Abstract
The first use of lentiviral vectors in humans involved transduction of mature T-cells with an human immunodeficiency virus (HIV)–derived env antisense (envAS) vector to protect cells from HIV infection. In that study, only a minority of the patient T-cell population could be gene-modified, raising the question of whether the altered cells could affect replicating HIV populations. We investigated this using humanized NOD/SCID IL-2Rγnull (hNSG) mice reconstituted with ~4–11% envAS-modified human T-cells. Mice were challenged with HIV-1NL4-3, which has an env perfectly complementary to envAS, or with HIV-1BaL, which has a divergent env. No differences were seen in viral titer between mice that received envAS-modified cells and control mice that did not. Using 454/Roche pyrosequencing, we analyzed the mutational spectrum in HIV populations in serum—from 33 mice we recovered 84,074 total reads comprising 31,290 unique sequence variants. We found enrichment of A-to-G transitions and deletions in envAS-treated mice, paralleling a previous tissue culture study where most target cells contained envAS, even though minority of cells were envAS-modified here. Unexpectedly, this enrichment was only detected after the challenge with HIV-1BaL, where the viral genome would form an imperfect duplex with envAS, and not HIV-1NL4-3, where a perfectly matched duplex would form.
Introduction
Highly active antiretroviral therapy fails to eliminate human immunodeficiency virus (HIV) completely from patients and often elicits drug-resistant variants, leading to interest in additional forms of therapy. Adoptive T-cell therapy using gene-modified T-cells is one such approach, in which T-cells are harvested from HIV-infected subjects, transduced ex vivo with genes that obstruct HIV replication, then reinfused back into patients. One type of anti-HIV gene encodes antisense RNA, which in several studies has been shown to inhibit HIV replication efficiently in tissue culture.1,2,3,4
In a phase I clinical trial preceding our study, an HIV-derived lentiviral vector encoding env antisense (envAS), called VRX496, was used to treat patients who failed two or more antiretroviral regimens.5 The trial was a success in that no adverse events were reported in this first-in-human use of lentiviral vectors.5,6 There was also significant reduction in viral loads and improvement of immune function in a subset of patients, even though only a fraction of autologous T-cells were modified with VRX496. This then raised the question of whether envAS modification of a minority of cells could influence HIV infection.
In this study, we have investigated the effect of envAS on HIV-1 populations in a setting where only a minority of cells were vector-modified. The vector VRX494 used here is the research counterpart of the clinically used VRX496, the only difference being that VRX494 has a green fluorescent protein (GFP) gene that allows convenient quantification of transduction (Figure 1a). Similar to the clinical vector, VRX494 is derived from HIV and directs envAS expression from the HIV long terminal repeat, so that following HIV infection of a vector-modified cell and expression of HIV tat, the antisense payload is expressed. In addition, cis signals are present in the vector to allow mobilization by HIV proteins provided in trans during infection, so that the vector can potentially spread from cell to cell. In a computational simulation, formation of such defective interfering particles, combined with antisense inhibition, inhibited HIV replication effectively.7
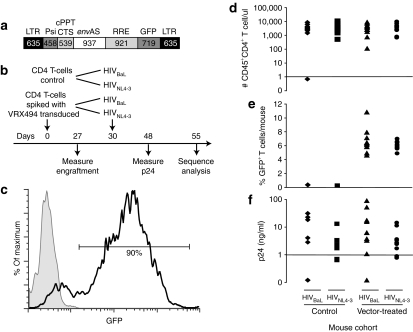
Figure 1.
Analysis of hNSG mice with ~4–11% VRX494 envAS-vector modified T-cells and challenged with HIVNL4-3 or HIVBaL. (a) The VRX494 vector. (b) Time line of mouse transplantation and infection. The hNSG mice were transplanted on day 0, then challenged with HIVNL4-3 or HIVBaL on day 30. (c) Flow cytometry analysis of transduction of CD4+ T-cells with envAS. Cells with high transduction levels were obtained as evidenced by GFP detection. (d) Analysis of the levels of T-cell engraftment. (e) Analysis of the levels of engraftment of VRX494-transduced cells, analyzed by sorting GFP+ cells. (f) Analysis of the level of HIV p24 antigen in serum at 48 days after infection. cPPT, central polypurine tract; CTS, central termination sequence; envAS, env antisense; GFP, green fluorescent protein; HIV, human immunodeficiency virus; hNSG, humanized NOD/SCID IL-2Rγnull; LTR, long terminal repeat; RRE, rev-responsive element.
The antisense RNA encoded by VRX494 can bind the HIV RNA genome, forming double-stranded RNA. In previous work, HIV infection of VRX494-modified cells in culture, followed by recovery of challenge virus sequences, showed (i) large deletions in most HIV genomes and (ii) a high frequency of A–G transitions in the envAS-targeted region, potentially the result of A–I editing by cellular double-stranded RNA adenosine deaminase (dsRAD; a member of the Adenosine Deaminase Acting on RNA, or ADAR, family of enzymes).4 Such modifications may help the virus evade antisense pressure by reducing the complementarity of the viral genome and the envAS. However, most of these deleted genomes would be replication defective, and a few A-to-G changes would be unlikely to significantly destabilize pairing with the ~900-bp antisense RNA used. Thus, it may be more likely that the accumulation of these sequences reports the action of cellular systems acting on the double-stranded sense–antisense RNA, but that these mutations do not confer reduced sensitivity to the antisense.4
Here, we report an analysis of HIV-1 populations following growth in the presence of envAS using a humanized NOD/SCID IL-2Rγnull (or hNSG) mouse model. Cohorts of mice were compared that were reconstituted with ~4–11% vector-modified T-cells or with unmodified T-cells to model the frequency of vector marking in the human phase I trial. We found no reduction in viral load, but we did detect a significant excess of challenge virus variants with enriched A-to-G transitions and deletions, documenting antisense pressure on HIV-1 populations even with VRX494-modification of only a minority of cells. Unexpectedly, the variants with enriched A–G transitions and deletions only accumulated significantly when the antisense was imperfectly matched to its target.
