Abstract
The stable introduction of therapeutic transgenes into human cells can be accomplished using viral and nonviral approaches. Transduction with clinical-grade recombinant viruses offers the potential of efficient gene transfer into primary cells and has a record of therapeutic successes. However, widespread application for gene therapy using viruses can be limited by their initially high cost of manufacture at a limited number of production facilities as well as a propensity for nonrandom patterns of integration. The ex vivo application of transposon-mediated gene transfer now offers an alternative to the use of viral vectors. Clinical-grade DNA plasmids can be prepared at much reduced cost and with lower immunogenicity, and the integration efficiency can be improved by the transient coexpression of a hyperactive transposase. This has facilitated the design of human trials using the Sleeping Beauty (SB) transposon system to introduce a chimeric antigen receptor (CAR) to redirect the specificity of human T cells. This review examines the rationale and safety implications of application of the SB system to genetically modify T cells to be manufactured in compliance with current good manufacturing practice (cGMP) for phase I/II trials.
Introduction
Nonviral, DNA-mediated gene transfer has been explored for gene therapy in order to avoid four major disadvantages of viral-based delivery systems: (i) Viral vectors are time-consuming and expensive to prepare in the quantities and at the high titers required for gene therapy. (ii) Preparations of viruses have risks of contamination by infectious agents, including replication-competent virus generated by recombination between vector and packaging functions.1 (iii) The viral vector may elicit unwanted cellular consequences, e.g., acute immune/inflammatory2 or neurotoxic3 responses. (iv) The efficient genetic propagation of viral vectors may constrain therapeutic cargo size and may require genetic motifs for regulation of vector replication. In contrast, preparations of nonviral DNA plasmid–based vectors are relatively inexpensive to purify, are largely nonimmunogenic, and have no hard constraints on sequences that can be delivered. The major problems with nonviral, plasmid-based systems are as follows: (i) low rates of delivery of the vectors to target-cell nuclei; (ii) low rates of integration of transgenes, generally with accompanying plasmid sequences that have a propensity to silence expression;4,5 and (iii) integration of multiple copies (concatemers) of the transgene, which also can silence their expression.5,6
These problems can be alleviated by using transposons, which do not depend on low rates of illegitimate recombination for integration of the sequences of interest. Sleeping Beauty (SB) is an example of a transposon system that can be adapted for human gene therapy.7,8,9 Since its creation in 1997, the SB transposon system has been characterized in >200 papers (PubMed—“Sleeping Beauty”). The SB system10 was developed (resurrected) to provide an efficient, nonviral method for introducing defined DNA sequences into vertebrate chromosomes. SB transposase is a synthetic enzyme that was constructed for mammalian use by step-by-step reverse engineering of extinct sequences found in salmonid fish.10,11,12 SB is a model vector system because the transposons insert more randomly than most viral vectors as SB transposons can integrate into any of ~2 × 108 TA sites in mammalian genomes. As a result of this unbiased widespread integration, they have been used for insertional mutagenesis, gene tagging, induction of neoplasias, and gene delivery to confer new desired functions to cells and tissues.13,14 Consequently, a body of knowledge has emerged that allows assessment of the risks of “random” SB-mediated insertions of genetic sequences and that can be used to justify adapting the SB system for gene therapy applications.
The SB system consists of two components—the transposon, composed of inverted terminal repeat sequences (IRs)—that sandwich a desired genetic cargo, and a SB transposase enzyme (Figure 1). The IRs and a hierarchical series of hyperactive transposases, SB10, SB11, SB100, etc.15,16,17,18 with increasing enzymatic activities have been developed to mediate transposition of transposons encoding therapeutic polypeptides. The ~230-base-pair (bp) IRs contain two shorter, nonidentical direct repeats (DRs) of 32 bp, hence the description of the inverted terminal repeats of SB transposons as IR/DRs.10 The requirement for four transposases for each transposition,19,20 but no more,15 may be a safety feature of this vector system. Thus, SB transposons overcome the principal problems cited above that are associated with other nonviral vectors.
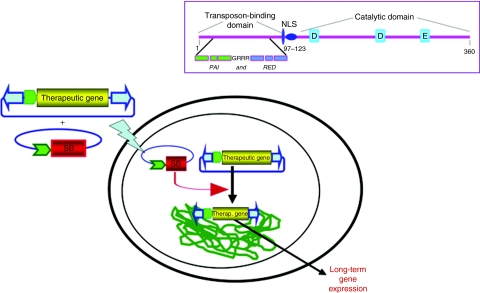
Figure 1.
SB transposon-mediated gene transfer into chromosomal DNA for long-term expression of a therapeutic gene. A SB transposon, with flanking inverted terminal repeats (ITRs, blue arrowheads), in a plasmid (blue line) provides only transient expression of a transgene (yellow) from a promoter (green) unless the transposon vector transposes into a host genome (green line) by SB transposase (red expression cassette). The lightning bolt represents any of several methods for delivery of the transposon system into a cell and its nucleus. The structure of SB transposase, which is composed of three functional domains, is diagrammed in the box. The transposase's a-helical rich N-terminal domain binds to the ITRs of the transposon, the nuclear localization sequence (NLS) directs import of the transposase into the nucleus, and the catalytic domain catalyzes the cut-and-paste transposition reaction and recognizes TA base pairs into which the transposon will be inserted. The N-terminal domain is further divided into two subdomains, PAI and RED, each of which has helix-turn-helix regions (green and blue boxes, respectively) that are separated by an arginine-rich sequence GRRR. Three signature amino acids (D, D, and E) in the catalytic domain are indicated by blue boxes.
The improved integration efficiency associated with SB transposition has led to two primary uses: (i) delivery of therapeutic genes and (ii) insertional mutagenesis, e.g., for the identification of oncogenes. In mice, SB transposons have been used to correct several genetic deficiencies including those for hemophilia B21,22 and hemophilia A,23,24,25 tyrosinemia type I,26 junctional epidermolysis bullosa,27 diabetes,28 Huntington disease,29 mucopolysaccharidosis I and VII30,31 as well as for treatment of a xenograft model for glioblastoma.32,33 Because the SB system can deliver genes (almost) randomly,34,35,36 investigators have used SB to identify pathways and genes in model organisms. These applications of the SB system can be erroneously seen as contradictory. One of the goals of this review is thus to resolve this apparent incongruity and to show how the translation of this efficient nonviral gene-transfer approach to therapeutic clinical application is not compromised by its use to discover the genetic origins of cancer.
