Abstract
Liposome:DNA is a promising gene therapy vector. However, this vector can elicit a systemic inflammatory response syndrome (SIRS). Prior reports indicate that liposome:DNA vectors activate Toll-like receptor (TLR)9. We hypothesized that liposome:DNA vectors also activate the cytosolic DNA-sensing pathway, which signals via interferon (IFN) regulatory factor (IRF)3. To test this, we treated dendritic cells (DCs) with liposome:DNA in vitro and found that IRF3 was phosphorylated independent of TLR9. To test the contribution of this pathway in vivo, we injected a liposome:DNA vector into wild-type (WT), TLR9-knockout (KO), IRF3-KO, and TLR9-IRF3-double-KO (DKO) mice. WT mice exhibited a systemic inflammatory response, evidenced by elevations in serum cytokines, serum enzyme changes indicating organ damage, hypothermia, and mortality. The cytokine response was reduced in TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice and all three groups survived. We found that IFN-γ-KO mice that receive liposome:DNA had a reduced cytokine response and 100% survival. CD11c+ and NK1.1+ cells produced IFN-γ and depleting CD11c+ cells reduced the cytokine response in mice injected with liposome:DNA. These findings may facilitate the development of immunologically inert gene therapy vectors and may provide general insight into the mechanisms of SIRS.
Introduction
Gene therapy is a promising therapeutic modality for many diseases. However, animal studies and clinical trials have revealed that gene therapy vectors activate the innate immune system, which can cause a systemic inflammatory response reminiscent of sepsis.1,2,3,4,5,6,7,8,9,10,11,12,13,14,15 This is a major barrier to the use of gene therapy in the clinic. Liposome:plasmid DNA complex is a promising nonviral gene therapy vector, which lacks viral proteins that may activate the adaptive immune system, but still elicits an innate immune response.1,3,4,5,7,11,12,14 There is a critical need to characterize the host innate immune response to viral and nonviral gene therapy vectors to facilitate the development of immunologically inert gene therapy vectors.
The innate immune system is the first line of defense against microbial invasion and is critical for immediate containment of pathogens and subsequent priming of adaptive immunity.16 An important component of innate immunity is the rapid response to conserved microbial motifs. Toll-like receptors (TLRs) are germline-encoded receptors, which recognize conserved microbial motifs and initiate a signal transduction cascade to produce an innate immune response.17 Of relevance to this study, TLR9 resides in endosomes and recognizes unmethylated CpG residues in DNA that is taken up via endocytosis, a common entry route for pathogens and gene therapy vectors.18,19 TLRs are expressed by antigen-presenting cells, such as dendritic cells (DCs) and macrophages, which play an important role in the innate immune response.17 Accumulating evidence indicates that TLRs are also expressed by a variety of other lymphocytes, including natural killer (NK) cells.20,21
In addition to the TLRs, alternative receptors in the cytosol recognize nucleic acids from invading pathogens, leading to activation of interferon (IFN) regulatory factors (IRFs),22,23 with similar consequences to TLR activation.22,24 The retinoic acid–inducible gene I (RIG-I) receptor-like helicases recognize cytosolic RNA, leading to activation of IRF3 and IRF7 (refs. 23,25,26). Cytosolic DNA can activate IRF3 in a similar manner,27,28 although the mechanisms of this pathway have not been fully elucidated. DNA-dependent activator of IRF is one of the receptors involved in this pathway.29,30 Furthermore, recent reports indicate that RNA polymerase III can synthesize RNA from certain cytosolic DNA templates, leading to activation of IRF3 via RIG-I (refs. 31,32). However, prior reports indicate that additional sensors of cytosolic DNA exist, which remain to be elucidated.32,33
Although the innate immune response is critical to pathogen containment, an excessive innate immune response can lead to systemic inflammatory response syndrome (SIRS).34,35 Clinically, SIRS is characterized by low blood pressure, elevated heart and respiratory rate, and alteration of body temperature and lymphocyte levels. In severe cases, hemodynamic changes, diffuse endothelial injury and apoptosis ensue, resulting in organ damage and mortality.
This report investigates the mechanisms of the innate immune response to a liposome:DNA vector. Prior studies indicate that TLR9 is partially responsible for the innate immune response to liposome:DNA.5,7,14,36,37,38,39,40 However, this response is not completely abolished in TLR9-knockout (KO) mice,38 indicating that a second pathway must be involved. We hypothesized that liposome:DNA vectors also activate the cytosolic DNA-sensing pathway, which signals via IRF3 (refs. 27,28). We used TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice to test this hypothesis. We found that liposome:DNA induced IRF3 phosphorylation in DCs, independent of TLR9. In mice injected with the liposome:DNA vector, the TLR9 and IRF3 pathways cooperated to induce proinflammatory cytokines. Liposome:DNA also induced hypothermia, elevations in serum enzymes indicative of organ damage and mortality (signs of SIRS), which required the presence of both TLR9 and IRF3. IFN-γ was critical for this response and was produced by splenic CD11c+CD11b+ and NK1.1+ cells. These findings have implications for nonviral gene therapy and are relevant to the understanding of SIRS in general.
Results
Liposome:DNA activated IRF3 independent of TLR9
Prior reports indicate that liposome:DNA vectors induce an innate immune response, SIRS, and mortality, and that this is partially due to CpG motifs in the plasmid DNA, which activate TLR9 (refs. 5,7,14,36,37,38,39,40). We hypothesized that plasmid DNA also activates the cytosolic DNA-sensing pathway, which signals via IRF3 (refs. 27,28).
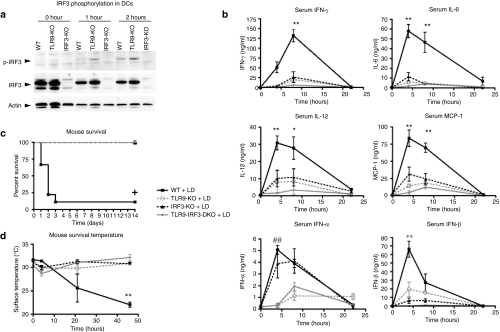
We first investigated whether IRF3 was activated by a liposome:DNA vector in vitro, and whether this required TLR9. We cultured wild-type (WT), TLR9-KO, and IRF3-KO splenic CD11c+ DCs in vitro with a liposome:DNA vector (containing DOTAP:cholesterol liposomes and plasmid DNA), then performed a western blot to quantify phosphorylation of IRF3 serine-396, which is indicative of IRF3 activation.41 IRF3 serine-396 was phosphorylated in both WT and TLR9-KO DCs after 1–2 hours of liposome:DNA treatment (Figure 1a). These data directly demonstrate that IRF3 was activated in DCs treated with liposome:DNA, independent of TLR9.
