Abstract
Exon skipping has demonstrated great potential for treating Duchenne muscular dystrophy (DMD) and other diseases. We have developed a drug-screening system using C2C12 myoblasts expressing a reporter green fluorescent phosphate (GFP), with its reading frame disrupted by the insertion of a targeted dystrophin exon. A library of 2,000 compounds (Spectrum collection; Microsource Discovery System) was screened to identify drugs capable of skipping targeted dystrophin exons or enhancing the exon-skipping effect by specific antisense oligomers. The 6-thioguanine (6TG) was effective for inducing skipping of both human dystrophin exon 50 (hDysE50) and mouse dystrophin exon 23 (mDysE23) in the cell culture systems and increased exon skipping efficiency (more than threefolds) when used in combination with phosphorodiamidate morpholino oligomers (PMO) in both myoblasts and myotubes. Guanine and its analogues were unable to induce detectable skipping of exon 23 when used alone but enhanced PMO-induced exon skipping significantly (approximately two times) in the muscles of dystrophic mdx mouse in vivo. Our results demonstrate that small-molecule compounds could enhance specific exon skipping synergistically with antisense oligomers for experimental therapy to human diseases.
Introduction
The dystrophin gene is one of the largest genetic loci and contains at least 79 exons distributed over 2.5 million base pairs (bp) on the p21 subregion of X chromosome.1 The primary transcript takes 16 hours to transcribe and produce a mature mRNA of 140 kbp in muscles. The 427 kd muscle form of dystrophin protein contains 3,685 amino acids, including four major domains: a N-terminal actin-binding domain, a spectrin-like rod domain, a cysteine-rich region, and a C-terminal domain.2 Dystrophin, the central component of the dystrophin-associated protein complex, links the cytoskeleton of muscle fibers with the extracellular matrix, stabilizes the sarcolemma, and protects muscle fibers from contraction-induced damage.3 Lack of dystrophin or defects in the function of the protein compromise the entire dystrophin-associated protein complex, leading to muscle wasting and degeneration.4,5
Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy are allelic disorders caused by mutations within the dystrophin gene.2,6 DMD is the most severe form of dystrophinopathy, caused by frame-shifting mutations, resulting in premature termination of dystrophin translation and, consequently, the absence of dystrophin protein. DMD affects ~1 in 3,500 boys, and there is no effective treatment currently.3,7,8 Becker muscular dystrophy, the milder form of dystrophinopathy with later onset, is caused by mostly in-frame deletions producing a shorter but partially functional dystrophin protein.7
In the past few years, antisense oligonucleotide (AON)-induced, specific exon skipping has been reported to restore dystrophin expression in cells and in muscles of animal models of DMD.8,9,10,11,12,13,14 This strategy aims to skip an exon or exon(s) adjacent to the mutation sites, so that the out-of-frame dystrophin mRNA is corrected to an in-frame transcript, which can then be translated into a shortened Becker muscular dystrophy-like protein.8 Two phase-I clinic trials of the therapy have now provided the proof of principle that intramuscularly delivered AONs were able to induce targeted skipping of human dystrophin exon 51 (hDysE51) with partial restoration of dystrophin production in the treated muscles of DMD patients.15,16 DMD is a systemic disease, affecting body-wide muscles; therefore, effective therapy relies on long-term systemic treatment. However, the long-term systemic efficacy of the exon skipping remains to be determined.
Several factors are critical for effective exon skipping, including the specific sequence selected for the targeted exon and the chemical nature and delivery efficiency of AONs. Currently, 2-O methyl phosphorothioate and phosphorodiamidate morpholino oligomer (PMO) are the most widely used for exon skipping in dystrophin gene and are being applied in clinical trials.15,16 Modifications of AONs with cell-penetrating peptides have led to significant improvement in the delivery of AONs and achieved short-term rescue of dystrophin expression in body-wide muscles, including the cardiac muscle with improved pathology and partial restoration of muscle functions.17,18 However, the efficacy and safety of the long-term use of positively charged peptides remain to be investigated. Here, we developed a drug-screening system with a green fluorescent phosphate (GFP)-based reporter in C2C12 mouse myoblasts to identify drugs capable of inducing and enhancing targeted exon skipping. We found that 6-thioguanine (6TG) exhibits positive effect on exon skipping, and, importantly, several analogues improve PMO-induced exon skipping (approximately two times) in mdx muscles in vivo.