Results
A mouse model for envAS pressure on HIV-1
Previous studies measuring the effect of HIV-1 antisense therapy have used T-cell populations in which the vast majority of viral target cells expressed the antisense RNA. However, in the therapeutic setting, although most of the cells that are infused express the antisense message, they are quickly diluted and persist as only a small percentage of the total.5 To study how T-cells transduced with the VRX494 envAS vector (Figure 1a) might alter viral populations in patients, we established an in vivo model that would mirror the published clinical trial,5 but allow infection with defined HIV challenge stocks. We took advantage of the ability of NOD/SCID IL-2Rγnull (NSG) mice to stably engraft human T-cells8 to model HIV-1 infection and therapy. We established four cohorts of 10 mice each as diagrammed in Figure 1b. Two control cohorts were engrafted with untransduced T-cells and challenged with either of two HIV-1 derivatives, HIVNL4-3 or an HIVNL4-3 derivative engineered to express the BaL envelope (henceforth “HIVBaL”). The other two cohorts were engrafted with untransduced T-cells spiked with ~4–11% of VRX494-transduced T-cells and challenged similarly (termed “vector-treated cohort” below).
The VRX494-transduced T-cells used in this study were generated in a manner that replicated the clinical trial. Freshly isolated primary human CD4+ T-cells were activated, transduced with a lentiviral vector expressing envAS and GFP (VRX494), expanded for 10 days, frozen, and stored in liquid nitrogen for several weeks. After thawing, GFP expression was measured by flow cytometry and these cells were found to be highly transduced (Figure 1c), as with the cells used for treating patients in the clinical trial.5 Mice in the control cohorts were injected with 10 million untransduced T-cells, whereas mice in the vector-treated cohorts were injected with 9 million untransduced T-cells and 1 million of VRX494 transduced cells (thus the target frequency of transduced cells was 10%).
After 27 days, we measured the engraftment for each of the 40 animals. One animal failed to engraft T-cells and was excluded from further analysis. For the remaining animals, similar levels of engraftment of total CD4+ T-cells were seen for all cohorts (Figure 1d). The two cohorts that received VRX494-transduced cells showed similar distributions for the proportion of engrafted cells transduced with GFP+ VRX494 (4–11%, Figure 1e).
Three days later (day 30 postinfusion of T-cells) we challenged each mouse with supernatants collected from 293 T-cells transfected with HIVBaL or HIVNL4-3. We note that this differs from the clinical trial in that the human subjects were HIV-infected before infusion of gene-modified cells, whereas here gene-modified cells were introduced into mice before HIV infection. After an additional 18 days (day 48 postinfusion), we measured viral replication by assaying p24 in the mouse serum (Figure 1f and Supplementary Table S1a). We did not observe a significant difference in the viral titers between the vector-treated and control cohorts. This is consistent with the fact that it took ≥6 months to see substantial differences in the viral load of patients treated with the clinical envAS vector (VRX496). After an additional 7 days (day 55 postinfusion), some of the mice (across all four cohorts) began to display the early stages of graft versus host disease and the experiment was terminated. Serum obtained from the peripheral blood at the time of killing was used for all subsequent analysis. Two mice died during the study, and four mice had undetectable viral loads by serum p24 assay, so a total of 33 mice were available for analysis.
Amplification and sequencing of viral quasi-species from mouse plasma
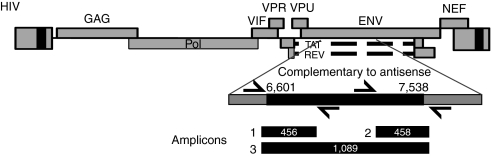
To investigate possible pressure on viral populations exerted by envAS, we studied the structure of viral populations using the 454/Roche pyrosequencing technology.9 HIV RNA was extracted from serum of each mouse, reverse transcribed, and PCR amplified according to the scheme in Figure 2. Three amplicons were designed covering the envAS-targeted region of the challenge viruses—two covering each side of the envAS target region, and a third spanning the two. The outermost primers annealed slightly outside each edge of the envAS-targeted region. The smaller two amplicons permit detection of A–G transitions and smaller deletions. The longer amplicon, at 1,037 bases in length, is too long for use in the 454/Roche sequencing procedure, which accommodates molecules of a maximum of ~500 bases in length. Thus, the longer amplicon could only yield sequences with large deletions.
Figure 2.
The HIVNL4-3 genome, showing the regions targeted by the VRX494 envAS, and the HIV env amplicons used in this study. The numbering in the env gene refers to the HIVNL4-3 genome. The envAS-targeted region extends from 6,601–7,538 (see blow-up). Three amplicons were designed to recover potential deletions and A→G changes. The relative positions and length of the amplicons (including 454 adapters and barcodes) are shown. Three amplicons were similarly designed in the homologous region of the HIVBaL genome. envAS, env antisense; HIV, human immunodeficiency virus.