Figure 1.
TLR9 and IRF3 induced serum cytokines, mortality, and hypothermia in response to liposome:DNA (LD) vector. (a) Western blot demonstrates that IRF3 serine-396 was phosphorylated (P-IRF3) in TLR9-KO and wild-type (WT) DCs, but not IRF3-KO DCs, following 1–2 hours of LD treatment in vitro. Total IRF3 and actin controls are also shown. (b) Serum IFN-γ, IL-6, IL-12, MCP-1, IFN-α, and IFN-β levels were reduced in TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice relative to WT mice injected with LD. Graphs show mean ± SE. (For all graphs P ≤ 0.0061 for 0–8 hours time points by two-way analysis of variance (ANOVA); Bonferroni post-test *P < 0.05 and **P < 0.01 for WT versus TLR9-KO, IRF3-KO and TLR9-IRF3-DKO; ##P < 0.01 for WT versus TLR9-KO and TLR9-IRF3-DKO and IRF3-KO versus TLR9-KO and TLR9-IRF3-DKO). (c) There was significant mortality in WT mice following injection of LD, whereas all TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice survived. Graph shows survival fractions (Kaplan–Meier method), +P < 0.0001 by Logrank test. (d) Surface body temperature dropped significantly in WT mice injected with LD, whereas TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice maintained normal temperature. Graph shows mean ± SE. P = 0.0316 by two-way ANOVA for 0- to 46-hour time points, Bonferroni post-test **P < 0.01 for WT versus TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO. DC, dendritic cells; IFN, interferon; IL, interleukin; IRF, IFN regulatory factor; KO, knockout; MCP, monocyte chemotactic protein; TLR, Toll-like receptor.
The TLR9 and IRF3 pathways contribute to the innate immune response after liposome:DNA vector administration in vivo
Next, we investigated the roles of TLR9 and IRF3 in the innate immune response to liposome:DNA vector in vivo. For this purpose, we intravenously injected WT, TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO (C57BL/6) mice with a liposome:DNA vector containing 50 µg of DNA or 5% dextrose vehicle. WT mice exhibited a rapid elevation in the serum proinflammatory cytokines IFN-γ, interleukin (IL)-6, IL-12, and the chemokine monocyte chemotactic protein-1 as well as the type I IFN-α and IFN-β (Figure 1b). In contrast, the tumor necrosis factor-α response was weaker (Supplementary Figure S1a). The cytokine responses were dose-dependent (Supplementary Figure S1b). In comparison to WT mice, TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice injected with the vector had a significant reduction in IFN-γ, IL-6, IL-12, monocyte chemotactic protein-1, and IFN-β (Figure 1b and Supplementary Figure S1b). There was also a trend toward a further reduction in TLR9-IRF3-DKO mice compared to TLR9-KO or IRF3-KO mice (Figure 1b and Supplementary Figure S1b). IFN-α levels were similar in the WT and IRF3-KO mice injected with the vector, whereas TLR9-KO and TLR9-IRF3-DKO mice injected with the vector showed a significant reduction compared to WT mice (Figure 1b). Proinflammatory cytokines remained at background levels throughout the experiment in mice of each genotype injected with 5% dextrose vehicle control (data not shown). These data indicate that both TLR9 and IRF3 contributed to the innate immune response to liposome:DNA.
The proinflammatory cytokine response to liposome:DNA required presence of plasmid DNA, as injection of liposomes in 5% dextrose vehicle or 5% dextrose vehicle alone, did not induce serum cytokines (Supplementary Figure S2a). Endotoxin was undetectable in our liposomes and was present at biologically insignificant levels in our plasmid DNA [<271 pg lipopolysaccharide (LPS)/50 µg DNA dose administered to a mouse]. To confirm that the endotoxin level in plasmid DNA was biologically insignificant, we intravenously injected mice with LPS mixed with liposomes: we did not observe an elevation in serum proinflammatory cytokines even at an LPS dose of 6 ng (20-fold higher than the endotoxin levels in our plasmid DNA preparations) (Supplementary Figure S2b). In fact, only a 50 µg intraperitoneal (i.p.) dose of LPS was sufficient to induce similar levels of IL-6 and IL-12 to liposome:DNA vector, and this dose of LPS failed to induce IFN-γ. Furthermore, to ensure that LPS did not synergize with plasmid DNA to induce proinflammatory cytokines, we injected mice with liposome:DNA supplemented with 6 ng LPS. These mice exhibited similar IL-6 levels to mice that received liposome:DNA only, at 4 hours (Supplementary Figure S2c). These data indicate that plasmid DNA, rather than endotoxin contamination, induced the innate immune response that we observed.
TLR9 and IRF3 induced mortality and signs of SIRS in mice injected with liposome:DNA vector
We found that 90% of the WT mice died within 3 days of liposome:DNA administration (Figure 1c); in contrast, 100% of the TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice survived >14 days. Death was preceded by a drop in surface body temperature in WT mice, which was not observed in the TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice (Figure 1d). We did not observe mortality or hypothermia in any of the genotypes following injection of 5% dextrose vehicle control (data not shown). In summary, the hypothermia and mortality induced by liposome:DNA required both the TLR9 and IRF3 pathways.
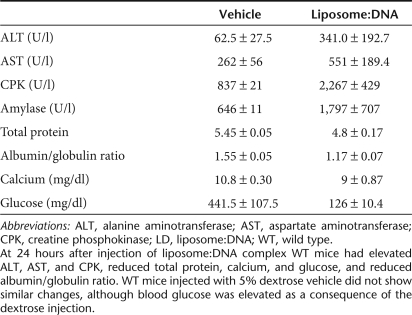
We observed alterations in several enzymes indicative of multiple organ failure in WT mice injected with liposome:DNA vector, these changes were not observed in the 5% dextrose vehicle controls (Table 1). These included elevations of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) (both are released during hepatocellular injury), elevation of creatine phosphokinase (indicative of heart, skeletal muscle, or brain damage), and elevation of amylase (secreted in pancreatic, liver, or small intestinal damage). At 24 hours after administration of liposome:DNA, WT mice also had a substantially reduced percentage of CD3+ cells in the blood, relative to naive mice (Supplementary Figure S3). These results provide evidence that systemic administration of liposome:DNA induced organ damage and lymphopenia, which are typical signs of SIRS.
Table 1.
