cell proliferation (26% over vector control) in AR-dependent cells. Similarly, BIPep significantly attenuated prostate cancer cell proliferation in the presence of androgen (31% reduction). By contrast, BIPep did not suppress BrdUrd incorporation in AR-negative (PC-3) cells, thus displaying specificity of action. In synopsis, these data provide evidence that disruption of the AR-BAF57 interaction can serve as a novel means to antagonize AR function in prostate cancer, and reveal SWI/SNF as a new therapeutic target in this disease.
Discussion
AR is the critical therapeutic target for disseminated prostate cancer and is effectively targeted through manipulation of the receptor LBD. However, recurrent tumors invariably overcome this point of intervention, and alternative strategies to target AR function therapeutically remain elusive (9, 10). The present study identifies abrogation of BAF57 recruitment as a novel means to inhibit AR activity based on several key observations herein. First, BAF57 is expressed in prostate cancer and elevated in a subset of tumors, indicating that the protein may continue to support AR activity in all stages of disease progression. Second, BAF57 is required specifically for the response to AR agonists but not therapeutic antagonists and is transiently recruited to sites of AR activity after androgen stimulation. These observations suggest that BAF57 is required at the early stages of AR function. Third, BAF57 impinges on a region of AR not currently targeted by cancer therapeutics and interacts with a motif known to be essential for transactivation potential. Fourth, the AR-binding site of BAF57 was used to provide proof of principle that abrogation of BAF57-AR association can block receptor function. Expression of this inhibitory peptide (BIPep) caused destabilization of AR residence on chromatin and suppressed AR function on clinically relevant target genes. BIPep was also sufficient to inhibit proliferation in AR-positive (but not AR-negative) prostate cancer cells. Together, these data provide conceptual evidence of a new means to hinder AR function and AR-dependent cellular proliferation in prostate cancer.
The observation that BAF57 expression is retained in human prostate cancer specimens and seems to be elevated in a subset of these tumors is critical to the development of this AR effector as a potential therapeutic target. Several AR cofactors are induced as a function of prostate cancer progression, including SRC1, SRC2, and ARA70, and it is thought that these events may contribute to the development of recurrent disease (1, 35, 36). Whether BAF57 belongs to this class of AR modulator can now be determined, as the antisera described are effective for immunohistochemical studies. Intriguingly, several reports have been published, which support the contention that tissue-specific alteration of SWI/SNF subunits confers differential effects in cancer. Although it is well established that the SNF5 subunit is a rhabdoid-specific tumor suppressor (37), recent studies have implicated loss of the ATPase subunit Brm in gastric cancer (38), and depletion of Brm facilitates carcinogen-induced lung adenoma formation (39). Although Brm can support AR activity in prostate cancer cells, it has been observed that it is frequently down-regulated in prostate cancer6 and is associated with the acquisition of proliferative advantage. By contrast, Brg1 may be induced in prostate cancer and is associated with invasive disease (40). These collective observations underscore the growing appreciation that SWI/SNF complexes seem to exist in a delicate balance and that alterations in the expression of individual subunits can result in significant cellular outcomes. The observation herein that BAF57 levels seem to be induced in a subset of cancers suggests that altered BAF57 expression may be required to support or trigger alterations in cooperating SWI/SNF subunits. Although this hypothesis remains to be tested, it is apparent from the present study that BAF57 is critical for agonist-induced AR function in the context of androgen-dependent tumor cells.
The observed early recruitment of BAF57 to the sites of AR activity and its subsequent displacement indicate that SWI/SNF may be necessary to facilitate initial events in transcriptional activation. This supposition agrees with the proposed mechanisms of active SWI/SNF remodeling function in that the complex is thought to progress along the nucleosome-bound DNA (away from the AREs; refs. 7, 41). Importantly, AR is able to induce recruitment of the ATPase (42), presumably through the BAF57 interaction motif as shown herein. It has been proposed that retention of AR at the PSA enhancer permits looping and contact with the proximal promoter, thus augmenting transcriptional output (33). The observation of BAF57 dismissal indicates that SWI/SNF may no longer be required after this looping event occurs, and future investigations will test this hypothesis directly. Given the dynamic interplay between AR and BAF57, the biochemical mechanisms that underpin BAF57 recruitment were delineated.