Potential for genotoxicity
The long-term introduction of a therapeutic gene can treat severe diseases. Recently, patients with X-linked adrenoleukodystrophy (ALD) who received infusions of hematopoietic stem cells (HSCs) genetically modified ex vivo with lentivirus, have demonstrated successful expression of the ALD gene.37 Furthermore, HSCs have been transduced with retroviruses to correct X-linked chronic granulomatous disease (gp91phox deficiency)38 as well as severe combined immunodeficiencies (SCID), such as adenosine deaminase-deficient SCID, wherein over 30 SCID patients have benefited from robust reconstitution of immune function.39 However, a single integration event can lead to genotoxicity as demonstrated by five reported cases of apparent T-cell leukemia in 20 patients treated for X-linked SCID, one of which resulted in death.40,41 These leukemias typically occurred ≥3 years after administration of therapeutic murine leukemia virus–derived retrovirus vectors into HSC,42,43 which validated theoretical fears that insertion of corrective, viral-based vectors could cause severe adverse effects. The linkage between the gene-transfer event and the subsequent leukemias was inferred because the expanded transformed cell populations derived from genetically modified HSC that harbored clonal integrations close to endogenous genes associated with cellular proliferation.44,45,46 Similarly, clinical trials for chronic granulomatous disease indicated the presence of dominant hematopoietic clones, mainly in the myeloid compartment, as defined by insertions in certain genetic loci.38 Although clonal dominance appeared to be due to a growth and/or survival advantage conferred by gene-activating or gene-suppressing effects attributed to the integrated retroviral vector, the cases of induced clonal dominance did not appear to lead to malignant transformation of affected cell clones in the treated chronic granulomatous disease patients.47 In contrast to the adverse events that occurred with genetic modification of HSC, there have been no apparent toxicities due to the viral-mediated transduction of T cells, and some patients are now symptom-free many years after adoptive immunotherapy.48 The relative safety of virally transduced T cells was emphasized in a murine study in which T cells and HSCs were genetically modified with γ-retroviral vectors that expressed T-cell oncogenes. Indeed, adoptive transfer of HSC with enforced expression of either LMO2, TCL1, or TrkA resulted in T-cell leukemia/lymphoma, whereas the infusion of similar genetically modified T cells was well tolerated.49
Mice are commonly used for initial testing of vectors for gene therapy, and in 2003, the American Society of Gene and Cell Therapy reviewed the literature for adverse events in mice following delivery of retroviral vectors.50 They found that of 5,436 mice followed for >50 days, and 4,846 for >3 months, there were 13 hematopoietic anomalies including six that involved malignant transformation. Moreover, it was noted that high doses of integrating murine leukemia virus vectors can cause leukemias following retroviral expression of MDR1 and selection of clones with mutated proto-oncogenes or other signaling genes.51 More recent studies indicate that induction of cancer or leukemia following integration of vectors involves an interplay between the genetics of the recipient cell, the vector, and the transgene and its transcriptional regulators.45,52,53 Lentiviral vectors with their preferred integration into transcribed genes may be less genotoxic than γ-retroviral vectors with their preference for integration next to transcriptional start sites and regulatory gene regions. However, lentiviral vectors rendered self-inactivating can show evidence of insertional transformation of hematopoietic cells.54 Other changes to the viral sequence may reduce genotoxicity, such as the addition of chicken HS4 insulator elements.55
Large animal models have also been used to assess the risk of genotoxicity from viral transduction. There was no evidence of leukemia in 42 rhesus macaques that underwent transplantation up to 7 years earlier with autologous CD34+ HSC transduced with murine leukemia virus–derived retroviral vectors that integrated into at least 702 distinct genomic loci.56,57 However, vector integration proximal to proto-oncogenes was observed, including 14 integrations (in nine of the primates) into the Mds1/Evi1 region. The over-representation of events in the Mds/Evi1 locus was likely due to the self-renewal and engraftment potential of CD34+ progenitor cells with integrations at this specific locus even though there was no evidence of ongoing in vivo clonal expansion of the Mds1/Evi1 populations. Likewise, there was a notable absence of abnormal hematopoiesis or leukemia in 23 baboons and 17 dogs whose CD34+ HSCs were treated ex vivo with significant levels of retroviral vectors.50,58
Viral vectors other than γ-retroviruses have been occasionally implicated in causing severe adverse effects due to insertional mutagenesis. In humans, there is a single report implicating HIV-1 in transformation of T cells into a rare lymphoma,59 which is surprisingly low based on the large numbers of HIV-1-infected individuals under medical treatment and the preference of HIV proviruses to integration into or near genes.60 A recent study analyzing lentiviral-integration sites from three patients who received CD4+ T cells transduced to express antisense env, demonstrated that the recombinant vector preferentially integrated into active transcriptional units, but there was no evidence of abnormal expansion due to aberrant vector-mediated insertional activation of proto-oncogenes.61 Another lentivirus, feline immunodeficiency virus, has been associated with a B-cell lymphoma, although the cause and effect was by implication rather than rigorously demonstrated.62
Other viral vectors have been implicated in causing severe adverse effects due to insertional mutagenesis. A study of 695 mice injected with adeno-associated virus vectors revealed that the frequency of hepatic tumors was 0.14% (a single lipoma in one animal).63 Given the background level of tumors in untreated mice and the absence of high levels of vector DNA observed in the single tumor, the authors concluded that treatment with an adeno-associated virus vector, which targets active transcriptional units,64,65 did not predispose recipients to tumor development. However, a later study of the incidence of tumorigenesis in long-term rodent studies, in which MPS VII mice were transfused with recombinant adeno-associated virus harboring a β-glucuronidase expression cassette, identified a common integration site that was associated with hepatocellular carcinomas and angiosarcomas.66 Moreover, in a study that employed equine infectious anemia lentivirus for delivery of factor IX to hemophilic neonatal mice, seven of 10 mice developed liver tumors.67 PCR amplification of the integration sites in eight of the equine infectious anemia virus–treated mice revealed 26 unique integration sites of which 20 were within 10 kbp of a RefSeq gene, and the Gene Ontology database indicated that 10 of the 20 genes encoded either a kinase or DNA-binding protein. These findings suggested particular sensitivity of neonates to integrating vectors, although the integration specificities of equine infectious anemia virus, which are unknown, could also have contributed to the development of adverse effects. In contrast, when a stripped-down herpes simplex virus vector devoid of viral genes was used to convey the SB transposon system into neuronal cells of newborn mice, integration and long-term expression were noted without obvious attendant adverse effects.68