Serum chemistry in wild-type mice at 24 hours after injection of a high dose of liposome:DNA vector or 5% dextrose vehicle
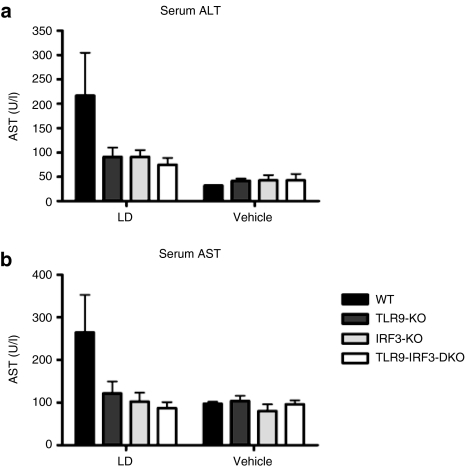
We next examined the importance of TLR9 and IRF3 in the aforementioned systemic response using AST and ALT as readouts of liver damage. There was a trend toward a reduced ALT and AST response in TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice relative to WT mice (Figure 2a,b).
Figure 2.
A trend toward reduced ALT and AST in mice lacking TLR9 or IRF3. There was a trend toward reduced serum (a) ALT and (b) AST in TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice versus WT mice 24 hours after injection of LD. AST and ALT remained at background levels in mice injected with 5% dextrose vehicle. Graphs show mean ± SE. ALT, alanine aminotransferase; AST, aspartate aminotransferase; IRF, interferon regulatory factor; LD, liposome:DNA; TLR, Toll-like receptor; WT, wild-type.
The IRF3 pathway reduced transgene expression
Prior reports indicate that inhibiting the innate immune response may increase transgene expression.5,37,42,43 Hence, we examined the impact of the TLR9 and IRF3 pathways on transgene expression. To determine the site of transgene expression, we injected WT, TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO intravenously with liposome:DNA containing 25 µg of pGL4.13 plasmid encoding luciferase or 5% dextrose vehicle, then examined the luciferase activity at 24 hours using in vivo imaging. We observed transgene expression in the lungs in all genotypes after injection of liposome:DNA, but not after injection of 5% dextrose vehicle (Supplementary Figure S4a). Next, we used a biochemical assay to quantify luciferase transgene expression in the lungs 24 hours after administering liposome:DNA containing 25 or 50 µg of pGL4.13. We observed a significantly higher level of luciferase in the lungs of IRF3-KO and TLR9-IRF3-DKO mice versus WT mice at the 25 µg dose and there was a trend toward increased luciferase levels in TLR9-KO mice versus WT mice (Supplementary Figure S4b). These data indicate that the IRF3 pathway reduced transgene expression.
IFN-γ plays a key role in the response to liposome:DNA
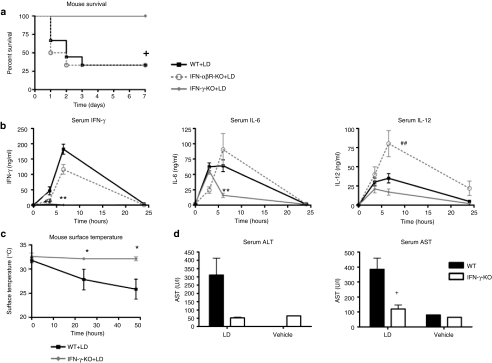
Next, we examined which cytokines were critical to the inflammatory response and mortality induced by liposome:DNA. As both IFN-α and IFN-β (type I IFNs) exhibited a rapid rise following liposome:DNA administration, we first examined the importance of this pathway. However, liposome:DNA induced similar mortality in IFN-αβ receptor (IFN-αβR)-KO and WT mice (Figure 3a). The IFN-αβR-KO mice had similar serum IFN-γ and IL-6 levels to WT mice, and significantly increased IL-12 levels (Figure 3b). We then examined the importance of IFN-γ in this model. Although this cytokine was released later than the type I IFNs, the levels of IFN-γ were the highest of all the cytokines measured (Figure 1b). We found that 100% of the IFN-γ-KO mice survived following liposome:DNA administration (Figure 3a). Furthermore, IFN-γ-KO mice had significantly reduced levels of serum IL-6, and a trend toward reduced serum IL-12 at the 6-hour time point (Figure 3a). The IFN-γ-KO mice maintained normal body temperature (Figure 3b) and had significantly reduced serum AST levels and a trend toward reduced ALT levels, relative to WT mice (Figure 3d).
Figure 3.
IFN-γ played a key role in the innate immune response to liposome:DNA (LD) vector complex: experiments using genetic knockouts. (a) There was significant mortality in wild-type (WT) and IFN-αβR-KO mice following injection of LD, whereas all IFN-γ-KO mice survived. Graph shows survival fractions (Kaplan–Meier method), +P = 0.0487 by Logrank test. (b) Serum IFN-γ and IL-6 levels were significantly reduced in IFN-γ mice versus WT mice, and there was a trend toward reduced IL-12, after injection of LD. IL-6 and IL-12 levels were similar in IFN-αβR-KO mice and WT mice, whereas IL-12 levels were significantly higher in IFN-αβR-KO mice, after injection of LD. For all graphs P ≤ 0.0387 for 0- to 6-hour time points by two-way analysis of variance (ANOVA), Bonferroni post-test **P < 0.01 for IFN-γ-KO versus WT. ##P < 0.01 for IFN-αβR-KO versus WT. (c) Surface body temperature dropped significantly in WT mice injected with LD, whereas IFN-γ-KO mice maintained normal temperature. P = 0.0033 by two-way ANOVA for 0- to 48-hour time points, Bonferroni post-test *P < 0.05 for IFN-γ-KO versus WT. (d) There was trend toward reduced serum ALT, and AST values were significantly lower in IFN-γ-KO mice versus WT mice at 24 hours after injection of LD. ALT and AST remained at background levels in mice injected with 5% dextrose vehicle. *P < 0.05 by t-test. (b–d) Graphs show mean ± SE. ALT, alanine aminotransferase; AST, aspartate aminotransferase; IFN, interferon; IL, interleukin; KO, knockout.
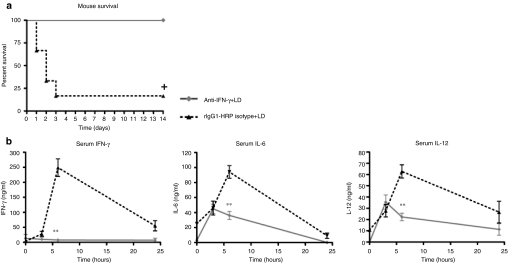
To verify this result, we treated WT mice with an IFN-γ-neutralizing antibody, or rat-IgG1 anti-horseradish peroxidase (rIgG1-HRP) isotype control, 18 hours before administering liposome:DNA. All (100%) the mice administered the IFN-γ neutralizing antibody survived, whereas 83% of the mice that received the isotype control died (Figure 4a). The mice that received the IFN-γ neutralizing antibody also had significantly reduced levels of IL-6 and IL-12 serum cytokines relative to mice administered the isotype control at the 6-hour time point (Figure 4b). Mice administered 5% dextrose vehicle after the IFN-γ neutralizing antibody or isotype control exhibited 100% survival and serum cytokines remained at background levels (data not shown). Together, these results demonstrate that IFN-γ played an important role in the serum cytokine response and SIRS that occurred following liposome:DNA administration.