Results
Drug screening for exon skipping of dystrophin gene in cell culture systems with GFP expression as reporter
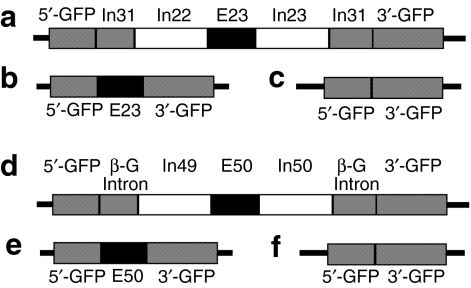
As shown in Figure 1, two GFP-reporter vectors were constructed for skipping of the hDysE50 and mouse dystrophin exon 23 (mDysE23). This system adopted the model established by Kole et al. that the coding sequence of the reporter gene GFP is split into two fragments by the insertion of the human β-globin intron sequence.19 Under normal circumstance, the intron sequences are faithfully spliced out and the two fragments of the GFP-coding sequence are joined together, which in turn leads to the restoration of the reading frame. For exon skipping in the dystrophin gene, the targeted exons, hDysE50 and mDysE23, together with their flanking intron sequences at both ends, were inserted into the β-globin intron sequence and the mouse dystrophin intron 31 sequence, respectively (Figure 1a–f). The presence of the dystrophin exons in the mRNA disrupts the reading frame of the GFP, and thus its expression. However, in the presence of effective AONs targeting the individual exon, the removal of the targeted dystrophin exons will restore the GFP-reading frame. The levels of GFP expression are, therefore, good indicators of the efficiency of the AONs. This system aims to identify drug(s) capable of inducing exon skipping by itself or enhancing AON-induced exon skipping.
Figure 1.
Graphic illustration of the GFP-reporter vectors for dystrophin exon skipping. (a) Linear vector structure shows that the GFP sequence is split into two regions, 5′-GFP and 3′-GFP, by the mouse dystrophin intron (In31) that contains the 213-bp mouse exon (E23), 3′ side of intron (In22), and 5′ side of intron (In23). (b) Expression of GFP is driven by MCK promoters. Without AON intervention, the vector expresses an out-of-frame GFP/exon 23 chimeric transcript (normal transcript). (c) Skipping exon 23 with AONs restores the GFP-reading frame and expression of GFP protein. (d) Linear vector structure shows that the GFP sequence is split into two regions, 5′-GFP and 3′-GFP, by the human β-globin intron sequence (β-G), which contains the 109-bp human dystrophin exon 50 (E50), flanked by intron 49 (In49) on the 5′ side and intron 50 (In50) on the 3′ side. (e) Expression of GFP is driven by a β-actin promoter. Without AON intervention, the vector expresses the out-of-frame GFP/exon 50 chimeric transcripts (normal transcripts). (f) Skipping exon 50 with AON restores the GFP-reading frame and expression of the protein. Other structures of the vectors are not shown here. AON, antisense oligonucleotide; GFP, green fluorescent phosphate; MCK, muscle creatine kinase.
Stable C2C12 myoblast cell lines expressing GFP/hDysE50 (C2C12E50) or GFP/mDysE23 (C2C12E23) transgenes were established by transfection of the cells with the expression vectors and selection with antibiotic G418. A total of 2,000 bioactive compounds were initially screened at the final concentration of 33 µmol/l in the C2C12E50 cells without AON treatment. These compounds included 1,040 US Food and Drug Administration-approved drugs, 800 compounds from a variety of natural resources, and 160 synthetic and natural substances (Spectrum collection; Microsource Discovery System; http://www.msdiscovery.com/index.html). Initial screening with fluorescence microscope identified 10 compounds with extremely strong signals (fluorescence was clearly visible with naked eye), 18 compounds with strong signal (easily identifiable under fluorescence microscope), 14 compounds with weak signal (slightly higher than the background observed in the control cells without drug treatment; see Supplementary Table S1). Many of the compounds were determined to be false positive or autofluorescent when similar levels of signal were found in the parental C2C12 cells without reporter gene. Nevertheless, one compound, 6TG, was identified as real positive with strong signal.
6TG induced hDysE50 skipping dose-dependently and synergized with PMO
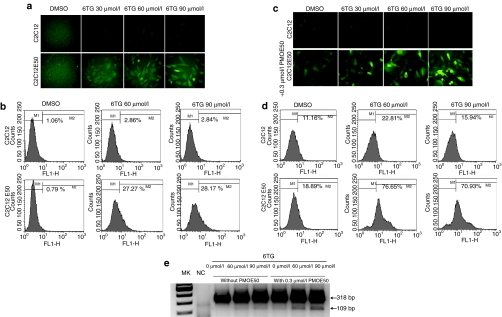
To further explore the effect of 6TG on exon skipping, cultured C2C12 and C2C12E50 cells were treated with 6TG at different concentrations for 48 hours. As shown in Figure 2a, GFP expression was enhanced dose-dependently in the C2C12E50 cells treated with 30, 60, and 90 µmol/l 6TG as judged by the increasing signal intensity when compared to the cells without 6TG treatment. No clear GFP signal was detected in the control C2C12 cells treated with 6TG. The induction of GFP expression by 6TG was further confirmed by quantitative flow cytometry analysis (Figure 2b), which showed the presence of <5% GFP+ cells in the control group (untreated C2C12E50 cells), but the same analysis showed 27.27 and 28.17% GFP+ cells in the treatment group, after 60- and 90-µmol/l 6TG treatment, respectively. However, the levels of GFP expression were generally low in most of the positive cells.