In summary, although many studies have demonstrated that activation of oncogenes can occur by recombinant viral vectors, it is most apparent for certain genes in selective circumstances typically involving the genetic manipulation of HSC,45,69,70 whereas transduction of T cells derived from the peripheral blood appears safe.71,72 These data are all associated with viral-based gene insertion approaches, and as yet, there are insufficient data to evaluate the relative risk of insertional mutagenesis associated with nonviral gene transfer. Significantly, it is evident from extensive sequencing of retroviral and lentiviral vector insertion sites that only a small subset of all the integrations near proto-oncogenes actually cause transformation.70,73 The same is likely to be true for nonviral gene-transfer approaches, such as those that use the SB system. That is, the adverse events appear to require an accompanying secondary genomic alteration, such as chromosomal rearrangements, that contribute additional “hits” required for uncontrolled cell proliferation.40,74 This is perhaps best exemplified by the lack of any report of clonal lymphoproliferation in patients treated for adenosine deaminase deficiency,39 despite the observation of a similar frequency of integration near LMO2 and other proto-oncogenes.47,75
Safety evaluation of the SB system in mouse models
Every integrating vector used at present has some genotoxic potential as well as potential for the treatment of human disease. Indeed, the same viral vectors used for delivery of therapeutic genes have also been used to identify cancer genes based upon their ability to induce neoplasias in precancerous laboratory mice after insertional mutagenesis.76 One approach to risk assessment of vectors used for gene delivery is to approximate the maximal tolerated cancer dose before cancer is initiated (or accelerated) in model animals. For reasons noted below, these experiments should be conducted in the presence of a relevant disease genotype. By using mice that are genetically predisposed to developing cancer, we can increase the number of cells at risk and/or the number of vector insertion mutations induced per cell in order to elucidate both particular integration loci and conditions that lead to adverse situations. Although one cannot reliably extrapolate to human cells using quantitative data from mouse modeling, it should be possible to compare the relative genotoxicities of vectors such as retroviral, lentiviral, and transposon plasmids.
For gene therapy, SB vectors may be introduced, often by electroporation, into cells ex vivo. This mode of delivery results in only one to a few stable integrations of the transgenic construct into a specific cell. Although single integrations per cell are desirable for gene therapy, this level of insertion is too low to permit comprehensive screening for integrations that can lead to the emergence of genotoxic events. However, by using transgenic mice designed to reveal oncogenesis from insertional mutagenesis, e.g., containing both concatemers of SB transposons together with an SB transposase gene, remobilization of transposons to many sites in practically every cell of an organ or tissue can be achieved, thereby mimicking delivery to far more cells that can be achieved by extracellular delivery.13,77 By studying these insertion sites present in tumors induced by SB, one can define those loci sensitive to insertional mutagenesis in the context of specific tissues, cell types and genetic background, and the effects of prolonged expression of the transposase can be determined over the lifetime of a mouse.
To identify genes in vertebrates involved with cancer, the T2/onc transposon was designed specifically to induce gain-of-function and/or loss-of-function mutations upon insertion either near or within endogenous genes. Thus, T2/onc is designed to inflict “worst-case” scenarios with respect to genotoxicity. T2/onc contains splice acceptor sites followed by polyadenylation signals in both orientations (Figure 2). Following integration into an intron, these elements can intercept upstream splice donors and elicit premature termination of the transcript. Between the two splice acceptors are sequences from the 5′LTR of the murine stem cell virus that contain a strong constitutive retroviral promoter and enhancer elements that are methylation-resistant and active in stem cells.78,79,80 Immediately downstream of the LTR is a splice donor site for splicing of a transcript initiated from the LTR into a neighboring gene. The T2/onc transposon is thus specialized to identify both tumor suppressors and oncogenes. In somatic cells of p19Arf−/− mice, remobilization of T2/onc transposons accelerated sarcoma development and in wild-type mice, it induced T-cell leukemia and medulloblastoma.81,82 In both the accelerated sarcomas and leukemias, T2/onc insertions were found to cluster near or within specific genes in multiple independent tumors. These clusters are designated common insertion sites and reflect selection for tumor cells carrying these rare events among the many random somatic insertion mutations that occurred in the hundreds of millions of cells of the SB transgenic mice. Common insertion sites associated with gastrointestinal tract adenoma/adenocarcinoma, medulloblastoma, sarcoma, leukemia, lung adenocarcinoma, and hepatocellular carcinoma have been identified.81,82,83,84 These studies provide important baseline information about what conditions can result in cancer induced by SB insertional mutagenesis. However, these studies by no means indicate that SB gene delivery must result in cancer. Most importantly, the controls that comprise either just transposons or just a transposase gene, which were included in all of the oncogene-screening experiments, indicate that neither SB transposons nor SB transposase alone has any appreciable toxicity in mice. This point is emphasized in the following paragraphs.
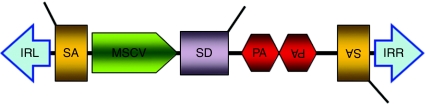
Figure 2.
The T2/onc transposon vector to introduce mutations. This DNA vector contains elements designed to elicit either transcriptional activation (MSCV 5′ LTR and splice donor, SD) or inactivation [splice acceptor (SA) and polyadenylation signal, pA] when landing within/near an endogenous gene. IRL and IRR denote the left and right inverted terminal repeats of the SB transposon, respectively. MSCV, murine stem cell virus.
There are significant differences between the oncogenic SB mouse models and patients undergoing the gene therapy made possible via the SB system. First, as described above, a vector specifically designed to activate or inactivate endogenous genes was used in the oncogenic mouse model. Although gene therapy vectors, such as DNA plasmids, contain a promoter, they are designed to express a transcript completely contained within the vector rather than generate chimeric transcripts with endogenous genes. Second, every cell in the transgenic mouse harbors multiple copies (often >100) of the SB vector. In the context of gene therapy, a limited number of cells would only have a single, or possibly a few insertions, of SB sequence per whole genome. Third, in the oncogene screens, the SB transposase enzyme is expressed continuously for the entire life span of the mouse, either in specific organs or in almost all cells, allowing constant remobilization of the vector and accumulation of multiple, cooperating insertion mutations to occur per cell. In the context of SB-mediated gene therapy, the transposase gene is generally delivered on a plasmid separate from the transposon and its expression is limited.85 Fourth, in several of the published oncogene screens with SB mouse models, the mice used were carrying cancer-predisposing mutations.