Figure 4.
IFN-γ played a key role in the innate immune response to liposome:DNA (LD) vector: experiments using a neutralizing antibody. (a) 100% of wild-type (WT) mice survived when administered an IFN-γ-neutralizing antibody (anti-IFN-γ) before the injection of a high-dose of LD vector (50 µg plasmid DNA), in contrast to counterparts administered rat-IgG1 anti-horseradish peroxidase (rIgG1-HRP) isotype control. Graph shows survival fractions (Kaplan–Meier method). +P = 0.0043 by Logrank test. (b) Serum IFN-γ, IL-6, and IL-12 cytokine levels were lower in WT mice that received an IFN-γ-neutralizing antibody versus rIgG1-HRP isotype, before injection of LD. Graphs show mean ± SE. P ≤ 0.01 by two-way analysis of variance for 0- to 6-hour time points, Bonferroni post-test **P ≤ 0.01. IFN, interferon; IL, interleukin.
Splenic CD11c+CD11b+ and NK1.1+ cells produce IFN-γ in response to liposome:DNA vector
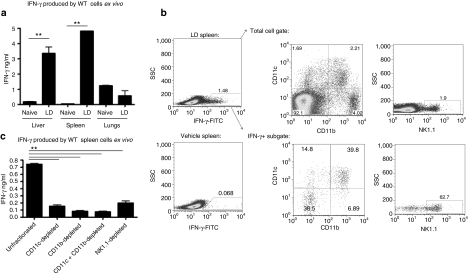
We performed further experiments to investigate which organ and cell type produced the IFN-γ following administration of liposome:DNA vector. We injected WT mice with the liposome:DNA vector, then harvested their livers, spleens, and lungs at 3 hours. A single-cell suspension was prepared from each organ and cultured for 18 hours, and then IFN-γ was quantified in the media. The liver and spleen cells from the mice injected with liposome:DNA vector produced significantly more IFN-γ ex vivo than cells from naive control mice (Figure 5a). In contrast, lung cells from mice injected with liposome:DNA vector failed to produce more IFN-γ than lung cells from naive control mice. These data indicate that the spleen and the liver produced IFN-γ in response to liposome:DNA.
Figure 5.
Liposome:DNA (LD) vector induced IFN-γ production by CD11c+CD11b+ and NK1.1+ spleen cells. (a) IFN-γ was produced ex vivo by liver and spleen cells from wild-type (WT) mice injected with LD. In contrast, lung cells from these mice failed to produce more IFN-γ than naive counterparts. P < 0.01 by one-way analysis of variance (ANOVA), Bonferroni post-test **P < 0.01. (b) Left panels: intracellular cytokine staining revealed an IFN-γ+ population in spleen cells from WT mice injected with LD (top). This IFN-γ+ population was not present in mice that received 5% dextrose vehicle (bottom). Center and right panels: the IFN-γ+ subgate predominantly contained CD11c+CD11b+ and NK1.1+ cells (39.8 and 62.7% respectively, bottom panels), whereas these cell types were rare in the total spleen cell gate (2.21 and 1.9% respectively, top panels). (c) Depletion of CD11c, CD11b, CD11c+CD11b, or NK1.1 populations abrogated IFN-γ production ex vivo by spleen cells from mice injected with LD. P < 0.0001 by one-way ANOVA, Bonferroni post-test **P < 0.01 unfractionated versus CD11c-depleted, CD11b-depleted, CD11c+CD11b-depleted, and NK1.1-depleted. (a and c) Graphs show mean ± SE.
We used intracellular cytokine staining to identify the cell type that produced cytokines in the spleen. We identified an IFN-γ+ population in spleen cells from WT mice that were injected with the liposome:DNA vector, which was not evident in mice injected with 5% dextrose vehicle (Figure 5b). This IFN-γ+ population was predominantly composed of CD11c+CD11b+ and NK1.1+ cells, although these cell types made up only a small fraction of the total spleen cell gate (Figure 5b). The IFN-γ+ population did not contain CD3+ T cells, CD19+B220+ B cells or CD1d-tetramer-binding NKT cells (data not shown).
We confirmed these results by harvesting spleen cells from WT mice injected with the liposome:DNA vector and magnetically depleting the CD11c+, CD11b+, and NK1.1+ subpopulations. The remaining cells were cultured for 18 hours and IFN-γ was quantified in the media. We observed that depletion of CD11c+ and/or CD11b+ cells significantly reduced IFN-γ levels (Figure 5c). Depletion of NK1.1+ cells also significantly reduced IFN-γ levels (Figure 5c). Cells isolated from WT mice injected with 5% dextrose vehicle control did not produce IFN-γ, either with or without magnetic depletion of the aforementioned populations (data not shown). These results demonstrate that splenic CD11c+CD11b+ and NK1.1+ cells produced IFN-γ following liposome:DNA administration.
CD11c+ cells contributed to cytokine production in response to in vivo liposome:DNA vector administration
We next examined the consequence of depleting CD11c+ or NK1.1+ cells in vivo. To deplete CD11c+ cells, we employed CD11c-DTR mice, which express the diphtheria toxin receptor under the control of the CD11c promoter. Diptheria toxin was administered to CD11c-DTR or WT control mice i.p. 18 hours before injection of liposome:DNA. To deplete NK cells, WT mice were administered an NK1.1-depletional antibody or mouse-IgG2a isotype control i.p. 18 hours before injection of liposome:DNA. Depletion of CD11c+ and NK1.1+ cells was confirmed by antibody staining and fluorescence-activated cell sorting analysis of spleen cells (data not shown).
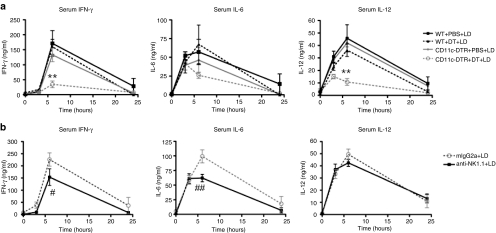
Depletion of CD11c+ cells inhibited the production of IFN-γ and IL-12 to a substantial degree and there was a trend toward reduced IL-6 production (Figure 6a). NK1.1+ cell depletion had only a minor effect on IFN-γ and IL-6 production, and IL-12 production was unaffected (Figure 6b). Cytokines remained at background levels in control mice administered 5% dextrose vehicle, regardless of CD11c+ or NK1.1+ depletion (data not shown). These data demonstrate that CD11c+ cells played a role in the systemic inflammatory cytokine response to liposome:DNA.