RNA from each serum sample was separately amplified and products gel-purified for each of the three amplicons. Gel regions corresponding both to full-length and shorter sequences were isolated to avoid selecting against potential deletions. To allow determination of sequences from many samples in pools, unique 8-nucleotide error-correcting barcodes10 were incorporated into each primer to index PCR products from each mouse.6,11,12,13 Following pyrosequencing, sequence reads were assigned to the source mouse by decoding the barcode.
We analyzed a total of 84,074 sequences from 33 mice across three amplicons. Reads were filtered to require perfect matches to the 5′ barcode and env primer,14 yielding 79,040 (or 94% of the total) sequences. Based on the distribution of read lengths recovered, additional filtering was performed to remove sequences with <220 bases, as short reads are reported to be error prone.15 A total of 68,642 (or 82% of the total) sequences remained after filtering. Finally, the sequence pools from each mouse were de-replicated and curated to yield a nonredundant set of 31,290 unique sequences. The distribution of these across the four study groups is summarized in Supplementary Tables S1a and S1b. Subsequent analysis was carried out using the unique viral variants (rather than also considering their frequencies of isolation). Analysis involved comparison of vector-treated and control animals, so that the comparative framework minimized the influence of error arising due to mutagenesis in the PCR procedure or in pyrosequence read determination.
Analysis of nucleotide changes in VRX494-transduced mice
HIV sequences were then analyzed for enrichment for A–G transitions and deletions, which were previously found to be associated with replication in the presence of the envAS.4 Each sequence was aligned to the reference HIV genome using pair-wise global alignment. A stringent gap opening penalty was selected to allow detection of deletions of up to ~900 bases. Use of a stringent gap opening penalty also allowed detection of multiple nearby mismatches, as can occur if envAS elicits clustered mutations. Two parsed multiple sequence alignments (MSAs), one for HIVBaL and one for HIVNL4-3, were constructed from the individual pair-wise global alignments.
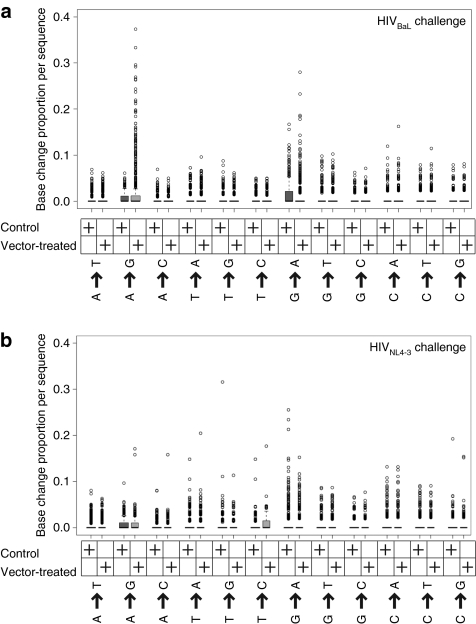
To investigate A–G transition rates, we trimmed off primer sequences, then calculated the proportion of nucleotide positions that changed from A-to-G for each sequence, yielding a value between 0 and 1. As controls, all other base-change proportions per sequence were also calculated. For each base-change, the distribution of proportions for the vector-treated and control groups were plotted (Figure 3a,b). The boxes in the figure indicate the middle two quartiles (interquartile range; in most cases the intervals are so close that the box appears to be a line at 0.0). Outliers, defined as sequences with base-change proportions beyond 1.5 times the interquartile range from the box edges, are plotted as open black circles.
Figure 3.
Box plots illustrating the types of base substitutions that accumulated during growth of HIV-1 in hNSG mice. (a) Base substitutions that accumulated during growth of HIVBaL in the vector-treated and control mice. (b) Base substitutions that accumulated during growth of HIVNL4-3 in the vector-treated and control mice. The boxes comprise all sequences with proportion changes in the middle two quartiles. Outliers, defined as sequences beyond 1.5 times the interquartile range, are plotted as open black circles. HIV, human immunodeficiency virus; hNSG, humanized NOD/SCID IL-2Rγnull.
As can be seen from the plots in Figure 3a, following challenge with HIVBaL, there were substantially more sequences with high proportions of A–G transitions in the vector-treated group compared to the control group. Multiple sequences in the vector-treated group had A–G change proportions of 0.1 or more (i.e., at least 10% of A changed to G), although the control group had none. No such effect was seen for any other base-change.
In contrast, we found that the A–G effect was not pronounced for HIVNL4-3 challenge (Figure 3b). The envAS was derived from the NL4-3 strain, so it is perfectly complementary to HIVNL4-3 but not to HIVBaL—thus one conjecture is that mismatches in the RNA/RNA hybrid altered the processing of the duplex. We return to this point in the discussion.