The SB screens have revealed certain features of the oncogenic SB mouse models that are important for cancer induction (unpublished analyses in the Largaespada lab): (i) Mice that lack SB transposons, but ubiquitously express SB transposase for their lifetime, do not show a higher rate of spontaneous cancer than do wild-type littermates. (ii) The presence of concatemers of T2/onc transposons does not appear to increase the rate of spontaneous cancer. (iii) A promoter within the T2/onc transposon is necessary for induction of cancer. When a line of mice with a concatemer of transposons with a splice acceptor, reporter gene, and polyadenylation signal sequence, but no promoter, was crossed with a line harboring the Rosa26-SB11 transgene, the double transgenic mice in which the promoterless transposon was mobilized, did not develop significantly more cancer than did control groups. Although further studies are needed to determine whether transposons with “standard gene therapy expression cassettes” can induce tumors when mobilized in a large number of cells over a long period of time, the evidence to date overwhelmingly supports the idea that SB transposons are inherently no more likely to cause adverse effects than other integrating (viral) vectors.
In another study to address the concern that random integration and extended expression of a SB transposase gene might lead to remobilization of inserted transposons in the same genomes, we have examined the relative levels of SB transposon genes and their duration of expression of SB transposase when delivered via plasmids in vivo to cells in livers of mice. Although there was some evidence of residual SB transposase protein expression, there was no evidence of continued transposition, probably because the level was too low to be effective.85 These initial studies suggest that delivery of the gene encoding the transposase does not represent a measurable risk to the recipient. Successful SB transposon delivery to the liver in dogs has been initiated, and although no adverse effects have been noted, it is too early to make any conclusions regarding safety in this large animal model.86 In aggregate, given their relatively random patterns of integration compared to retroviral and lentiviral vectors, SB transposons would appear less likely to insert in or close to loci that are associated with proliferative disease and thereby should have a relatively lower genotoxic potential.
Strategies for enhancing targeted integration
Transposition of SB transposons appears to be unregulated per se, which could be the reason why the estimated ~300,000 DNA transposons in human genomes (~75,000 Tc1/mariner-type87) and ~112,000 DNA transposons in mouse genomes (~1,000 Tc1/mariner-type88) lack active transposase genes. There is an apparent powerful evolutionary selection against autonomous elements. Although the spread of transposons in host organisms is poorly understood,89,90,91 it is clear that transposition by retrotransposons is ongoing as about 100 of the estimated 500,000 LINE-1 (L1) mobile elements have the potential to hop in human cells.92,93,94 These retrotransposons are estimated to be responsible for about 0.1% of de novo mutations in humans, some of which lead to genetic diseases. In mice, the mutational rate from LINE elements is about 100-fold higher than in humans.95 Thus, introduction of one or a few (SB) transposons into a small subset of cells for the purposes of gene therapy amounts to an extremely tiny fraction of the number of total, active transposons in humans.
Nevertheless, there are additional steps that can be taken to increase confidence in the use of transposon vectors. The first is shielding of transcriptional regulators within the expression cassette.96 A second approach is to employ site-directed integration of therapeutic genes into “safe havens” to minimize genotoxicity due to insertional mutagenesis. At present, no such vectors have been published that are both efficient and suitable for application to human gene therapy. Like nearly all viral vectors, DNA-type transposon vectors, such as SB, integrate throughout the mammalian genome as evidenced by their roughly 20% contribution to mammalian genomes.87,97 Vector systems that have some preference for gene site-specific integration include the phiC31 phage system.98,99,100 However, phiC31 recombinase directs integration into preferred sites that differ in various cell types and, most important, phiC31-mediated recombination has been associated in some studies with a relatively high rate of induction of chromosomal translocations.101,102,103 Chromosomal translocations are associated with cancer, and therefore, until this potential form of genotoxicity can be eliminated, the phiC31 system probably will not be attractive for human gene therapy. In contrast, SB transposons have not been reported to induce chromosomal translocations in culture,103,104 umbilical cord blood-derived CD34+ HSCs,105 or in genetically modified human T cells.106,107,108 Alternative targeting strategies that involve site-directed mutagenesis using designer zinc-finger nucleases are under development, but the degree of off-target effects in human genomes is largely unknown.109
Several groups have proposed using modified transposases for site-specific targeting.110 However, there are three problems with the current approaches: (i) Conferring a selection for the transposase on top of a random activity may not preclude the transposon from also integrating into untargeted genomic sites. At this time, transposases with site-specific activity have not been as effective as transposons that have not efficiently inserted into chromosomal sites to which they have been targeted.111,112,113 (ii) Site specificity requires a minimal recognition signature sequence of about 16 bp. However, there are >2,000 sites across the human genome that vary by 2 bp. Because a variation by 2 bp is unlikely to reduce the affinity of integration below 1%, the odds are that a given recombinase will, more often than not, direct integration into a related site with a similar nucleotide sequence (this expectation is confirmed by phiC31-mediated recombination into pseudo-att sites in the human genome101). (iii) Recombinases in general have complex structures with recognition domains that are an integral part of the catalytic domain. Therefore, genetic modification of a recombinase to improve recognition of desired nucleotide sequences is likely to lead to impaired enzymatic activity, as has been found with modified SB transposase enzymes discussed above. The quest to develop targeting vectors for site-specific integration remains the “holy grail” of systems that stably deliver therapeutic transgenes. Nevertheless, this general approach to targeting will need to account for the likelihood that modifying a recombinase for one purpose may undermine other desired activities. As a result, developing site specificity is likely to be arduous and expensive, especially as each transposase or construct will likely need extensive testing.