Figure 6.
CD11c+ cells played a major role in proinflammatory cytokine production in vivo. (a) Depletion of dendritic cells in CD11c-DTR mice via diphtheria toxin (DT) treatment reduced IFN-γ and IL-12 production and there was a trend toward reduced IL-6 production, in comparison to control mice [CD11c-DTR mice treated with phosphate-buffered saline (PBS) and wild-type (WT) mice treated with DT or PBS], following injection of LD. For IFN-γ and IL-12 graphs P ≤ 0.0055 by two-way analysis of variance (ANOVA) for 0- to 6-hour time points, Bonferroni post-test **P < 0.01 for CD11c-DTR+DT versus CD11c-DTR+PBS, WT+DT, and WT+PBS. (b) Depletion of NK1.1+ cells in WT mice via administration of NK1.1 depletion antibody (anti-NK1.1) reduced IFN-γ and IL-6 production to a small degree and did not affect IL-12 production, in comparison to control mice [administered mouse-IgG2a (mIgG2a) isotype control], following injection of LD vector. For IFN-γ and IL-6, P ≤ 0.04 by two-way ANOVA for 0- to 6-hour time points, (IL-12 = ns), Bonferroni post-test #P < 0.05 and ##P < 0.001 for anti-NK1.1 versus mIgG2a. Graphs show mean ± SE. IFN, interferon; IL, interleukin; LD, liposome:DNA.
Discussion
Liposome:DNA vectors elicit a strong innate immune response, which is problematic for nonviral gene therapy. Following injection of the liposome:DNA vector into WT mice, we observed elevations in proinflammatory cytokines accompanied by signs of SIRS including hypothermia, serum enzyme changes indicating organ damage and death (Figures 1 and 2, Table 1). These observations are consistent with prior studies.15
Prior reports indicate that the innate immune response to liposome:DNA is partially, but not entirely, dependent on CpG residues in plasmid DNA, which activate TLR9 (refs. 4,5,7,14,15,36,37,38,39,40). This report demonstrates that a separate IRF3-dependent pathway also contributes to this response. We provide direct evidence that IRF3 is activated by liposome:DNA vector in vitro, independent of TLR9. IRF3 serine-396 became phosphorylated in WT and TLR9-KO DCs treated with liposome:DNA in vitro (Fig. 1a).
In future studies, it will be important to determine whether activation of IRF3 by liposome:DNA involves DNA-dependent activator of IRF, or RNA polymerase III, as these proteins play a role in the innate immune response to cytosolic DNA.29,30,31,32 Furthermore, these reports indicate that there are additional unidentified cytosolic DNA receptors, which play a redundant role with DNA-dependent activator of IRF and RNA polymerase III in sensing cytosolic DNA in vivo. It will be important to elucidate these receptors in future studies.
The TLR9 and IRF3 pathways cooperate to induce an innate immune response and SIRS in vivo. Serum cytokines were substantially elevated in WT mice injected with liposome:DNA vector, whereas the cytokine levels were lower in TLR9-KO, IRF3-KO and TLR9-IRF3-DKO mice (Figure 1b). Furthermore, WT mice injected with liposome:DNA exhibited signs of SIRS, including hypothermia, elevations in serum enzymes indicating organ damage and mortality (Figure 1 and 2, Table 1). In contrast, all the TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice treated with liposome:DNA survived. Hypothermia was attenuated in these mice and there was a trend toward reduced ALT and AST levels. Collectively, these data indicate that both the TLR9 and IRF3 pathways must be activated for liposome:DNA to induce a maximal proinflammatory response and SIRS. Prior reports indicate that the innate immune response can compromise transgene expression.5,37,42,43 We observed increased transgene expression in IRF3-KO and TLR9-IRF3-DKO mice, relative to WT mice, after administration of liposome:DNA (Supplementary Figure S4). Thus, inhibiting IRF3 might be a useful strategy to reduce the innate immune response, prevent SIRS and increase transgene expression in nonviral gene therapy.
Type I IFNs (IFN-α and IFN-β) are induced by IRFs and play an important role in host defense against viruses.44 However, IFN-αβR-KO mice exhibited similar mortality, similar IFN-γ and IL-6, and increased IL-12 levels relative to WT mice, following injection of liposome:DNA (Figure 3a,b). These data indicate that type I IFN signaling is not critical to the innate immune response to liposome:DNA, or the associated mortality. Of relevance to this result, the Tyro3, Axl, and Mer (TAM) receptors form a complex with the IFN-αβ receptor and act as negative regulators of the TLR response.45 We speculate that TAM ligands might be produced during the host response to liposome:DNA and activate TAM receptors. This might explain the increased IL-12 cytokine production in IFN-αβR-KO mice, as the TAM inhibitory pathway should also be abrogated in these mice.
IFN-γ plays a critical role in the innate immune response to liposome:DNA. Mice lacking IFN-γ had reduced levels of proinflammatory cytokines, reduced serum ALT and AST and did not exhibit mortality following the injection of liposome:DNA (Figures 3 and 4). These data imply that inhibiting IFN-γ might be a useful strategy to reduce the innate immune response and prevent SIRS in nonviral gene therapy.
Interestingly, the effect of IFN-γ on the production of other proinflammatory cytokines did not become evident until the 6-hour time point (Figures 3 and 4). IFN-γ is known to signal in a positive feedback loop with IL-12, each cytokine inducing the other.46 We speculate that the serum proinflammatory cytokines are initially elevated as a direct consequence of TLR9 and IRF3 activation, then IFN-γ and IL-12 act in a positive feedback loop to maintain and increase cytokine production.
We demonstrated that IFN-γ is produced by spleen and liver cells derived from mice injected with liposome:DNA (Figure 5a). Our experiments indicate that splenic CD11c+CD11b+, and NK1.1+ cells produce IFN-γ (Figure 5b). Ex vivo, depletion of either subset reduced IFN-γ to near background levels (Figure 5c). In vivo, depletion of CD11c+ cells substantially reduced the levels of proinflammatory cytokines in mice treated with liposome:DNA, whereas depletion of NK cells caused only a minor reduction in cytokine levels (Figure 6). We speculate that an interaction between DCs and NK1.1+ cells is required to elicit the full levels of IFN-γ that we observed. Perhaps either (i) DC-mediated IFN-γ production drives NK cell IFN-γ production, or (ii) DCs produce another molecule (possibly IL-12), which drives both DC and NK-mediated IFN-γ production. Our data imply that inhibiting CD11c+ cells may be a viable strategy to prevent cytokine production in nonviral gene therapy. Further investigation is required to elucidate the full interplay of CD11c+ and NK1.1+ cells and the role of IL-12 in this system.