We also found that viral sequences enriched in G–A transitions were present at a relatively high frequency following both HIVNL4-3 and HIVBaL infections, though independently of whether or not envAS was present (Figure 3a,b). High rates of G–A transitions are well documented during HIV infection, and are due to reverse transcriptase errors16 and the C–T deamination activity of APOBEC family enzymes.17,18,19
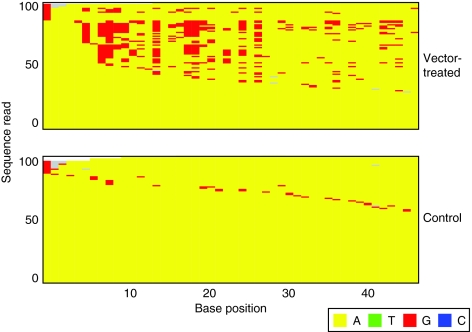
We next investigated whether the enrichment for A–G transitions in vector-treated samples was statistically significant. We defined informative sequences as those that had at least one A–G transition. We then calculated the probability of an A–G transition over all informative sequences from both vector-treated and control groups, because according to our null hypothesis there is no difference between the two groups. Using the A–G transition probability, we assigned a binomial P value to each informative sequence based on the number of A–G events as a fraction of the total A positions examined. Thus, we could define an A–G enrichment score for each informative sequence as the negative logarithm (−log10) of their P values. The lower the P value of a sequence, the less likely the observed frequency of A–G transitions occurring by chance, and the higher the A–G enrichment score. Sequences with statistically significant enrichment levels have P values of ≤0.05 (A–G enrichment score ≥1.301). Figure 4 depicts a representative portion of the MSA, showing only A positions in the region and comparing the most A–G enriched sequences for vector-treated and control samples following HIVBaL challenge.
Figure 4.
Comparison of the 100 sequences with the greatest enrichment of A–G transitions from the VRX494-treated and control mice challenged with HIVBaL. The most enriched sequences from viruses grown in vector-treated mice (top) are compared to the most enriched sequences from controls (bottom). The bases were color-coded as indicated at the bottom of the figure. Only base positions that were A (in yellow) in the starting viral stock are shown, those substituted with G are shown in red. Grey indicates sequence gaps.
As an initial statistical scan of the data, we performed a one-sided Fisher's exact test to determine whether there was a significant excess of sequences enriched in A–G transitions in the vector-treated versus the control group (pooled over all mice). For HIVBaL challenge, we obtained a highly significant trend that persisted at least up to A–G enrichment scores as high as 4 (data not shown). This confirms our observations (Figures 3a and 4) that there are sequences with significantly higher proportions of A–G transitions primarily in the HIVBaL vector-treated group. No strongly significant trend was observed when we repeated our analysis for other base-changes. A weak excess of sequences with G–A changes was seen, but this did not survive the more stringent test described below.
For the HIVNL4-3 challenge, the A–G effect was weak and showed no statistical significance above an enrichment score of 1.3 (data not shown). This confirms that in this case there is no preferential occurrence of sequences with high rates of A–G changes in the vector-treated group. No other types of base-change showed significant enrichment following HIVNL4-3 challenge.
As a control for the A–G effect, we examined the frequency of A-to-G transitions outside the region targeted by envAS. There are 20 A sites present in the regions between the edges of the PCR primers used for sequence isolation and the env region complementary to envAS. These were compared to the 243 A sites in the env region targeted by envAS and analyzed in depth using the shorter two amplicons. Only 1 of 20 control sites (5%) was enriched in A-to-G transitions in the vector-treated compared to the control group (as determined by Fisher's exact test with Type I error of 0.05). In contrast, 70 of the 243 envAS-targeted sites (28%) were enriched in the vector-treated group relative to the control. The difference achieved significance (P = 0.01) by one-sided Fisher's exact test. Thus, we conclude that the region of the HIVBaL genome complementary to envAS had an elevated vector-induced frequency of A-to-G changes compared to flanking nontargeted regions of the viral genome.
Sequence features at A-to-G transitions
We investigated whether A-to-G changes were associated with any nearby sequence motifs in the HIV RNA. We aligned sites of A-to-G transitions at the affected base, and used WebLogo (http://weblogo.berkeley.edu/) to scan for conserved sequence features. No strongly conserved motifs were detected, though a weak preference for a 5′ A or T residue was seen (data not shown).
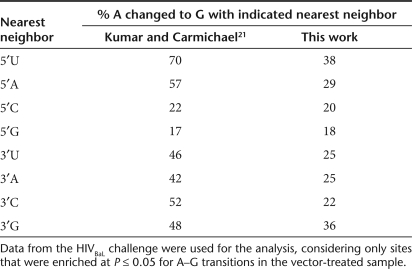
We then analyzed the proportions of bases changed as a function of nearest neighbors and found a bias (Table 1). Previous studies have suggested that dsRAD acts on A residues with 5′ neighbors in order of preference U and A greater than C and G.20,21 We found a relatively higher proportion of modified A residues 3′ of an U or A compared to 3′ of a G or C (Table 1). This suggests that dsRAD is responsible for modifying A to I in our experiments, leading to substitution with G. However, in the published study no preference for 3′ neighbors was found, whereas for unknown reasons we found a preference for G residues.
Table 1.
Proportions of A residues converted to G with the indicated 5′ and 3′ nearest neighbor nucleotides
We further asked whether there was a relation between the frequency of A-to-G substitutions and the extent of complementarity of the target genome with envAS. For this, we classified sites in the HIVBaL template into those that were complementary to the corresponding antisense position and those that were not. Likewise, we classified the A sites in HIVBaL as ones that underwent significant transition to G or not. We then compared the extent of sequence complementarity and fraction of sites with A-to-G transitions (using a sliding window of 25 bases). These were positively correlated with a highly significant P value (P < 0.0001) for Spearman's rank correlation coefficient. This is consistent with earlier reports of preferential RNA editing by dsRAD within perfectly matched regions of partially mismatched templates.
Comparing sequences enriched in A–G transitions among the groups of mice
A more rigorous statistical approach involves treating each mouse as a single measurement instead of analyzing all sequences pooled over a cohort of mice. For the analysis over pooled sequences described above, a single anomalous mouse might contribute sequences that dominated the behavior of the pool, particularly in a study of rare sequence variants, whereas trends reproducible over many mice are of greater interest.