Transposition of chimeric antigen receptor (CAR) using SB system
Although there may be a variety of strategies to improve the SB transposon system, it is ready for testing its therapeutic potential in human lymphocytes. T cells are an appropriate target for first-in-human application of the SB system because they have been genetically modified using viral72 and nonviral114 gene transfer hundreds of times without any reported serious genotoxic effects in vivo.71 One candidate transgene for transpositional insertion is a CAR to redirect T-cell specificity to a cell surface antigen independent of the major histocompatibility complex. The prototypical CAR consists of an extracellular antigen-binding domain, typically derived from the scFv sequence of a monoclonal antibody that is attached to an immunoglobulin Fc region and fused to one or more intracellular signaling endodomains. The endodomains usually include CD3-ζ so that the T cell can be activated for CAR-dependent cytotoxicity to specifically kill tumor cells. The inclusion of one or more chimeric co-stimulatory domain(s), such as CD28, within the CAR endodomain enables it to deliver a sustained proliferative signal upon binding cell surface antigen. As stated earlier, the major advantage of viral transduction is the improved integration efficiency compared with nonviral approaches such as electrotransfer of a DNA plasmid. This enhanced integration efficiency can minimize the time (and associated costs) incurred for propagation of clinical-grade T cells ex vivo prior to infusion, while potentially preserving a central memory phenotype115 and/or naive phenotype116 that is/are associated with improved in vivo persistence. Indeed, the prolonged numeric expansion ex vivo may not only cause differentiation of the T cells, but could lead to replication-senescence thereby curtailing their therapeutic potential.117 However, T cells that have undergone viral-mediated gene transfer almost always still undergo rounds of replication for typically up to 3 weeks, by crosslinking CD3 with OKT3 to achieve clinically sufficient numbers for infusion. In addition to consideration of their relative integration efficiencies, the decision to use a clinical-grade recombinant viral vector to genetically modify human T cells takes into account the time to produce, package, and release the viral vectors, the financial cost of these vectors, scheduling their manufacture at the limited number of viral production facilities operating in compliance with current good manufacturing practice (cGMP), and the current need to undertake expensive replication-competent retrovirus testing for each patient-specific genetically modified T-cell product. In contrast, approaches to nonviral gene transfer appear to offer several advantages, especially for early-phase human trial design that include a greater number of vendors that can produce clinical-grade plasmids for ex vivo gene transfer at reduced cost. Indeed, as transgenes, such as CARs, are being refined and tumor targets defined, the ability to develop and then redevelop clinical-grade vectors at reasonable cost is an advantage of DNA plasmids.
The generation of transposon/transposase systems, such as SB, and their introduction into mammalian cells by electroporation have enabled investigators to significantly improve the integration efficiency of transgenes in T cells and HSC.106,107,108,118,119,120 To achieve a population of T cells that stably expresses an introduced CAR transgene, we have developed artificial antigen-presenting cells (aAPC) that can (i) selectively propagate T cells in a CAR-dependent manner, and (ii) serve as a culturing environment to use pharmacological agents to reprogram T cells. Our aAPC were derived from K562 cells and genetically modified to express desired T-cell co-stimulatory molecules, such as CD86, CD137L, and a membrane-bound variant of IL-15 (mIL-15). Because the therapeutic potential of T cells in vivo depends on their persistence after adoptive transfer, infusion into patients of genetically modified T cells that are preselected ex vivo for a propensity to proliferate in response to antigen is clinically appealing. Figure 3 summarizes our current strategy to use the SB system to express CAR in human T cells and propagate CAR+ T cells by recursive additions of γ-irradiated aAPC in the presence of soluble cytokines. By combining (i) the Nucleofector system to electrotransfer a CAR as SB transposon and a hyperactive transposase (SB11) gene, on separate supercoiled DNA plasmids, into quiescent T cells derived from peripheral or umbilical cord blood with (ii) outgrowth of CAR+ T cells on aAPC, we can currently manufacture clinically sufficient populations of clinical-grade CAR+ T cells within 3–4 weeks after electroporation, some of which express markers of central memory phenotype and naive phenotype, and exhibit desired effector functions. This technology is robust, cost-effective, and is to be readily undertaken in compliance with cGMP for phase I/II trials using standard operating procedures to manufacture CAR+ T cells that have specificity for CD19 or other tumor-associated antigens. As transposon/transposase systems, such as SB, are used in compliance with cGMP, a comparison can be undertaken to assess the relative expense of nonviral gene versus viral gene transfer that factors in the costs of (i) vector production, (ii) time to culture clinically sufficient numbers of cells, and (iii) release testing.
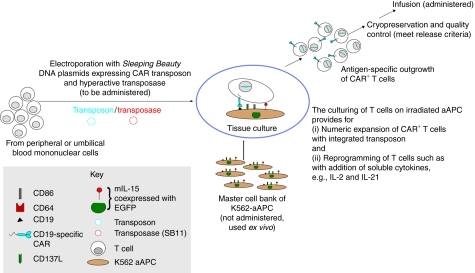
Figure 3.
Schematic for the electroporation of SB system and subsequent selective propagation of CAR+ T cells on aAPC. The culturing of T cells on aAPC leads to their expansion as well as provides a window in which pharmacologic agents can be added to reprogram T cells to exhibit desired properties (e.g., those associated with a central memory or naive phenotypes). The peripheral blood mononuclear cells can be collected by steady-state apheresis or a simple blood draw. The release testing before human application is performed on the cryopreserved product manufactured in compliance with current good manufacturing practice for phase I/II trials. aAPC, artificial antigen-presenting cells; EGFP, enhanced green fluorescent protein; SB, Sleeping Beauty.
Transition of SB system to clinical trials
The first-in-human clinical application of the SB system, and indeed apparently the first to use a transposon system in general, will use circulating human T cells that have been genetically modified to express a CD19-specific CAR. To ensure patient safety, criteria have been established to minimize the infusion of T cells with potential for harm or aberrant function (Table 1). The release of clinical-grade T cells for early-phase trials includes measurement of viability, sterility, and establishing a “chain of custody.” Sterility is assessed for up to 14 days by measuring the bacterial and fungal burden according to the United States Pharmacopeia. The presence of mycoplasma is determined by PCR and the endotoxin burden is assessed using the limulus amebocyte lysate method. A “chain of custody” is ensured by adherence to cGMP and is validated by matching the HLA typing with the donor. In addition, it is necessary to measure the expression of the CAR transgene in the genetically modified T-cell product, generally by flow cytometry with antibody against the exodomain (e.g., Fc region) and/or western blot assay using a CD3-ζ-specific monoclonal antibody. Because we are including γ-irradiated aAPC in the culturing process, flow cytometry is also used to establish that there is not an excess of K562 cells remaining in the numerically expanded T-cell product. For the first-in-human application of the SB system, we have instituted an additional test that ensures there is no autonomous T-cell growth upon withdrawal of antigen and exogenous cytokine(s). Indeed, the ex vivo culturing period serves as a test to monitor for the emergence of T cells with deleterious properties. A PCR test will also be used as a research assay (in-process testing) to validate the absence of integrated SB transposase in the genomes of the T-cell product to be infused. As gene therapies transition from pilot studies to later-phase trials, additional information regarding the T-cell product will be needed, such as a measure of potency (CAR-dependent killing of target cells) and ability of the cryopreserved product to be successfully thawed after storage. In addition, there will hopefully be other opportunities for clinical application of the SB system such as ex vivo introduction of other transgenes into T cells to enhance function and gene transfer into HSC. For instance, preliminary studies on the potential of SB transposons to treat Fanconi anemia have been initiated.121
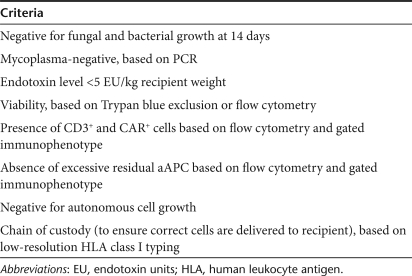
Table 1.