As has been well documented, AR contains a zinc finger DBD that requires the CTE domain to permit binding to DNA (3, 26). A closer examination of the AR CTE uncovered this region as an important region for BAF57 binding and activation of AR. Interestingly, there is precedence for a requirement of the CTE to confer the binding of HMG DBD-containing proteins (HMGB1 and SOX9) to AR (43, 44). Moreover, this region of AR can serve as a platform for interaction with Ubc9, a SUMO-1 conjugating enzyme (45). As previous studies showed that BAF57 can cooperate with Ubc9 for AR activation (13), it will be intriguing to determine how Ubc9 may influence the ability of SWI/SNF to bolster AR activity. The DBD/hinge domain within the estrogen receptor (ER) has also been shown to be a primary region for BAF57 interaction, indicating a possible trend in BAF57-nuclear receptor binding (46). Because our data show that the AR-binding region of BAF57 (amino acids 1–145) is able to reduce AR residence at AREs, it is possible that binding of the BAF57 fragment to the CTE is sufficient to block either DNA binding or agonist-induced stabilization on DNA. Although additional investigation will be necessary to challenge this hypothesis, the current data strongly support ablation of the AR-BAF57 interaction as a means to disrupt AR-DNA association.
To move further toward the goal of inhibiting the AR-BAF57 interaction, the region of AR binding within BAF57 was defined. The PR and HMG domains of BAF57 emerged as the primary binding sites for AR, as deletion of these regions elicited a loss in AR binding. Interestingly, the observed binding to AR differs from that of ER association with BAF57, wherein truncation of either the NH2 or COOH terminus of BAF57 caused significant disruption of ER binding (46). Similarly, the PR/HMG domain of BAF57 is also dispensable for association with BRG1/BRM (47) and the BAF155 subunit of SWI/SNF, as this interaction was dependent on the coiled-coil and NHRLI domains of BAF57 (30). This latter observation may have implications for AR, as BAF155 (and its murine homologue, SRG3) has been shown to regulate expression of specific SWI/SNF components, including BAF57, and is thought to serve as a critical regulator of SWI/SNF subunit levels (30, 48). As would be expected, therefore, SRG3 is important for AR regulation (15).
Given the necessary role for the BAF57 NH2 terminus in AR binding and the ability of this region to disrupt AR recruitment, the utility of the AR-binding motif (BIPep) as a means to suppress AR activity was examined. As shown, AR activity was significantly attenuated and proliferation of AR-positive prostate cancer cells was diminished. Importantly, introduction of BIPep caused no reduction of endogenous full-length BAF57 protein expression, as shown in Fig. 6A. This is in contrast to previous observations that showed a decrease in wild-type BAF57 (wtBAF57) expression on BAF57 mutant introduction (30, 47). Therefore, effects observed on addition of this AR-binding region are not a result of decreased wtBAF57 expression. Taken together, these data present the BAF57-AR interface as a viable medium for disrupting AR signaling. These findings gain significance, as recent research has validated the feasibility of developing small molecules to effectively disrupt protein-protein interaction in cancer. For example, the Nutlin compounds were developed to block MDM2 interaction with p53, consequently stabilizing the p53 tumor suppressor and inducing cell cycle arrest and apoptosis (49). Furthermore, expression of the NH2-terminal region of AR has been shown to block the growth of prostate cancer (50). Additionally, targeting BAF57 could potentially be useful in combination with the commonly used therapy of AR antagonists (e.g., bicalutamide), as examination of BAF57 and SWI/SNF function on antagonist treatment revealed no functional requirement for the complex in antagonist-mediated repression (Supplementary Fig. S2). These findings are also consistent with analyses of SWI/SNF function in living cells, wherein the Brm ATPase was shown to be recruited to sites of AR function in the presence of agonist but not antagonist (42). The present observations provide much impetus to explore disruption of the AR-BAF57 interaction through small-molecule inhibitors and/or peptide exposure to impede prostate cancer proliferation.