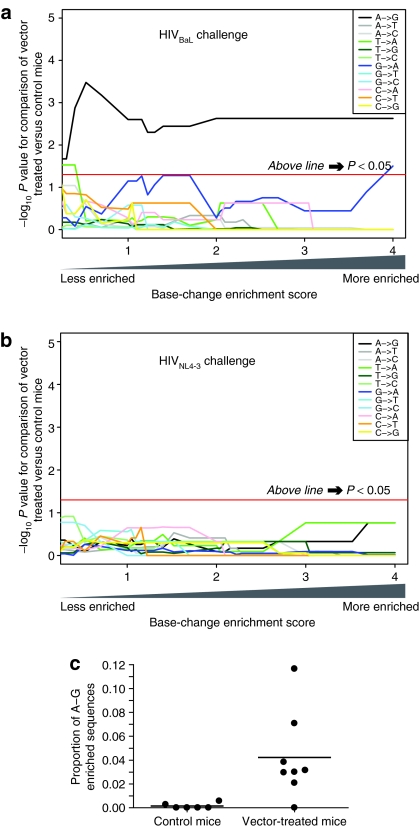
Figure 5a shows a comparison of the enrichment scores for base substitutions in the sequence reads, comparing vector-treated and control mice after challenge with the HIVBaL virus (one-sided Mann–Whitney comparison of means). The x axis shows the enrichment score, and the y axis shows the P value for the mean sequence excess comparing the vector-treated mice to control mice at the indicated enrichment score. For A–G substitutions (Figure 5a, black line), there is a significant excess of enriched sequences (above the horizontal red line) regardless of whether only high levels of enrichment (right side) or both low and high levels (left side) are used in the statistical analysis.
Figure 5.
Statistical analysis of base substitution frequencies in vector-treated and control mice. For the statistical analysis, each mouse was treated as an individual measure of proportions. (a) Comparison of vector-treated and control mice after HIVBaL challenge. (b) Comparison of vector-treated and control mice after HIVNL4-3 challenge. In each panel, the x axis indicates the extent of enrichment per sequence for each base substitution used in the analysis, so that at any indicated enrichment score, only sequences with at least that score were considered. Progressing from left to right indicates analysis of increasingly high levels of substitution. The y axis indicates the –log10 P value from the Mann–Whitney nonparametric comparison of means (one-sided) for the excess in vector-treated cohort compared to control cohort. The analysis was carried out for each of the 12 base substitutions. The horizontal red line indicates the threshold for achieving statistical significance at P values ≤0.05 (above is significant). (c) Scatter plot showing the proportions of sequences with enrichment scores >1.3 (P values <0.05) of A–G transitions for each mouse in the HIVBaL group. HIV, human immunodeficiency virus.
The remaining 11 transitions and transversions were also compared. None showed a consistently significant trend for sequences enriched for substitutions in the vector-treated cohort. Only T–A and G–A substitutions achieved a low level of significance at specific enrichment scores but were nonsignificant at most x values and are of questionable biological importance.
The analysis was repeated for the experiment with the HIVNL4-3 challenge virus (Figure 5b). None of the 12 substitutions achieved significance. Thus, the low level of significance seen for A-to-G transitions when all HIVNL4-3 sequences were pooled was not confirmed in the more rigorous analysis in which each mouse was treated as a single measurement.
A representation of the excess of sequences with A–G enrichment among vector-treated mice in the HIVBaL challenge group is depicted in Figure 5c, using the enrichment level of P < 0.05 (score of >1.301) as an example. Most of the control mice had few or no sequences passing this A–G enrichment threshold, whereas the vector-treated mice had an average of ~4.5% passing the threshold (P = 0.005).
Frequency of deletions after challenge of the vector-modified cells
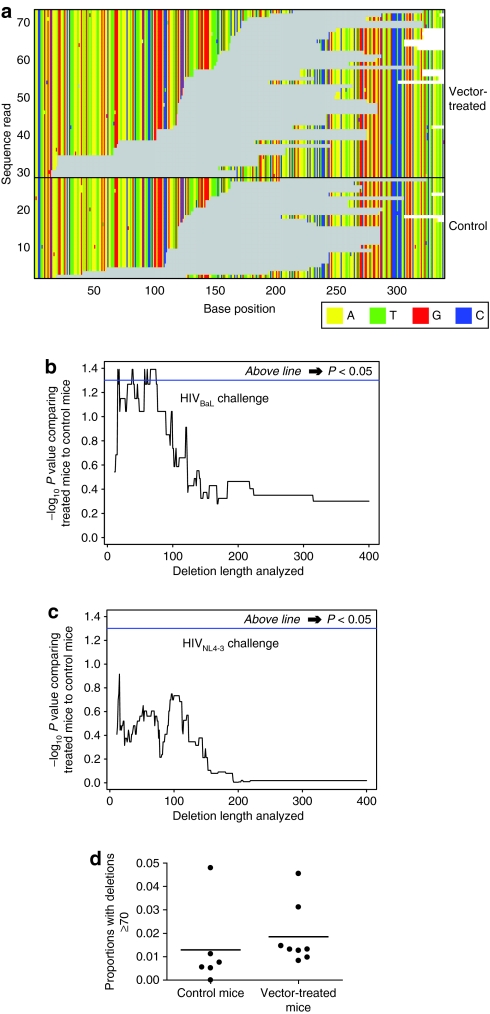
Studies of antisense inhibition of HIV in tissue culture, where most cells contained the VRX494 vector, indicated that deletions also accumulated,4 so we analyzed the frequencies of deletions here. The distribution of sequences with deletions of at least 70 bases is shown in Figure 6a for one of the HIVBaL amplicons. Inspections suggested that the number of sequences with deletions is greater in the vector-treated mice than in the controls.