Suggested criteria for release of clinical-grade CAR+ T cells, after SB-mediated gene transfer, for phase I/II trials
Clinical trial design infusing CD19-specific CAR+ T cells
The first clinical trial approved by NIH-OBA (no. 0804–922) to infuse autologous CAR+ T cells genetically modified with the SB system will enroll patients with advanced B-lineage lymphoid malignancies who are undergoing autologous HSC transplantation using a standard-of-care conditioning regimen and who remain at high risk of relapse. The T cells will be administered within a few days of infusing autologous HSC, during which time the research participants are expected to be lymphopenic. This will test whether the adoptively transferred T cells can undergo homeostatic expansion and whether the immunosuppression can prevent immune-mediated rejection of the genetically modified T cells that express a potentially immunogenic transgene (e.g., the murine scFv region of the CAR). Similar to other trials using retrovirus to transduce T cells, the CAR employs “second-generation” technology as it activates T cells in a CD19-dependent manner via CD28 and CD3-ζ, for killing, proliferation, and cytokine production.122 The recipients will be eligible to receive low-dose IL-2 in the event that there is insufficient persistence or number of infused T cells. Correlative studies will be undertaken to assess the persistence of the infused T cells by Q-PCR using CAR-specific primers, and if there are sufficient numbers of recovered T cells, by flow cytometry using a CAR-specific antibody. Bone marrow and lymph node biopsies will reveal whether the infused T cells can migrate to these sites of malignant disease. In addition, studies will reveal if recipients mount a deleterious humoral and/or cellular immune response against the introduced CAR.
Conclusion
Gene-transfer clinical trials that use either viral or nonviral vectors have inherent risks that are chiefly associated with genotoxicity arising from insertional mutagenesis. These risks must be carefully evaluated by investigators and regulators alike, against the potential benefit that delivery of a therapeutic transgene can provide.41,123 Currently, large amounts of money, staff, and time are required to design and implement a gene therapy trial due to legal barriers, indemnification of the genetically modified product, third-party payer systems, and regulatory hurdles. Often these protocols are single institution, phase I studies that are powered to provide information on safety and feasibility, but not prove efficacy. Although there have been some gene therapy successes, the number of trials is limited because only a few institutions and pharmaceutical companies can support the necessary infrastructure and costs. The human application of gene therapy in countries with first-world technology, as well as nations with developing economies, would be extended if barriers were lowered. Transposon-based gene therapy offers one approach to reducing costs of the vector system without any evident compromise on patient wellbeing. Given the track record of safety regarding the infusion of genetically modified T cells and the urgent need to provide new therapy for children and adults at risk of death from advanced malignancies as well as reduce the morbidity from current therapies, we plan to test the first-human application of the SB system in autologous T cells that have been electroporated ex vivo to express a CD19-specific CAR. We expect this trial will provide the necessary data to evaluate the safety of this novel gene-transfer approach that could lower the threshold to undertaking gene therapy trials using the SB system in the future.
Acknowledgments
We thank Elena L. Aronovich for helpful discussions. This project was supported by NIH grants P01HD32652 and R01DK082516 to P.B.H., R01CA113636 to D.A.L., and RO1CA124782 and R01CA120956 to L.J.N.C.
REFERENCES
- Kay MA, Glorioso JC., and , Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- Lozier JN, Csako G, Mondoro TH, Krizek DM, Metzger ME, Costello R, et al. Toxicity of a first-generation adenoviral vector in rhesus macaques. Hum Gene Ther. 2002;13:113–124. doi: 10.1089/10430340152712665. [DOI] [PubMed] [Google Scholar]
- Graham A, Walker R, Baird P, Hahn CN., and , Fazakerley JK. CNS gene therapy applications of the Semliki Forest virus 1 vector are limited by neurotoxicity. Mol Ther. 2006;13:631–635. doi: 10.1016/j.ymthe.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Selker EU. Gene silencing: repeats that count. Cell. 1999;97:157–160. doi: 10.1016/s0092-8674(00)80725-4. [DOI] [PubMed] [Google Scholar]
- Bestor TH. Gene silencing as a threat to the success of gene therapy. J Clin Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák Z., and , Ivics Z. Sleeping Beauty transposition: biology and applications for molecular therapy. Mol Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Hackett PB, Ekker SC, Largaespada DA., and , McIvor RS. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- VandenDriessche T, Ivics Z, Izsvák Z., and , Chuah MK. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood. 2009;114:1461–1468. doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH., and , Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Ivics Z., and , Hackett PB. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio) Mol Gen Genet. 1995;247:312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z, Minter A., and , Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CM., and , Largaespada DA. Insertional mutagenesis in mice: new perspectives and tools. Nat Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- Ivics Z., and , Izsvák Z. A whole lotta jumpin' goin' on: new transposon tools for vertebrate functional genomics. Trends Genet. 2005;21:8–11. doi: 10.1016/j.tig.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, et al. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Yant SR, Park J, Huang Y, Mikkelsen JG., and , Kay MA. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baus J, Liu L, Heggestad AD, Sanz S., and , Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Khare D, Behlke J, Heinemann U, Plasterk RH., and , Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- Cui Z, Geurts AM, Liu G, Kaufman CD., and , Hackett PB. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z., and , Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T., and , Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- Liu L, Mah C., and , Fletcher BS. Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice using an endothelial-targeted Sleeping Beauty transposon. Mol Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kren BT, Unger GM, Sjeklocha L, Trossen AA, Korman V, Diethelm-Okita BM, et al. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J Clin Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Held PK, Noll M, Morcinek N, Al-Dhalimy M, Finegold M, et al. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol Ther. 2002;6:759–769. doi: 10.1006/mthe.2002.0812. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Lin Q, Yant SR, Keene D, Kay MA., and , Khavari PA. Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 2003;10:1099–1104. doi: 10.1038/sj.gt.3301978. [DOI] [PubMed] [Google Scholar]