In summary, the robust expression of BAF57 in a multitude of prostate cancer specimens and its dependence on the SWI/SNF complex strongly suggest that the integrity of BAF57 function is maintained in prostate cancer as a means to support AR activity. Specificity of action was revealed in that BAF57 seems selectively required for the response to agonist (but not antagonist) and acts rapidly at AR target sites on receptor activation. Disruption of this event, made possible through dissection of the AR-BAF57 interaction surfaces, proved sufficient to disrupt AR residence on chromatin, AR-dependent gene expression, and AR-positive prostate cancer cell proliferation. Combined, these data show that disruption of the BAF57-AR interaction may prove an effective, novel means to assist in suppressing AR function and cellular outcome in prostate cancer cells.
Supplementary Material
Acknowledgments
Grant support: R01CA116777 and R21CA121402 (K.E. Knudsen), Fonds voor Wetenschappelijk Onderzoek-Vlaanderen grant 1.5.065.05 (F. Claessens), and Department of Defense grant W81XWH-06-1-0098 (K.A. Link). A. Haelens is a postdoctoral fellow of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen.
We thank Dr. E. Knudsen, Dr. L. Morey, and M. Schiewer for critical reading of the manuscript; Drs. S. Khan, J. Zhang, C. Price, and B. Weissman for critical discussions; Dr. B. Weissman for the BRG1i and BRMi expression plasmids; and Dr. T. Archer for the BAF57ΔPR and BAF57ΔHMG constructs.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Shen et al., submitted for publication.
References
- 1.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann AO, Blok LJ, de Ruiter PE, et al. Mechanisms of androgen receptor activation and function. J Steroid Biochem Mol Biol. 1999;69:307–13. doi: 10.1016/s0960-0760(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 3.Schoenmakers E, Alen P, Verrijdt G, et al. Differential DNA binding by the androgen and glucocorticoid receptors involves the second Zn-finger and a C-terminal extension of the DNA-binding domains. Biochem J. 1999;341:515–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Haelens A, Verrijdt G, Callewaert L, et al. DNA recognition by the androgen receptor: evidence for an alternative DNA-dependent dimerization, and an active role of sequences flanking the response element on transactivation. Biochem J. 2003;369:141–51. doi: 10.1042/BJ20020912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–15. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Heinlein CA, Chang C. Androgen receptor (AR) coregulators: an overview. Endocr Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 7.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 8.Urnov FD, Wolffe AP. Chromatin remodeling and transcriptional activation: the cast (in order of appearance) Oncogene. 2001;20:2991–3006. doi: 10.1038/sj.onc.1204323. [DOI] [PubMed] [Google Scholar]
- 9.Balk SP. Androgen receptor as a target in androgen-independent prostate cancer. Urology. 2002;60:132–8. doi: 10.1016/s0090-4295(02)01593-5. discussion 8–9. [DOI] [PubMed] [Google Scholar]
- 10.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–61. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 11.Huang ZQ, Li J, Sachs LM, Cole PA, Wong J. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and mediator for transcription. EMBO J. 2003;22:2146–55. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–92. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 13.Link KA, Burd CJ, Williams E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–15. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall TW, Link KA, Petre-Draviam CE, Knudsen KE. Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem. 2003;278:30605–13. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 15.Hong CY, Suh JH, Kim K, et al. Modulation of androgen receptor transactivation by the SWI3-related gene product (SRG3) in multiple ways. Mol Cell Biol. 2005;25:4841–52. doi: 10.1128/MCB.25.12.4841-4852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 17.Wetherill YB, Petre CE, Monk KR, Puga A, Knudsen KE. The xenoestrogen bisphenol A induces inappropriate androgen receptor activation and mitogenesis in prostatic adenocarcinoma cells. Mol Cancer Ther. 2002;1:515–24. [PubMed] [Google Scholar]
- 18.Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403–9. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 19.Klein KA, Reiter RE, Redula J, et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med. 1997;3:402–8. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 20.Sadar MD. Androgen-independent induction of prostate-specific antigen gene expression via cross-talk between the androgen receptor and protein kinase A signal transduction pathways. J Biol Chem. 1999;274:7777–83. doi: 10.1074/jbc.274.12.7777. [DOI] [PubMed] [Google Scholar]