Figure 6.
Frequency of deletions in HIV-1 challenge viruses grown in the presence of vector-treated cells or controls. (a) Illustration of the numbers and locations of deletions in HIVBaL from the vector-treated and control groups corresponding to the 5′-end of amplicon 2. Gray indicates sequence gaps. Deletions of ≥70 bases were plotted. (b) Analysis of the significance of the difference in deletion frequencies between vector-treated and control mice after HIVBaL challenge. The x axis shows the length of deletions included in the analysis, so that at any indicated value only deletions of that length or greater were included in the analysis. The y axis shows the P value for the comparison of means between vector-treated and control groups calculated using the nonparametric Mann–Whitney test (one-sided). Each mouse was treated as a single measurement of proportions. (c) As in b, but analysis of the HIVNL4-3 challenge group. (d) Comparison of proportions for deletions ≥70 bases, for the control and vector-treated mice. Each mouse is shown as a point.
We thus calculated the significance, again treating each mouse as a single measure, of the proportion of deleted viruses (Figure 6b,c). The x axis indicates the length of deletion used in the analysis—progressing to the right indicates restricting the analysis to incrementally longer deletions. The y axis shows the statistical significance of testing for the excess of sequences in vector-treated mice versus controls at the indicated deletion length. Segments of the curve above the horizontal blue line indicate statistical significance. For HIVBaL challenge, some data sets with deletions in the range of ~10–100 bases showed significant enrichment in the vector-treated group, though others did not, indicating that the enrichment of deletions achieved marginal significance (Figure 6b). No such significant enrichment in deletions was seen with HIVNL4-3 challenge (Figure 6c). The enrichment in deletions for HIVBaL sequences, using deletions of ≥70 bases for analysis, is shown in Figure 6d, where each mouse is plotted individually. As can be seen, the difference in means is significant (P = 0.04), but the effect slight. We conclude that there is a significant though modest increase in the frequency of deletions in the HIVBaL but not the HIVNL4-3 challenged vector-treated mice.
We next checked whether variants with deletions ≥70 bases were also enriched for A–G transitions, as both features potentially arise from envAS pressure. We performed Fisher's exact test to establish whether there was an excess of deletion-harboring variants among the population of sequences significantly enriched for A–G. We did not observe any significant enrichment or depletion. Thus, the A–G transitions and deletions appear to accumulate independently.
Correlation between the extent of T-cell modification and the effects of VRX494 envAS treatment
Each cohort of mice showed variation in the final extent of T-cell modification by the VRX494 vector following engraftment, allowing us to investigate possible correlations with the frequency of A-to-G transitions or deletions. No significant correlation was observed between modification frequencies and enrichment of sequences with high frequencies of A–G transitions (for Spearman's rank correlation coefficient). However, a modest but significant correlation (P = 0.04) was seen for vector modification (measured at day 48) and deletion frequency for HIVBaL challenged mice. Significance was achieved when deletions of about ≥70 bp were studied, which we know are enriched in vector-treated mice, but not with shorter lengths (data not shown). We thus conclude that a possible modest positive correlation could be seen where increased vector modification was associated with increased deletion frequency.
Discussion
Here, we report modeling the effects of an HIV envAS vector on HIV challenge virus under conditions where cells harboring the envAS construct were a minor component of the cell population. This setting is of interest because studies of cells in culture, where all cells contain the envAS, show strong inhibition of HIV replication by the envAS,4 but trials to date in human subjects have achieved vector marking in only a minority of circulating T-cells. Despite this, some patients showed intriguing alterations in disease parameters.5 We thus sought to study in detail the effects on HIV populations when only a minority of cells contained the envAS using hNSG mice. We found a significant enrichment compared to controls for sequences enriched for A–G transitions and deletions in mice harboring the envAS in the experimental infections with the HIVBaL challenge virus.
However, unexpectedly, we also found that A–G transitions were only enriched in samples from the HIVBaL infections, and not HIVNL4-3 infections. The origin of the difference is unknown. According to one idea, the different co-receptor usage of the two challenge viruses might have resulted in the infection of two different cell types possibly resulting in different treatment of double-stranded RNA, but at present we have no evidence in favor of this view. A more attractive model is that double-stranded RNA is processed differently depending on whether the duplexes were perfectly matched or contained mismatches. The envAS was derived from HIVNL4-3, so RNA duplexes formed with HIVNL4-3 RNA would form perfectly paired duplexes, although those with HIVBaL would contain a mixture of duplex and mismatched regions. Favored action at duplex regions near mismatches, and the observed RNA nearest neighbor preferences, are consistent with action of the dsRAD enzyme.20,21 However, dsRAD is also able to act on perfectly matched duplexes, so it is unclear why no A-to-G transitions accumulated in the HIVNL4-3 infection.
The difference in processing could be a consequence of differential initial attack, in which only RNA duplexes containing mismatches are substrates for the duplex-processing enzyme. However, another possibility is that the perfect duplexes are attacked but degraded more quickly, so that no HIV sequences with perfect matches survive to allow detection even by deep sequencing. We presently have no basis for favoring either model.