- He CX, Shi D, Wu WJ, Ding YF, Feng DM, Lu B, et al. Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J Gastroenterol. 2004;10:567–572. doi: 10.3748/wjg.v10.i4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, ZJ, Kren, BT, Wong, PY, Low, WC., and , Steer, CJ. Sleeping Beauty-mediated down-regulation of huntingtin expression by RNA interference. Biochem Biophys Res Commun. 2005;329:646–652. doi: 10.1016/j.bbrc.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, Erickson DC, et al. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronovich EL, Bell JB, Khan SA, Belur LR, Gunther R, Koniar B, et al. Systemic correction of storage disease in MPS I NOD/SCID mice using the Sleeping Beauty transposon system. Mol Ther. 2009;17:1136–1144. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlfest JR, Lobitz PD, Perkinson SG., and , Largaespada DA. Integration and long-term expression in xenografted human glioblastoma cells using a plasmid-based transposon system. Mol Ther. 2004;10:260–268. doi: 10.1016/j.ymthe.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Ohlfest JR, Demorest ZL, Motooka Y, Vengco I, Oh S, Chen E, et al. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the Sleeping Beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol Ther. 2005;12:778–788. doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Daigle B, Garrison BA, Burgess SM, et al. Nonrandom insertion site preferences for the SB transposon in vitro and in vivo. Mol Ther. 2004;9:S309–S310. [Google Scholar]
- Geurts AM, Hackett CS, Bell JB, Bergemann TL, Collier LS, Carlson CM, et al. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 2006;34:2803–2811. doi: 10.1093/nar/gkl301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and , Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin CT, Alexander IE., and , Kerridge I. Accepting risk in clinical research: is the gene therapy field becoming too risk-averse. Mol Ther. 2009;17:1842–1848. doi: 10.1038/mt.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol Ther. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Tsukahara T, Yoshino K, Kojima K, Agawa H, Yamashita Y, et al. Identification of a high incidence region for retroviral vector integration near exon 1 of the LMO2 locus. Retrovirology. 2009;6:79. doi: 10.1186/1742-4690-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehse B., and , Roeder I. Insertional mutagenesis and clonal dominance: biological and statistical considerations. Gene Ther. 2008;15:143–153. doi: 10.1038/sj.gt.3303052. [DOI] [PubMed] [Google Scholar]
- June CH, Blazar BR., and , Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat Rev Immunol. 2009;9:704–716. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newrzela S, Cornils K, Li Z, Baum C, Brugman MH, Hartmann M, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- Kohn DB, Sadelain M, Dunbar C, Bodine D, Kiem HP, Candotti F, American Society of Gene Therapy (ASGT) et al. American Society of Gene Therapy (ASGT) ad hoc subcommittee on retroviral-mediated gene transfer to hematopoietic stem cells. Mol Ther. 2003;8:180–187. doi: 10.1016/s1525-0016(03)00212-0. [DOI] [PubMed] [Google Scholar]
- Modlich U, Kustikova OS, Schmidt M, Rudolph C, Meyer J, Li Z, et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood. 2005;105:4235–4246. doi: 10.1182/blood-2004-11-4535. [DOI] [PubMed] [Google Scholar]
- Shou Y, Ma Z, Lu T., and , Sorrentino BP. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci USA. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NB, Bottero V, Schmidt M, von Kalle C., and , Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Xiong D, Stamatoyannopoulos G., and , Emery DW. Genomic and functional assays demonstrate reduced gammaretroviral vector genotoxicity associated with use of the cHS4 chromatin insulator. Mol Ther. 2009;17:716–724. doi: 10.1038/mt.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiem HP, Sellers S, Thomasson B, Morris JC, Tisdale JF, Horn PA, et al. Long-term clinical and molecular follow-up of large animals receiving retrovirally transduced stem and progenitor cells: no progression to clonal hematopoiesis or leukemia. Mol Ther. 2004;9:389–395. doi: 10.1016/j.ymthe.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Herndier BG, Shiramizu BT, Jewett NE, Aldape KD, Reyes GR., and , McGrath MS. Acquired immunodeficiency syndrome-associated T-cell lymphoma: evidence for human immunodeficiency virus type 1-associated T-cell transformation. Blood. 1992;79:1768–1774. [PubMed] [Google Scholar]
- Brady T, Agosto LM, Malani N, Berry CC, O'Doherty U., and , Bushman F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS. 2009;23:1461–1471. doi: 10.1097/QAD.0b013e32832caf28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G, et al. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther. 2009;17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty J, Terry A, MacDonald J, Gault E, Cevario S, O'Brien SJ, et al. Feline immunodeficiency virus integration in B-cell lymphoma identifies a candidate tumor suppressor gene on human chromosome 15q15. Cancer Res. 2002;62:7175–7180. [PubMed] [Google Scholar]
- Bell P, Wang L, Lebherz C, Flieder DB, Bove MS, Wu D, et al. No evidence for tumorigenesis of AAV vectors in a large-scale study in mice. Mol Ther. 2005;12:299–306. doi: 10.1016/j.ymthe.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Nakai H, Montini E, Fuess S, Storm TA, Grompe M., and , Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- Nakai H, Wu X, Fuess S, Storm TA, Munroe D, Montini E, et al. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J Virol. 2005;79:3606–3614. doi: 10.1128/JVI.79.6.3606-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- Themis M, Waddington SN, Schmidt M, von Kalle C, Wang Y, Al-Allaf F, et al. Oncogenesis following delivery of a nonprimate lentiviral gene therapy vector to fetal and neonatal mice. Mol Ther. 2005;12:763–771. doi: 10.1016/j.ymthe.2005.07.358. [DOI] [PubMed] [Google Scholar]
- Bowers WJ, Mastrangelo MA, Howard DF, Southerland HA, Maguire-Zeiss KA., and , Federoff HJ. Neuronal precursor-restricted transduction via in utero CNS gene delivery of a novel bipartite HSV amplicon/transposase hybrid vector. Mol Ther. 2006;13:580–588. doi: 10.1016/j.ymthe.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Gene Therapy Expert Group of the Committee for Proprietary Medical Products (CPMP); European Agency for the Evaluation of Medical Products Insertional mutagenesis and oncogenesis: update from non-clinical and clinical studies. Gene Therapy Expert Group of the Committee for Proprietary Medical Products (CPMP), European Agency for the Evaluation of Medical Products—June 2003 meeting. J Gene Med. 2004;6:127–129. doi: 10.1002/jgm.466. [DOI] [PubMed] [Google Scholar]