The CD11c+CD11b+ subset in the spleen represents the myeloid DC lineage. The authors of a prior study concluded that macrophages contribute to the cytokine response induced by liposome:DNA, as treating these mice with gadolinium chloride inhibited cytokine production in response to this vector.12 However, prior reports show clodronate liposomes, an agent with very similar properties to gadolinium chloride, deplete certain splenic DC populations as well as macrophages.47,48 Hence, the relative roles of DCs and macrophages in the innate immune response to liposome:DNA requires further investigation in future studies.
Because liposome:DNA induced clinical symptoms of SIRS, this may be an appropriate model for further research into the mechanisms of SIRS and sepsis. Commonly used mouse models of sepsis include cecal ligation and puncture, administration of large doses of LPS, and administration of CpG to D-galactosamine-sensitized mice. We propose that administering liposome:plasmid DNA to mice may be an appropriate model of sepsis, and should be investigated in more detail in future studies. It will also be interesting to determine the role of cytosolic DNA and IRF3 in other models of sepsis in future studies.
This report investigates how the TLR9 and IRF3 pathways synergize during the in vivo host response to foreign DNA. We also present evidence that DNA-induced IRF3 activation can play a crucial role in SIRS. Our results imply that the IRF3 pathway represents a novel target for inhibition in the quest to create safer gene therapy approaches for clinical application. Similar strategies may prove useful in other forms of SIRS, including sepsis.
Materials and Methods
Liposome:DNA preparation. pGL4.13 plasmid DNA was prepared with an endotoxin-free maxiprep kit (QIAGEN, Valencia, CA). To make the liposomes, DOTAP methyl-sulfate (Avanti polar Lipids, Alabaster, AL) and cholesterol (Sigma-Aldrich, St Louis, MO) were mixed in a 1:1 molar ratio in chloroform:methanol (3:1). The solvent was evaporated and then the lipid film was hydrated with 5% dextrose and extruded twice through 0.2 µm Anotop-10 filters (Whatman, Piscataway, NJ). Plasmid DNA diluted in 5% dextrose was added to the liposomes drop-wise (1:14 weight ratio). A volume of 250 µl (containing 50 µg plasmid DNA) was injected into the tail vein of each mouse. E-Toxate reagent (Sigma-Aldrich) was used to quantify endotoxin. LPS from Escherichia coli 0111:B4 (Sigma-Aldrich) was used for control experiments.
Animals. All mice used in the study were on a C57BL/6 background. WT and IFN-γ-KO mice were obtained from The Jackson Laboratory (Bar Harbor, ME). TLR9-KO mice and IFN-αβR-KO mice were a gift from Dr Iwasaki (Yale University, USA). IRF3-KO mice were a gift from Dr Medzhitov (Yale University, USA), IRF3-KO mice were used with the kind permission of Dr Taniguchi (University of Tokyo, Japan). These strains were intercrossed to generate the TLR9-IRF3-DKO mice. All mice were housed under specific pathogen-free conditions. The Yale University Institutional Animal Care and Use Committee approved the use of animals in this study.
Depletional and neutralizing antibodies. To deplete NK cells, mice were injected i.p. with 500 µg NK1.1 depletional antibody (clone PK136) or mIgG2a isotype control (Bio X Cell, West Lebanon, NH). To neutralize IFN-γ, mice were injected i.p. with 2 mg anti-IFN-γ (clone R4-6A2) or rIgG1-HRP isotype control (Bio X Cell).
Serum preparation, ELISA, and blood chemistry. Blood was collected by retro-orbital bleed and spun down in BD microtainer serum separator tubes (Becton Dickinson, Franklin Lakes, NJ). Cytokine levels were analyzed by enzyme-linked immunosorbent assay (ELISA) (Becton Dickinson and PBL Biomedical Laboratories, Piscataway, NJ). The rIgG1-HRP isotype control created an artifact in these ELISA assays, which use HRP chemistry. To correct for this, the ELISAs were repeated without the detection antibody and this background signal was subtracted. ALT and AST were measured with Infinity GPT/GOT reagents (Thermo Scientific, Waltham, MA). Blood from mice utilized for creatine phosphokinase, amylase, ALT, and AST was collected after killing by carbon dioxide asphyxiation before cessation of cardiac function by cardiac puncture and assays were performed by Antech Diagnostics (Irvine, CA).
Primary cell culture. Spleen cells were dissociated from the capsule by mastication between glass slides. Lungs and livers were chopped up and digested with collagenase A (Sigma-Aldrich). Red blood cells were lysed and the cells were filtered as previously described.49,50 For intracellular cytokine staining, 2 × 106 cells were seeded in each well of a 96-well plate in growth media (RPMI, 10% fetal calf serum, 1% penicillin/streptomycin) with 1 µl/ml GolgiPlug (Becton Dickinson). At 4 hours, the cells were collected, incubated with FcBlock (eBioscience, San Diego, CA), and then stained with antibodies to cell surface markers (eBioscience). The cells were fixed and permeabilized, then stained with IFN-γ-FITC or isotype control (eBioscience) on ice for 30 seconds. For the depletional study, cells were stained with phycoerythrin (PE)-labeled antibodies to CD11c, CD11b, CD19, B220, CD3, NK1.1, and CD1d-tetramer (eBioscience). PE-labeled cells were depleted with the EasySep PE-selection kit (StemCell, Vancouver, BC). A volume of 5 × 105 cells were cultured in growth media for 18 hours before analysis of supernatant cytokine levels by ELISA. Spleen DCs were purified using the EasySep mouse CD11c-PE selection kit (StemCell). DCs (1 × 106) were cultured in growth media in a 96-well plate with 25 µl liposome:DNA (containing 5 µg DNA).
Western blots. Protein was extracted in 50 mmol/l Tris–HCl, 150 mmol/l NaCl, 1% Nonidet-p40, 5 mmol/l orthovanadate, 1% deoxycholate, pH 8.0 + Complete EDTA-free protease inhibitor (Roche, Indianapolis, IN) for 20 seconds on ice. Ten micrograms of each protein extract was mixed with loading dye and incubated at 95 °C for 10 seconds, then separated on a 12% NuPage Novex Bis–Tris Mini Gel (Invitrogen, Carlsbad, CA). The proteins were transferred onto an Immobilon-P PVDF membrane (Millipore, Billerica, MA), then incubated with primary antibodies against phosphorylated IRF3 serine-396 (Cell Signaling Technology, Davers, MA), total IRF3 (Abcam, Cambridge, MA) or Actin 1–19 (Santa Cruz, Santa Cruz, CA) and secondary antibodies (eBioscience) in 5% milk, Tris-buffered saline + 0.05% tween-20. Luminol chemiluminescent reagent (Santa Cruz) was used to detect signal.