The origins of the deletions in the HIVBaL challenge virus seen here and in a previous study4 are unclear. Not only must the RNA duplex be cleaved, it must be rejoined to form the internal deletion detected by sequencing. One candidate mechanism involves cleavage by a double-stranded specific ribonuclease (RNAse III) such as Drosha, followed by copy-choice polymerization by reverse transcriptase, so that template switching from a broken RNA template to a site elsewhere on a second HIV RNA would yield the observed internal deletion. We did not observe enrichment for deletions following HIVNL4-3 challenge, so one model would hold that the RNAse III involved acts preferentially on mismatched dsRNA. However, we note that the same results would be obtained if products of cleavage of perfectly matched RNA were rapidly degraded. No positive correlation was seen between the deletions and A–G transitions, indicating that the enzyme systems responsible for RNA cleavage and RNA editing are likely acting independently.
It is surprising that modification of only 4–11% of cells in the mice could have caused detectable alterations in the HIVBaL population, and similarly surprising to observe possible alterations in disease parameters in human subjects after gene therapy with a similar envAS vector.5 However, it is possible that mobilization of the VRX494 envAS vector augmented the effect. The VRX494 vector DNA contains intact long terminal repeats, the cis sites needed for RNA packaging, reverse transcription, and integration, and also sites needed for response to Tat and Rev. Thus, if a cell harboring the integrated VRX494 provirus were infected by HIV, the VRX494 provirus could be transcribed, then the RNA exported from the nucleus, packaged, and released. Infection into a naive cell, followed by reverse transcription and integration, would achieve spread of the VRX494 envAS vector between cells. Such conditional replication of the vector could expand the proportion of vector-containing cells and increase the likelihood of accumulating the observed mutations in the challenge viruses.
Are the A-to-G transitions and deletions viral escape mutations? This is an important question and the answer is not fully clarified here, but at present it seems unlikely that they confer escape. The large deletions in HIV envelope are sure to be env-minus, and a few A-to-G transitions in an ~900-bp duplex are not likely to destabilize RNA pairing significantly. More likely the mutations that accumulate in HIVBaL are by-products of the action of cellular RNA-modifying enzymes that inhibit viral replication. The modified RNAs with deletions could persist in the population by complementation during co-infection with wild-type viruses, as they were found in viral particles in serum. They may also be continually arising de novo in the infected cell populations during ongoing viral replication, or else complementation may be efficient enough to maintain modified RNAs in the population for multiple generations.
The experimental protocol used here differed from the clinical trial in the order of gene modification versus HIV infection. In the clinical trial, gene-modified cells were reinfused into patients with preexisting HIV infection. In our hNSG mouse model, the cells were first gene-modified and mice were infected afterward. The composition of preexisting viral populations can affect disease course,22 and this may be modified by prior treatment history. Thus, it would be of potential interest to compare effects of infection first followed by envAS modification in the hNSG model.
To summarize, we observe significant effects of envAS pressure in vivo when only 4–11% of HIV-1 target CD4+ T-cells are gene-modified. The effects were only detectable with divergent env target sequence, suggesting action of dsRNA-modifying enzymes acting on imperfectly matched sequences. As envAS has been shown to control HIV-1 replication and has also been deemed safe in gene therapeutic clinical trails, this represents a promising antiviral agent.
Materials and Methods
Transduction and culture of primary human CD4+ T-cells. All human-related studies were done per Declaration of Helsinki Protocols. Primary human CD4+ T-cells were obtained from the University of Pennsylvania CFAR Immunology Core under an institutional review board approved protocol and stimulated with anti-CD3 and anti-CD28 antibody–coated beads as previously described.23 The following day the activated T-cells were either left alone (untransduced), or transduced with VRX494 (ref. 24), expanded and frozen.
Infection of hNSG mice. NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ stock) were obtained from the Jackson Laboratory (Bar Harbor, ME) and bred and housed in the University of Pennsylvania Xenograft and Stem Core Lab. Before injection, each expanded cell population was thawed and transduced cells were mixed with untransduced T-cells so that the final concentration of transduced T-cells was ~10%. The mixed cells were washed in phosphate-buffered saline and 10 million cells were injected intravenously into each mouse. A total of 20 animals received cells containing 10% transduced cells with VRX494, whereas 20 animals received only the untransduced cells. The HIV-1-based vector VRX494 used in this study encodes an envAS complementary to the HIVNL4-3 env gene. After 20 days, the number and percentage of GFP+, CD4+ T-cells within the NSG mouse peripheral blood was measured using TruCount beads as per the manufacturer's recommendation (BD Biosciences, San Jose, CA). One mouse in the control group showed poor engraftment and was excluded from further studies. For both control and VRX494-treated groups, the engraftment data (and additionally the engrafted proportions of VRX494-modified cells for the treated group) were used to randomize mice between the HIVNL4-3 and HIVBaL challenge cohorts. Each group of mice was then challenged with the supernatants of 293 T-cells transfected with molecular clones of HIV-1 expressing either the HIVNL4-3 or HIVBaL env sequence [pNL4-3, a gift of VIRxSYS (Gaithersburg, MD) and pWT/BaL, obtained from the NIH AIDS Research & Reference Reagent Program (Germantown, MD), respectively]. HIV-1 infection of reconstituted mice was performed at Bioqual (Rockville, MD) under an approved animal protocol.