- Scobie L, Hector RD, Grant L, Bell M, Nielsen AA, Meikle S, et al. A novel model of SCID-X1 reconstitution reveals predisposition to retrovirus-induced lymphoma but no evidence of gammaC gene oncogenicity. Mol Ther. 2009;17:1031–1038. doi: 10.1038/mt.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini C, Grez M, Traversari C, Ciceri F, Marktel S, Ferrari G, et al. Safety of retroviral gene marking with a truncated NGF receptor. Nat Med. 2003;9:367–369. doi: 10.1038/nm0403-367. [DOI] [PubMed] [Google Scholar]
- Ciceri F, Bonini C, Stanghellini MT, Bondanza A, Traversari C, Salomoni M, et al. Infusion of suicide-gene-engineered donor lymphocytes after family haploidentical haemopoietic stem-cell transplantation for leukaemia (the TK007 trial): a non-randomised phase I-II study. Lancet Oncol. 2009;10:489–500. doi: 10.1016/S1470-2045(09)70074-9. [DOI] [PubMed] [Google Scholar]
- Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Kool J, Berns A., and , van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- Starr TK., and , Largaespada DA. Cancer gene discovery using the Sleeping Beauty transposon. Cell Cycle. 2005;4:1744–1748. doi: 10.4161/cc.4.12.2223. [DOI] [PubMed] [Google Scholar]
- Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D., and , Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ., and , Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- Lu M, Zhang N, Maruyama M, Hawley RG., and , Ho AD. Retrovirus-mediated gene expression in hematopoietic cells correlates inversely with growth factor stimulation. Hum Gene Ther. 1996;7:2263–2271. doi: 10.1089/hum.1996.7.18-2263. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG., and , Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ., and , Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JB, Aronovich EA, Schreifels JM, Clifford AN, Hoekstra ND, McIvor RS, et al. 2008Expression of Sleeping Beauty transposase in mouse liver following hydrodynamic delivery Mol Ther 16suppl. 1): S312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett PB, Urness M, Bell JB, Olson ER, Aronovich EL,, Cressman E, et al. 2008Use of non-invasive balloon catheters placed under fluoroscopic guidance for delivery of the Sleeping Beauty transposon system under pressure to the liver of dogs Mol Ther 16suppl. 1): S312 [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, International Human Genome Sequencing Consortium et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P., and , Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Lohe AR, Moriyama EN, Lidholm DA., and , Hartl DL. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol Biol Evol. 1995;12:62–72. doi: 10.1093/oxfordjournals.molbev.a040191. [DOI] [PubMed] [Google Scholar]
- Shapiro JA., and , von Sternberg R. Why repetitive DNA is essential to genome function. Biol Rev Camb Philos Soc. 2005;80:227–250. doi: 10.1017/s1464793104006657. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr, and , Goodier JL. LINE drive. retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA., and , Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN., and , Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisko O, Schorn A, Rolfs F, Devaraj A, Miskey C, Izsvák Z, et al. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther. 2008;16:359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B., and , Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS., and , Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA., and , Calos MP. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, et al. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Liu J, Jeppesen I, Nielsen K., and , Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Engler JA, Xu H, Cherry AM., and , Kay MA. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM., and , Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Huang X, Nodland SE, Mátés L, Ma L, Izsvák Z, et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wilber AC, Bao L, Tuong D, Tolar J, Orchard PJ, et al. Stable gene transfer and expression in human primary T cells by the Sleeping Beauty transposon system. Blood. 2006;107:483–491. doi: 10.1182/blood-2005-05-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Guo H, Kang J, Choi S, Zhou TC, Tammana S, et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- Coates CJ, Kaminski JM, Summers JB, Segal DJ, Miller AD., and , Kolb AF. Site-directed genome modification: derivatives of DNA-modifying enzymes as targeting tools. Trends Biotechnol. 2005;23:407–419. doi: 10.1016/j.tibtech.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Yant SR, Huang Y, Akache B., and , Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S., and , Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- Voigt K, Izsvák Z., and , Ivics Z. Targeted gene insertion for molecular medicine. J Mol Med. 2008;86:1205–1219. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- Till BG, Jensen MC, Wang J, Chen EY, Wood BL, Greisman HA, et al. Adoptive immunotherapy for indolent non-Hodgkin lymphoma and mantle cell lymphoma using genetically modified autologous CD20-specific T cells. Blood. 2008;112:2261–2271. doi: 10.1182/blood-2007-12-128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C., and , Riddell SR. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J Clin Invest. 2008;118:294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs CS, Borman ZA, Cassard L, Gattinoni L, Spolski R, Yu Z, et al. Adoptively transferred effector cells derived from naive rather than central memory CD8+ T cells mediate superior antitumor immunity. Proc Natl Acad Sci USA. 2009;106:17469–17474. doi: 10.1073/pnas.0907448106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Zhou J, Hathcock KS, Robbins P, Powell DJ, Jr, Rosenberg SA, et al. Persistence of tumor infiltrating lymphocytes in adoptive immunotherapy correlates with telomere length. J Immunother. 2007;30:123–129. doi: 10.1097/01.cji.0000211321.07654.b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi T, Holt NG, Hollis RP, Ge S, Cannon PM, Crooks GM, et al. Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells, using the Sleeping Beauty transposon system. Hum Gene Ther. 2009;20:1607–1626. doi: 10.1089/hum.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák Z, Chuah MK, Vandendriessche T., and , Ivics Z. Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods. 2009;49:287–297. doi: 10.1016/j.ymeth.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, et al. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland KA, Olson ER, Aronovich EL, Hackett PB, Blazar BR, Tolar JR, et al. 2009Analysis of Sleeping Beauty transposons for correction of Fanconi anemia in lymphoblastoid cell lines Mol Ther 16suppl. 1): S119 [Google Scholar]
- Kowolik CM, Topp MS, Gonzalez S, Pfeiffer T, Olivares S, Gonzalez N, et al. CD28 costimulation provided through a CD19-specific chimeric antigen receptor enhances in vivo persistence and antitumor efficacy of adoptively transferred T cells. Cancer Res. 2006;66:10995–11004. doi: 10.1158/0008-5472.CAN-06-0160. [DOI] [PubMed] [Google Scholar]
- Editorial Gene therapy deserves a fresh chance. Nature. 2009;461:1173. doi: 10.1038/4611173a. [DOI] [PubMed] [Google Scholar]