Figure 2.
GFP expression in C2C12E50 cells after 6TG and PMOE50 treatment. (a) Fluorescence microscope images and (b) FACS analysis of the C2C12 and C2C12E50 cells treated with DMSO, 30, 60, and 90 µmol/l 6TG only. (c) Fluorescence microscope images and (d) FACS analysis of the C2C12 and C2C12E50 cells treated with DMSO, 30, 60, and 90 µmol/l 6TG in the presence of 0.3 µmol/l PMOE50. M2 is the population of cells gated for the expression of GFP in b and d. (e) RT-PCR for exon 50 skipping in the C2C12E50 cells. The 318-bp bands are the chimeric transcripts containing exon 50; the 109-bp bands are the transcripts with exon 50 skipped that are clearly detected only in the cells treated with PMOE50, together with 60- and 90-µmol/l 6TG. AON, antisense oligonucleotide; DMSO, dimethyl sulfoxide; FACS, fluorescence-activated cell sorting; GFP, green fluorescent phosphate; MK, size marker; NC, C2C12 cells as negative controls; RT, reverse transcription; 6TG, 6-thioguanine.
We then examined whether 6TG might enhance the exon-skipping effect of AONs. Cells were treated with low concentration of specific PMO, PMOE50 (5′–AACUUCCUCUUUAACAGAAAAGCAUAC–3′), targeting hDysE50. The PMOE50 sequence has been selected through the screening of oligonucleotide sequences in the C2C12E50 cells (data not shown). As shown in Figure 2c, PMOE50 alone at the dose of 0.3 µmol/l induced detectable but very low level of GFP expression. We, therefore, treated the cells with 6TG at increasing doses in combination with the 0.3-µmol/l PMOE50. It is interesting that the expression level of GFP was greatly increased in the C2C12E50 cells treated with both 6TG and PMOE50 (Figure 2c). Only 18.89% cells were gauged as GFP+ when treated with 0.3-µmol/l PMOE50 alone, whereas 76.65 and 70.93% cells were GFP+ (more than threefolds increase) when 60 and 90 µmol/l 6TGs were used in combination with the PMOE50, respectively (Figure 2d). Reverse transcription (RT)-PCR showed that only the unskipped chimeric transcripts containing both GFP and the hDysE50 sequence (product length of 318 bp) were present in the untreated control cells. The GFP transcripts with hDysE50-skipped (product length of 109 bp) were detected in the cells after treatment of 0.3 µmol/l PMOE50 (Figure 2e). The signal of this 109 bp GFP transcript (indicative of hDysE50-skipping) was clearly enhanced in the cells treated with 60 and 90 µmol/l 6TG in the presence of 0.3 µmol/l PMOE50. Our results, therefore, demonstrated a synergistic effect of 6TG and low concentration of PMOE50 on skipping of hDysE50 in vitro.
6TG induced mDysE23 skipping in differentiated C2C12E23 cells
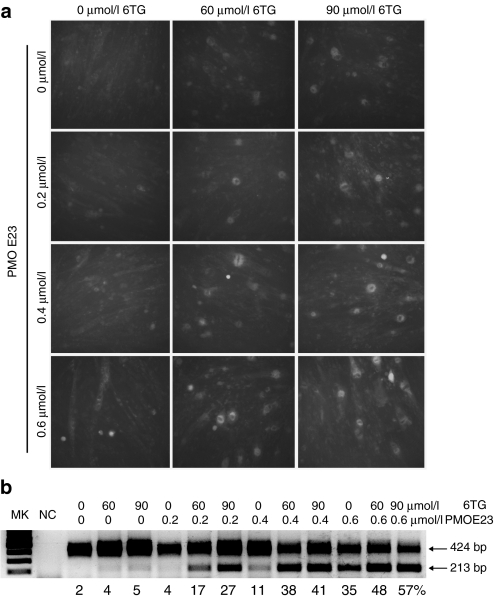
The C2C12E23 reporter construct used a muscle creatine kinase promoter to drive the expression of GFP, thus allowing us to test the potential of drugs for exon skipping in differentiating or differentiated myotubes. Cells reaching ~70% confluence were incubated in the differentiation media for 3 days and then treated with 6TG and/or PMOE23. Expression of GFP was examined 3 days later. As shown in Figure 3a, weak signal of GFP was detected in the myotubes treated with 6TG, but the intensity was enhanced in the presence of PMOE23. The GFP intensity increased with increasing concentration of 6TG (60 and 90 µmol/l) or PMOE23 (0.2, 0.4, 0.6 µmol/l). The 6TG also enhanced PMOE23-induced exon skipping effect in a dose-dependent manner with increasing GFP intensity from 60 to 90 µmol/l. RT-PCR analyses confirmed the increased percentage of the transcripts with mDysE23-skipping in the cells treated with both 6TG and PMOE23 as compared to the cells treated with PMOE23 alone (Figure 3b).