- 21.Edwards M, Twin J, Wilkinson S. New technique to assess the axilla for breast cancer metastases using cell separation technology. ANZ J Surg. 2002;72:655–9. doi: 10.1046/j.1445-2197.2002.02518.x. [DOI] [PubMed] [Google Scholar]
- 22.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. Update on the Gleason grading system for prostate cancer: results of an international consensus conference of urologic pathologists. Adv Anat Pathol. 2006;13:57–9. doi: 10.1097/01.pap.0000202017.78917.18. [DOI] [PubMed] [Google Scholar]
- 23.Allred DC, Clark GM, Elledge R, et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst. 1993;85:200–6. doi: 10.1093/jnci/85.3.200. [DOI] [PubMed] [Google Scholar]
- 24.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67:4514–23. doi: 10.1158/0008-5472.CAN-06-1701. [DOI] [PubMed] [Google Scholar]
- 27.Yuan X, Lu ML, Li T, Balk SP. SRY interacts with and negatively regulates androgen receptor transcriptional activity. J Biol Chem. 2001;276:46647–54. doi: 10.1074/jbc.M108404200. [DOI] [PubMed] [Google Scholar]
- 28.He B, Kemppainen JA, Wilson EM. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem. 2000;275:22986–94. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 29.Sun Z, Pan J, Balk SP. Androgen receptor-associated protein complex binds upstream of the androgen-responsive elements in the promoters of human prostate-specific antigen and kallikrein 2 genes. Nucleic Acids Res. 1997;25:3318–25. doi: 10.1093/nar/25.16.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–27. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186:136–45. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Lin B, Ferguson C, White JT, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4. [PubMed] [Google Scholar]
- 35.Taplin ME, Balk SP. Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence. J Cell Biochem. 2004;91:483–90. doi: 10.1002/jcb.10653. [DOI] [PubMed] [Google Scholar]
- 36.Hu YC, Yeh S, Yeh SD, et al. Functional domain and motif analyses of androgen receptor coregulator ARA70 and its differential expression in prostate cancer. J Biol Chem. 2004;279:33438–46. doi: 10.1074/jbc.M401781200. [DOI] [PubMed] [Google Scholar]
- 37.Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 38.Yamamichi N, Inada K, Ichinose M, et al. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67:10727–35. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
- 39.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–6. [PubMed] [Google Scholar]
- 40.Sun A, Tawfik O, Gayed B, et al. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67:203–13. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- 41.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–54. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 42.Klokk TI, Kurys P, Elbi C, et al. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol Cell Biol. 2007;27:1823–43. doi: 10.1128/MCB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verrijdt G, Haelens A, Schoenmakers E, Rombauts W, Claessens F. Comparative analysis of the influence of the high-mobility group box 1 protein on DNA binding and transcriptional activation by the androgen, glucocorticoid, progesterone and mineralocorticoid receptors. Biochem J. 2002;361:97–103. doi: 10.1042/0264-6021:3610097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67:528–36. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 45.Poukka H, Aarnisalo P, Karvonen U, Palvimo JJ, Janne OA. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J Biol Chem. 1999;274:19441–6. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Pedrero JM, Kiskinis E, Parker MG, Belandia B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem. 2006;281:22656–64. doi: 10.1074/jbc.M602561200. [DOI] [PubMed] [Google Scholar]
- 47.Chi TH, Wan M, Zhao K, et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–9. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 48.Sohn DH, Lee KY, Lee C, et al. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J Biol Chem. 2007;282:10614–24. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- 49.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 50.Quayle SN, Mawji NR, Wang J, Sadar MD. Androgen receptor decoy molecules block the growth of prostate cancer. Proc Natl Acad Sci U S A. 2007;104:1331–6. doi: 10.1073/pnas.0606718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.