Luciferase transgene expression. For in vivo detection of luciferase activity, mice were injected with 150 mg D-luciferin/kg body weight (AnaSpec, Fremont, CA) i.p. 5 minutes before imaging with a Xenogen IVIS 50 Bioluminescent imaging system (Caliper Life Sciences, Hopkinton, MA). For the biochemical assay, mice were killed and their lungs were harvested, homogenized, and spun down; supernatants were collected and the Luciferase assay system (Promega, Madison, WI) was used according to the manufacturer's instructions. Luciferase values were normalized to protein levels, determined with the Pierce BCA protein assay (Thermo Scientific), according to the manufacturer's instructions.
Statistical analysis. All experiments were performed on at least three separate occasions with n ≥ 3/experiment. Graphs show representative or pooled data from these experiments. For comparison of one variable between two groups, we employed a two-tailed t-test, assuming unequal variance between groups. For comparison of one variable between three or more groups, we employed a one-way analysis of variance. For comparison of two or more groups over time, we employed a two-way repeated measures analysis of variance. A Bonferroni post-test was performed to assess pair-wise differences. P ≤0.05 was considered significant, ≤0.01 highly significant.
SUPPLEMENTARY MATERIALFigure S1. Further characterization of cytokine production in wild-type, TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice injected with liposome:DNA.Figure S2. Plasmid DNA was required to elicit the serum cytokine response, and endotoxin contamination was not a factor.Figure S3. Wild-type mice became lymphopenic after liposome:DNA administration.Figure S4. IRF3 reduced luciferase transgene expression in the lungs.
Supplementary Material
Further characterization of cytokine production in wild-type, TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice injected with liposome:DNA.
Plasmid DNA was required to elicit the serum cytokine response, and endotoxin contamination was not a factor.
Wild-type mice became lymphopenic after liposome:DNA administration.
IRF3 reduced luciferase transgene expression in the lungs.
Acknowledgments
This work was supported by NIH R01AI064660, R01AG028082, K02AG033049, and AHA 0940006N (D.R.G.); NIH T32 HL007778-12(fellowship support for W.E.W.), and NIH 2P30DK034989 (Yale Liver Center pilot feasibility grant awarded to W.E.W.). We thank Anushree Shirali, Heather Stout-Delgado, Hua Shen, Wei Du, and Lauren Cohn for valuable discussions. We thank Maria Volkova, April Kalinowski, Ravi Singh, Miriam Alonso, and Jonathan Alderman for their assistance with experimental techniques. We thank Dr Flavell for use of the IVIS imaging system. We thank Akiko Iwasaki for the TLR9-KO and IFN-αβR-KO mice and Ruslan Medzhitov for the IRF3-KO mice.
REFERENCES
- Scheule RK, St George JA, Bagley RG, Marshall J, Kaplan JM, Akita GY, et al. Basis of pulmonary toxicity associated with cationic lipid-mediated gene transfer to the mammalian lung. Hum Gene Ther. 1997;8:689–707. doi: 10.1089/hum.1997.8.6-689. [DOI] [PubMed] [Google Scholar]
- McMahon JM, Wells KE, Bamfo JE, Cartwright MA., and , Wells DJ. Inflammatory responses following direct injection of plasmid DNA into skeletal muscle. Gene Ther. 1998;5:1283–1290. doi: 10.1038/sj.gt.3300718. [DOI] [PubMed] [Google Scholar]
- Freimark BD, Blezinger HP, Florack VJ, Nordstrom JL, Long SD, Deshpande DS, et al. Cationic lipids enhance cytokine and cell influx levels in the lung following administration of plasmid: cationic lipid complexes. J Immunol. 1998;160:4580–4586. [PubMed] [Google Scholar]
- Yew NS, Wang KX, Przybylska M, Bagley RG, Stedman M, Marshall J, et al. Contribution of plasmid DNA to inflammation in the lung after administration of cationic lipid:pDNA complexes. Hum Gene Ther. 1999;10:223–234. doi: 10.1089/10430349950019011. [DOI] [PubMed] [Google Scholar]
- Li S, Wu SP, Whitmore M, Loeffert EJ, Wang L, Watkins SC, et al. Effect of immune response on gene transfer to the lung via systemic administration of cationic lipidic vectors. Am J Physiol. 1999;276 5 Pt 1:L796–L804. doi: 10.1152/ajplung.1999.276.5.L796. [DOI] [PubMed] [Google Scholar]
- Alton EW, Stern M, Farley R, Jaffe A, Chadwick SL, Phillips J, et al. Cationic lipid-mediated CFTR gene transfer to the lungs and nose of patients with cystic fibrosis: a double-blind placebo-controlled trial. Lancet. 1999;353:947–954. doi: 10.1016/s0140-6736(98)06532-5. [DOI] [PubMed] [Google Scholar]
- Tan Y, Li S, Pitt BR., and , Huang L. The inhibitory role of CpG immunostimulatory motifs in cationic lipid vector-mediated transgene expression in vivo. Hum Gene Ther. 1999;10:2153–2161. doi: 10.1089/10430349950017149. [DOI] [PubMed] [Google Scholar]
- Norman J, Denham W, Denham D, Yang J, Carter G, Abouhamze A, et al. Liposome-mediated, nonviral gene transfer induces a systemic inflammatory response which can exacerbate pre-existing inflammation. Gene Ther. 2000;7:1425–1430. doi: 10.1038/sj.gt.3301240. [DOI] [PubMed] [Google Scholar]
- Schnell MA, Zhang Y, Tazelaar J, Gao GP, Yu QC, Qian R, et al. Activation of innate immunity in nonhuman primates following intraportal administration of adenoviral vectors. Mol Ther. 2001;3 5 Pt 1:708–722. doi: 10.1006/mthe.2001.0330. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chirmule N, Gao GP, Qian R, Croyle M, Joshi B, et al. Acute cytokine response to systemic adenoviral vectors in mice is mediated by dendritic cells and macrophages. Mol Ther. 2001;3 5 Pt 1:697–707. doi: 10.1006/mthe.2001.0329. [DOI] [PubMed] [Google Scholar]
- Loisel S, Le Gall C, Doucet L, Ferec C., and , Floch V. Contribution of plasmid DNA to hepatotoxicity after systemic administration of lipoplexes. Hum Gene Ther. 2001;12:685–696. doi: 10.1089/104303401300057405. [DOI] [PubMed] [Google Scholar]
- Sakurai F, Terada T, Yasuda K, Yamashita F, Takakura Y., and , Hashida M. The role of tissue macrophages in the induction of proinflammatory cytokine production following intravenous injection of lipoplexes. Gene Ther. 2002;9:1120–1126. doi: 10.1038/sj.gt.3301784. [DOI] [PubMed] [Google Scholar]