Figure 3.
GFP expression in C2C12E23 cells treated with 6TG in differentiation media. The cells were cultured in differentiation medium for 3 days, and myotube formation was observed. (a) Fluorescence detection for GFP expression. The cells in the left column were treated with 0, 0.2, 0.4, and 0.6 µmol/l PMOE23 only. The cells in the middle and right columns were treated with 60 and 90 µmol/l 6TG, together with the concentration of PMOE23 marked on the left side of the panel. Strongest GFP expression was observed in the cells treated with both 0.6-µmol/l PMOE23 and 90 µmol/l 6TG (bottom-right column). (b) RT-PCR for exon 23 skipping. Three different doses of 6TG were used in combination with 0, 0.2, 0.4, and 0.6 µmol/l of PMOE23. The 424 bp bands are the transcripts containing exon 23; the 213 bp bands are the transcripts with exon 23 skipped. The numbers listed under the gel image are relative percentage of the exon 23 skipped mRNA (213 bp) to the total levels of unskipped mRNA (424 bp) based on the density measurement by NIH ImageJ. GFP, green fluorescent phosphate; RT, reverse transcription; 6TG, 6-thioguanine.
Synergistic effect of guanine analogues on PMO-induced dystrophin exon skipping
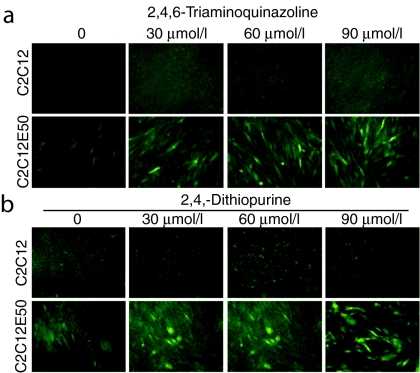
To search for more potent compounds for inducing exon skipping, we tested 19 guanine analogues at three concentrations in the C2C12E50 systems. Similar to the control cells, none of the 19 compounds was able to generate a clear GFP signal in the cells treated at any dose (Supplementary Table S2). However, in the presence of 0.3 µmol/l PMOE50, five analogues—guanine hydrochloride, 2-amino-6-hydroxy-8-mercaptopurine, 6-thioxopurine, 2,4,6-triaminoquinazoline, and 2,6-dithiopurine produced strong GFP signal indicating effective hDysE50-skipping (Supplementary Figure S1). As shown in Figure 4a,b, 2,4,6-triaminoquinazoline and 2,6-dithiopurine enhanced hDysE50-skipping in a dose-dependent manner. The same 19 guanine analogue compounds were further tested in the C2C12E23 cells in combination with the use of 0.5 µmol/l PMOE23. The five compounds, that is, guanine hydrochloride, 8-methyl-9H-purine-6-thiol, 2,4,6-triaminoquinazoline, 2,6-dithiopurine, and 2,8-dimercapto-6-hydroxypurine also demonstrated enhanced levels of GFP expression, when compared to the cells treated with PMOE23 alone (Supplementary Figure S1b). However, the GFP signals in these treated cells were less intense than those in the cells treated with both 6TG and PMOE23.
Figure 4.
Synergistic effect of guanine analogues and PMOE50 on inducing exon 50 skipping. C2C12 (as control cells) and C2C12E50 were treated with (a) 2,4,6-triaminoquinazoline and (b) 2,6-dithiopurine at three different doses. All samples were treated with 0.5 µmol/l PMOE50.
6TG and guanine analogues enhanced PMO-induced mDysE23 skipping in muscles of mdx mice in vivo
We next examined whether guanine and its analogues are able to induce and enhance exon skipping in the absence and presence of PMOE23 in vivo by intramuscular injection to the tibialis anterior (TA) muscles of the mdx mice. Two doses at 1 and 10 µg were first examined for each of the guanine analogues listed in the Supplementary Table S2. None of the compounds alone at either dosage resulted in clearly detectable levels of dystrophin protein in the treated muscles by western blots (data not shown). Immunohistochemistry showed that the number of dystrophin-positive fibers in the muscles treated with the compounds was not significantly different from that expressed in the control group—saline-treated TA muscles (data not shown). Significant muscle damage (necrotic fibers in the injection sites) was observed with the 6-mercapto-9h-purin-8-ol, 2-amino-6-hydroxy-8-mercaptopurine, and 2-amino-6-methylmercaptopurine at the dose of 1 µg; thus, no further test was conducted with the three compounds. The rest of the compounds were further examined by coinjection of 2 µg PMOE23 into the TA muscles. This amount of PMOE23 induced an average of 800 dystrophin-positive fibers in one cross-section (maximum <1,000) of the TA muscle of mdx mice compared with <100 revertant fibers (maximum) in control TA muscles (Figure 5a,b). The number of dystrophin-positive fibers increased in the muscles treated with PMOE23 in combination with each of the four compounds: guanine hydrochloride, 6TG, 2,6-dithiopurine, and 2,4,6-triaminoquinazoline. In particular, significant increase in the number of dystrophin-positive fibers was detected in the muscles treated with 2,4,6-triaminoquinazoline and 6TG at 10 µg and guanine at both 1 and 10 µg in combination with 2 µg of PMOE23 (Figure 5b). The number of dystrophin-positive fibers reached nearly 2,000 in one cross-section in the muscles treated with guanine and remained approximately two times higher than that in muscles treated with PMOE23 alone (Figure 5a,b). This synergistic effect was further supported by the observation that the levels of dystrophin mRNA with mDysE23 skipped were clearly higher in the combinatorial treated muscle samples (Figure 5c). Western blot also revealed a more than twofold increase in the amount of dystrophin protein in the samples treated with both PMOE23 and 6TG (or guanine) when compared to the samples treated with PMOE23 only (Figure 5d).
Figure 5.
Synergistic effect of guanine analogues and PMOE23 on dystrophin expression in TA muscles of mdx mice. (a) Immunohistochemistry for dystrophin expression. Control, untreated TA muscle; PMOE23 only, muscle treated with 2 µg PMOE23 only; The rest of the muscles were treated with both 2-µg PMOE23 and following compounds at 1- and 10-µg concentration. (b) Number of dystrophin-positive fibers after treatment with 2-µg PMOE23 with and without guanine analogues. The numbers of dystrophin-positive fibers were counted in a single cross-section. * and **, significant (P < 0.05) and very significant (P < 0.01) by Student's t-tests (n = 4) when compared to the PMOE23 treatment only. (c) RT-PCR demonstrates the skipping of mDysE23. E22–E23–E24 indicates normal dystrophin transcripts, and E22–E24 indicates transcripts with mDysE23 skipped. mdx control, saline-treated TA muscle of the mdx mouse. (d) Western blots demonstrate the expression of dystrophin protein. Dys, dystrophin detected with monoclonal antibody Dys 1. α-actin was used as a loading control. 2,6-dith, 2,6-dithiopurine; guanine, guanine hydrochloride; RT, reverse transcription; 2,4,6-tri, 2,4,6-triaminoquinazoline; 6TG, 6-thioguanine.
Discussion
Alternative pre-mRNA splicing is an important mechanism for regulating gene expression in higher eukaryote. Over 80% of genes have alternative forms, the major source of proteasome diversity in humans. Splicing abnormality has been established as a common mechanism responsible for a number of human diseases from cystic fibrosis to cancer to muscular dystrophy.20,21,22,23 Modulation of splicing and correction of erroneous splicing are thus potential approaches for treating relevant human diseases. Recent experiments have used modified oligonucleotides to inhibit cryptic exons or to activate exon inclusion.24 Furthermore, specific exon(s) can be skipped by oligonucleotides to restore reading frame disrupted by nonsense and frame-shift mutations. This new approach has been proved to be highly effective for restoration of dystrophin expression in both mouse and dog models of human DMD by local and systemical injections of AONs.17,25,26 Phase-I clinic trials have also provided the proof of principle that exon skipping is able to restore dystrophin expression in muscles of DMD patients.15,16 Treating DMD with antisense therapy requires high efficiency of exon skipping to achieve therapeutic effect. Factors known to be critical include efficacy of selected oligonucleotide sequence targeting specific exons, chemical nature of the antisense oligomers, and delivery efficiency. Here, we reported a new approach that has the potential to enhance exon skipping when used in conjunction with specific AONs. The 6TG demonstrated enhanced exon-skipping efficiency in a GFP-reporter cell culture system in vitro. More important, the combined use of PMO with guanine or 6TG showed significant enhancement of specific dystrophin exon skipping, leading to a twofold increase in dystrophin production in the treated muscles of mdx mice when compared to using PMO alone. This result provides evidence that small molecules could be used as adjuvant for enhancing specific exon skipping in experimental therapy.