- Raper SE, Chirmule N, Lee FS, Wivel NA, Bagg A, Gao GP, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Sakurai F, Kawabata K, Sasaki T, Koizumi N, Huang H, et al. Comparison of gene expression efficiency and innate immune response induced by Ad vector and lipoplex. J Control Release. 2007;117:430–437. doi: 10.1016/j.jconrel.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Kawabata K, Sakurai F, Nakagawa S., and , Mizuguchi H. Innate immune response induced by gene delivery vectors. Int J Pharm. 2008;354:9–15. doi: 10.1016/j.ijpharm.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Ezekowitz RAB., and , Hoffmann JA. Innate immunity. Curr Opin Immunol. 1996;8:1–2. doi: 10.1016/s0952-7915(96)80096-3. [DOI] [PubMed] [Google Scholar]
- Takeda K., and , Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- Krieg AM. Direct immunologic activities of CpG DNA and implications for gene therapy. J Gene Med. 1999;1:56–63. doi: 10.1002/(sici)1521-2254(199901/02)1:1<56::aid-jgm5>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- Sivori S, Falco M, Della Chiesa M, Carlomagno S, Vitale M, Moretta L, et al. CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc Natl Acad Sci USA. 2004;101:10116–10121. doi: 10.1073/pnas.0403744101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KN, Leung B, Kwong M, Zarember KA, Satyal S, Navas TA, et al. APC-independent activation of NK cells by the Toll-like receptor 3 agonist double-stranded RNA. J Immunol. 2004;172:138–143. doi: 10.4049/jimmunol.172.1.138. [DOI] [PubMed] [Google Scholar]
- Honda K., and , Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, et al. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., and , Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K., and , Fujita T. Cytoplasmic recognition of RNA. Adv Drug Deliv Rev. 2008;60:841–846. doi: 10.1016/j.addr.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Stetson DB., and , Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB., and , Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA., and , Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Pinsky MR. Sepsis and multiple organ failure. Contrib Nephrol. 2007;156:47–63. doi: 10.1159/000102070. [DOI] [PubMed] [Google Scholar]
- Rittirsch D, Flierl MA., and , Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8:776–787. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan G, Stevenson BJ, Davidson DJ., and , Porteous DJ. Bacterial DNA is implicated in the inflammatory response to delivery of DNA/DOTAP to mouse lungs. Gene Ther. 2000;7:384–392. doi: 10.1038/sj.gt.3301097. [DOI] [PubMed] [Google Scholar]
- Yew NS, Zhao H, Wu IH, Song A, Tousignant JD, Przybylska M, et al. Reduced inflammatory response to plasmid DNA vectors by elimination and inhibition of immunostimulatory CpG motifs. Mol Ther. 2000;1:255–262. doi: 10.1006/mthe.2000.0036. [DOI] [PubMed] [Google Scholar]
- Zhao H, Hemmi H, Akira S, Cheng SH, Scheule RK., and , Yew NS. Contribution of Toll-like receptor 9 signaling to the acute inflammatory response to nonviral vectors. Mol Ther. 2004;9:241–248. doi: 10.1016/j.ymthe.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Hodges BL, Taylor KM, Joseph MF, Bourgeois SA., and , Scheule RK. Long-term transgene expression from plasmid DNA gene therapy vectors is negatively affected by CpG dinucleotides. Mol Ther. 2004;10:269–278. doi: 10.1016/j.ymthe.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Hyde SC, Pringle IA, Abdullah S, Lawton AE, Davies LA, Varathalingam A, et al. CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol. 2008;26:549–551. doi: 10.1038/nbt1399. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W., and , Fujita T. Control of IRF-3 activation by phosphorylation. J Interferon Cytokine Res. 2002;22:73–76. doi: 10.1089/107999002753452674. [DOI] [PubMed] [Google Scholar]
- Gruneich JA, Price A, Zhu J., and , Diamond SL. Cationic corticosteroid for nonviral gene delivery. Gene Ther. 2004;11:668–674. doi: 10.1038/sj.gt.3302214. [DOI] [PubMed] [Google Scholar]
- Qin L, Ding Y, Pahud DR, Chang E, Imperiale MJ., and , Bromberg JS. Promoter attenuation in gene therapy: interferon-gamma and tumor necrosis factor-alpha inhibit transgene expression. Hum Gene Ther. 1997;8:2019–2029. doi: 10.1089/hum.1997.8.17-2019. [DOI] [PubMed] [Google Scholar]
- Zuniga EI, Hahm B., and , Oldstone MB. Type I interferon during viral infections: multiple triggers for a multifunctional mediator. Curr Top Microbiol Immunol. 2007;316:337–357. doi: 10.1007/978-3-540-71329-6_16. [DOI] [PubMed] [Google Scholar]
- Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB., and , Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Ma X, Chow JM, Gri G, Carra G, Gerosa F, Wolf SF, et al. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenen PJ, Radosevic K, Voerman JS, Salomon B, van Rooijen N, Klatzmann D, et al. Heterogeneity of mouse spleen dendritic cells: in vivo phagocytic activity, expression of macrophage markers, and subpopulation turnover. J Immunol. 1998;160:2166–2173. [PubMed] [Google Scholar]
- Nikolic T, Geutskens SB, van Rooijen N, Drexhage HA., and , Leenen PJ. Dendritic cells and macrophages are essential for the retention of lymphocytes in (peri)-insulitis of the nonobese diabetic mouse: a phagocyte depletion study. Lab Invest. 2005;85:487–501. doi: 10.1038/labinvest.3700238. [DOI] [PubMed] [Google Scholar]
- Walker WE., and , Goldstein DR. Neonatal B cells suppress innate toll-like receptor immune responses and modulate alloimmunity. J Immunol. 2007;179:1700–1710. doi: 10.4049/jimmunol.179.3.1700. [DOI] [PubMed] [Google Scholar]
- Walker WE, Nasr IW, Camirand G, Tesar BM, Booth CJ., and , Goldstein DR. Absence of innate MyD88 signaling promotes inducible allograft acceptance. J Immunol. 2006;177:5307–5316. doi: 10.4049/jimmunol.177.8.5307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Further characterization of cytokine production in wild-type, TLR9-KO, IRF3-KO, and TLR9-IRF3-DKO mice injected with liposome:DNA.
Plasmid DNA was required to elicit the serum cytokine response, and endotoxin contamination was not a factor.
Wild-type mice became lymphopenic after liposome:DNA administration.
IRF3 reduced luciferase transgene expression in the lungs.