High throughput screening has been widely employed to discover potential new applications of existing drugs or small compounds. In this study, we applied a GFP-based reporter system in C2C12 myoblasts to screen a drug library for enhancing exon skipping in the dystrophin gene. Expression of the reporter GFP is disrupted by the insertion of hDysE50 or mDysE23 and their adjacent intron sequences. The specificity of the vector for targeted hDysE50- and mDysE23-skipping was confirmed by the use of selected AONs as shown in the present study. Among the 2,000 compounds screened, only 6TG showed clearly enhanced GFP expression. However, the activity of exon skipping with 6TG alone was limited only to the GFP/hDysE50 reporter system. This is likely because the expression levels of reporter GFP are higher with the actin promoter in the C2C12E50 than with the muscle creatine kinase promoter in the C2C12E23. This together with the inability of the drug to induce detectable mDysE23-skipping in vivo suggests a very limited efficiency when 6TG and its analogues are used alone. However, when combined with low doses of specific PMOs, targeted exon skipping was significantly enhanced. Our results, therefore, demonstrate that the reporter system is most effective for identifying drugs that can enhance exon skipping induced by specific AONs rather than induce exon skipping on their own.
The mechanism by which 6TG and guanine alone induced exon skipping and enhanced PMO-induced exon skipping is not clear. The 6TG is an antimetabolite used in chemotherapy for cancer treatment. The compound is metabolized to deoxy-6-thioguanosine 5′-triphosphate (dthioGTP) via the purine salvage pathway initiated by hypoxanthine–guanine phosphoribosyltransferase and subsequently incorporated into DNA.27 Recent studies also suggest that 6TG alters the structure and stability of DNA duplex and inhibits quadruplex DNA formation.28,29,30,31 It is, therefore, possible that the incorporated 6TG in the proliferating C2C12 cells in culture could alter the structures of the dystrophin gene, leading to enhanced alternative splicing that would otherwise occur spontaneously at low levels. This may also explain the difficulty in achieving high levels of exon skipping in the C2C12E23 myoblasts cultured in differentiation media that significantly inhibit cell proliferation. This possible mechanism implies that the enhanced exon skipping is likely to be nonspecific to the dystrophin gene, consistent with its limited effect observed for specific skipping of mDysE23 and hDysE50. However, this is not supported by the fact that 6TG and guanine enhanced PMO-induced antisense effect in muscles in vivo, as myonuclei are not subjected to DNA turnover. The 6TG and guanine are small nucleobases; therefore, it is possible that they may interact with the bases of the PMO, leading to improved delivery efficiency, transportation from cytoplasm to nucleus, and interaction with the target sequences. Nevertheless, the low efficiency in inducing exon skipping by the drugs alone may be desirable for their therapeutic applications, which in turn implies that such drugs are unlikely to cause severe disruption of splicing in other genes, and hence, side-effects. Further studies are clearly required to elucidate the mechanism(s). In summary, our results suggest that small molecules could have profound effect on AONs-mediated alternative splicing and provide rationale for further large-scale screening to identify more potent drugs as exon-splicing modulators for experimental therapy.
Materials and Methods
Vector constructions and cell culture. The construction of the hDysE50/GFP-reporter vector was based on the procedure reported previously.19 The hDysE50 flanked by 600 bp of its respective intronic sequences (intron 49 on the 5′ side and intron 50 on the 3′ side) was amplified from DNA of human myoblasts. The sequence was inserted into the middle of the β-globin intron sequence that was placed inside the coding sequence of GFP gene under the control of actin promotor. For construction of the mDysE23/GFP-reporter vector, the entire mouse dystrophin intron 31 was placed inside the GFP-coding sequence under the control of a muscle creatine kinase promoter in the mammalian expression vector pEGFP-N3. The mDysE23, together with its two flanking intronic sequences (900 bp on each side), was amplified from mouse myoblasts and inserted into the mouse dystrophin intron 31 by an EcoRV restriction site. C2C12 mouse myoblasts were transfected with the vectors by electroporation, and the transfected cells were selected by geneticin (G418). The transfected cells were then maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, Carlsbad, CA) with 10% heat-inactivated fetal bovine serum (Gibco) and 1% penicillin/streptomycin at 37 °C with 5% CO2.
Myoblast culture, differentiation, and transfection. C2C12 cells were seeded in a 6-well plate at a density of 3 × 105 per well and cultured in DMEM supplemented with 10% fetal bovine serum over night. To induce differentiation, C2C12 cells were cultured in differentiation medium, which consisted of DMEM, 2% heat-inactivated horse serum, 2 µmol/l insulin, and 0.25-µmol/l dexamethasone, for 3 days. The cells were then treated with the small-molecule compounds with or without PMO for 72 hours in the differentiation medium. For plasmid transfection, C2C12 cells were electroporated with the linearized expression constructs and selected in DMEM supplemented with 1 mg/ml G418. The selected clones were maintained in DMEM supplemented with 200 µg/ml G418. For delivery of PMO to C2C12 cells, Endoporter (GeneTools, Philomath, OR) was added into the cultural medium according to the manufacture's instruction. PMOE23 sequence targeting mDysE23 has been described previously.17
Screening of 2,000 bioactive compounds. The screening of 2,000 bioactive compounds (Microsource Discovery System) was conducted with C2C12 hDysE50 reporter cells. In addition, 1 × 104 cells per well were seeded in the black Greiner F-bottom/chimney 96 well plates with clear bottom. The 2,000 compounds were loaded at a final concentration of 33 µmol/l. GFP expression was initially visualized under a fluorescence microscope 24 hours after treatment. C2C12 cells without hDysE50 reporter expression and hDysE50 reporter cells treated with 1% dimethyl sulfoxide were used as controls
Visualization of GFP by fluorescence microscopy and flow cytometric analysis. Cells were fixed with 4% paraformaldehyde, washed two times with phosphate-buffered saline, and visualized using an Olympus IX71 (Olympus, Center Valley, PA), an inverted, fluorescent microscope. Digital images were taken using the Olympus DP Controller and DP Manager software. Cells were also examined using flow cytometry to quantitatively gauge the GFP expression level. Cells were washed with phosphate-buffered saline (1×) and released from culture vessel with 0.05% trypsin-EDTA, neutralized by fetal bovine serum, pelleted by centrifugation, and then resuspended in 1 ml phosphate-buffered saline. Samples were run on a fluorescence-activated cell sorting Calibur flow cytometer (BD, Franklin Lakes, NJ), and 2 × 104 cells were counted and analyzed with CellQuest Pro (BD) software package.
RT-PCR analysis for cell culture. For RT-PCR analysis, cells were initially washed twice with phosphate-buffered saline, and RNA was extracted with TriZol reagent (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. RNA was stored at –80 °C for later use. RT-PCR was performed with RT-Fidelitaq MasterMix (USB, Cleveland, OH) to amplify the sequence of interest. Also, 100 ng of template RNA was used for each 25-µl RT-PCR. The primer sequences for the RT-PCR were eGFP5 (5′-CAGAATTCTGCCAATTGCTGAG-3′) and eGFP3′ (5′-TTCTTCAGCTTGTGTCATCC-3′). The conditions were 43 °C for 15 minutes, 94 °C for 2 minutes, then cycled 30 times at 94 °C for 30 seconds, 65 °C for 30 seconds, and 68 °C for 1 minute. The products were examined by electrophoresis on a 2% agarose gel.
In vivo delivery and RT-PCR. mdx mice aged 6–8 weeks were used with four samples for each experimental group. Experimental protocols were approved by the Institutional Animal Care and Use Committee, Carolinas Medical Center (Charlotte, NC). The PMOE23 (+07-18; 5′-GGCCAAACCTCGGCTTACCTGAAAT-3′) against the boundary sequences of exon and intron 23 of dystrophin gene (GeneTools) were used. For intramuscular injections, 2 µg PMOE23 with or without drugs was used in saline for each TA muscle. The muscles were examined 2 weeks later. Total RNA was extracted after dissection, and 100 ng of RNA template was used for a 50 µl RT-PCR with the Stratascript One-Tube RT-PCR System (Stratagene, Santa Clara, CA). The primer sequences for the RT-PCR were Ex20Fo, 5′ (CAGAATTCTGCCAATTGCTGAG-3′) and Ex26Ro (5′-TTCTTCAGCTTGTGTCATCC-3′) for amplification of mRNA from exons 20 to 26.
Antibodies and immunohistochemistry. Serial sections were cut from the treated TA muscles, together with control muscles. The sections were stained with a rabbit polyclonal antibody P7 for the detection of dystrophin protein as described previously.17 Polyclonal antibodies were detected by goat anti-rabbit Igs Alexa 594 (Invitrogen). Protein extraction and western blot were done as described previously.17 Briefly, the membrane was probed with NCL-DYS1 monoclonal antibody against dystrophin rod domain (Vector Laboratories, Burlingame, CA). The bound primary antibody was detected by HRP-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA) and the ECL Western Blotting Analysis System (Perkin-Elmer, Waltham, MA). The intensity of the bands with PCR-amplified products obtained from the treated mdx mice muscles was measured and compared with that from normal muscles of C57BL mice (ImageJ software; National Institutes of Health, Bethesda, MD), and α-actin was detected by rabbit antiactin antibody (Sigma, St Louis, MO) and used as a sample loading control.
SUPPLEMENTARY MATERIALFigure S1. Guanine analogue-induced exon skipping.Table S1. Summary of the first-round screening results with 2,000 bioactive compounds in C2C12E50 myoblasts.Table S2. List of guanine analogues tested.
Supplementary Material
Guanine analogue-induced exon skipping.
Summary of the first-round screening results with 2,000 bioactive compounds in C2C12E50 myoblasts.
List of guanine analogues tested.
Acknowledgments
This work was supported by Carolinas Medical Center (Charlotte, NC); US and Army Medical Research, Department of Defense (W81XWH-05-1-0616). We thank Yiumo Michael Chan, McColl–Lockwood Laboratory for muscular dystrophy research for critical reading, and Xiao Xiao, Division of Molecular Pharmaceutics, Eshelman School of Pharmacy, University of North Carolina (Chapel Hill, NC), for providing the MCK promoter for our expression vector.
REFERENCES
- Hoffman EP., and , Kunkel LM. Dystrophin abnormalities in Duchenne/Becker muscular dystrophy. Neuron. 1989;2:1019–1029. doi: 10.1016/0896-6273(89)90226-2. [DOI] [PubMed] [Google Scholar]
- Koenig M, Monaco AP., and , Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH., Jr, and , Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Michalak M., and , Opas M. Functions of dystrophin and dystrophin associated proteins. Curr Opin Neurol. 1997;10:436–442. doi: 10.1097/00019052-199710000-00014. [DOI] [PubMed] [Google Scholar]
- Straub V., and , Campbell KP. Muscular dystrophies and the dystrophin-glycoprotein complex. Curr Opin Neurol. 1997;10:168–175. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C., and , Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, et al. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C., and , Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007;15:1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- McClorey G, Moulton HM, Iversen PL, Fletcher S., and , Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- Gebski BL, Mann CJ, Fletcher S., and , Wilton SD. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet. 2003;12:1801–1811. doi: 10.1093/hmg/ddg196. [DOI] [PubMed] [Google Scholar]
- Bremmer-Bout M, Aartsma-Rus A, de Meijer EJ, Kaman WE, Janson AA, Vossen RH, et al. Targeted exon skipping in transgenic hDMD mice: A model for direct preclinical screening of human-specific antisense oligonucleotides. Mol Ther. 2004;10:232–240. doi: 10.1016/j.ymthe.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Dunckley MG, Manoharan M, Villiet P, Eperon IC., and , Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Moulton HM, Iversen PL, Jiang J, Li J, Li J, et al. Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer. Proc Natl Acad Sci USA. 2008;105:14814–14819. doi: 10.1073/pnas.0805676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lu Q., and , Wood M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in mdx mice. Mol Ther. 2008;16:38–45. doi: 10.1038/sj.mt.6300329. [DOI] [PubMed] [Google Scholar]
- Sazani P, Kang SH, Maier MA, Wei C, Dillman J, Summerton J, et al. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001;29:3965–3974. doi: 10.1093/nar/29.19.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP., and , Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F., and , Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Modrek B., and , Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Sammeth M, Foissac S., and , Guigó R. A general definition and nomenclature for alternative splicing events. PLoS Comput Biol. 2008;4:e1000147. doi: 10.1371/journal.pcbi.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK., and , Lutz GJ. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29:7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S, et al. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Li Y, Morcos PA, Doran TJ, Lu P., and , Lu QL. Octa-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx mice. Mol Ther. 2009;17:864–871. doi: 10.1038/mt.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEPAGE GA. Basic biochemical effects and mechanism of action of 6-thioguanine. Cancer Res. 1963;23:1202–1206. [PubMed] [Google Scholar]
- Somerville L, Krynetski EY, Krynetskaia NF, Beger RD, Zhang W, Marhefka CA, et al. Structure and dynamics of thioguanine-modified duplex DNA. J Biol Chem. 2003;278:1005–1011. doi: 10.1074/jbc.M204243200. [DOI] [PubMed] [Google Scholar]
- Bohon J., and , de los Santos CR. Structural effect of the anticancer agent 6-thioguanine on duplex DNA. Nucleic Acids Res. 2003;31:1331–1338. doi: 10.1093/nar/gkg203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohon J., and , de los Santos CR. Effect of 6-thioguanine on the stability of duplex DNA. Nucleic Acids Res. 2005;33:2880–2886. doi: 10.1093/nar/gki572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathias VM, Sawicki MJ., and , Bolton PH. 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res. 1999;27:2860–2867. doi: 10.1093/nar/27.14.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Guanine analogue-induced exon skipping.
Summary of the first-round screening results with 2,000 bioactive compounds in C2C12E50 myoblasts.
List of guanine analogues tested.