Amplification and deep sequencing of HIV quasi-species. Total RNA was extracted from mouse plasma samples using the Illustra RNAspin kit (GE Healthcare, Buckinghamshire, UK) with RNA carrier added to improve extraction efficiency. Composite primers (Supplementary Table S2) made of 454 sequencing adapters, barcodes, and HIVenv primers, 5′–3′ in that order, were used to amplify HIV-1 RNA by One-step reverse transcriptase-PCR (Qiagen, Valencia, CA) using RNasin RNase inhibitor (Promega, Madison, WI) and a touch-down protocol (with a total of 30 cycles of amplification) as follows:
1× (30 minutes at 50 °C), 1× (15 minutes at 95 °C), 10× (1 minute at 94 °C; 1 minute at 58–53 °C, 0.5 °C iterative decrement; 1 minute at 72 °C) (starting with a temperature of 58 °C and reducing it successively by 0.5 °C to reach 53 °C), 20× (1 minute at 94 °C; 1 minute at 53 °C; 1 minute and 15 seconds at 72 °C), 1× (10 minutes at 72 °C; maintained at 4 °C). Amplified products were gel-purified separately for each mouse-amplicon combination and pooled following DNA quantitation by Quant-iT PicoGreen dsDNA Assay kit (Molecular Probes, Invitrogen, Eugene, OR). Pooled sequences were pyrosequenced using the 454/Roche platform at the University of Pennsylvania. A total of 84,074 raw reads were recovered. These have been deposited with National Center for Biotechnology Information's Sequence Read Archive (SRA) under submission accession SRA010586.3. The raw reads have accessions SRR033739.1–SRR033739.84074.
Bioinformatics. Pyrosequencing reads were barcode-decoded for assignment to source mouse samples. Sequence reads were filtered for exact match to primers. To remove sequences shorter than the read length distribution peak, that are known to have high 454 error rates, additional filtering was carried out to select only sequences ≥220 bases long. Reference genomes corresponding to the HIVNL4-3 and HIVBaL infection stocks were obtained from National Center for Biotechnology Information's nucleotide database (http://www.ncbi.nlm.nih.gov/nuccore/), accession IDs M19921 (version: M19921.1, GI:328415) and M63929 (version: M63929.1, GI:326765). Viral infection stocks were Sanger-sequenced and verified that they matched the respective National Center for Biotechnology Information sequences for the genomic region analyzed. Sequences were then aligned to the reference HIV-1 genome using the Needleman–Wunsch pair-wise global alignment algorithm with a match score of 5, and gap opening and extension penalties of 20 and 0.5. A few sequences from HIVNL4-3 challenge group had a higher alignment score to the HIVBaL reference and vice-versa; these were classified as apparent sequence crossovers and were excluded from further analysis. For remaining sequences in HIVNL4-3 and HIVBaL group, two MSAs were created, one for each group, by parsing the individual pair-wise global alignments within each group. Finally, primer sequences were trimmed from each sequence because these are not part of the viral genome actually sampled.
All statistical programming and tests were performed in the R computing framework (http://www.r-project.org/). One-sided statistical tests were used for analysis of the A–G substitutions and deletions because a previous report established a directional hypothesis (increased frequency after treatment).4
A–G analysis: For each MSA, base positions having >50% change from the reference sequence base over all other sequences (discounting MSA gaps) were excluded, by reasoning that the preferred base reported by pyrosequencing could likely be the correct base at these positions. For the HIVBaL MSA, of the 994 positions sampled by our amplification scheme, 22 positions were excluded, of which 17 were in regions of high coverage (>1,000 reads per position) corresponding to the shorter two amplicons. For HIVNL4-3, all 40 positions excluded (out of 991) were in the low coverage region between the shorter two amplicons (<30 reads per position). For each base-change, overall probability of the event was obtained from the pool of all informative sequences (see text). Using this, a binomial distribution based P value was assigned per sequence based on the observed proportion of base-change in a sequence. This P value was termed the enrichment level and the –log10 of this was called the enrichment score (see text). For each base-change, excess within the vector-treated cohort, of sequences with a minimum enrichment was statistically inferred. For the pooled analysis, i.e., taking vector-treated group as a whole, this was done by Fisher's exact test. For un-pooled analysis, Mann–Whitney test was done (see text). In each case, statistical significance was tested with enrichment scores up to 4 (P value 0.0001) and displayed as –log10 of the P value (Figure 5a,b).
Investigating sequence features for A–G modifications:
1. Conserved motifs—To research sequence motifs on the HIV-1 genome associated with A–G transitions, for informative sequences, sites with enriched changes in the vector-treated group were chosen. Sites were selected at an enrichment corresponding to P value ≤0.05 by Fisher's exact test. Reference genome positions 10 bases upstream and downstream of each site were aligned and WebLogo was used for information content analysis to detect conserved motifs.
2. Neighbor base rules—All instances of XA and AX dinucleotides in the envAS target region were tabulated, X ε {A, T, G, C}. Then for each X, in both cases of X being 5′ or 3′ to A, the proportion of instances the A was a statistically preferred site for transitioning to G (as determined in (1) with Fisher's exact test) was calculated.
3. Genomic distribution in relation to complementarity—The fraction of sites in the HIVBaL template that were complementary to envAS was calculated across the envAS-targeted region with a sliding window of 25 bases. Similarly, of all A sites the fraction of A sites statistically preferred for transitioning to G (as determined in (1) with Fisher's exact test) was calculated. The correlation between the two quantities was estimated by Spearman's rank correlation coefficient.
Deletion analysis: Small deletions can arise by chance among viral quasi-species or during sample work up, so meaningful deletions were qualified as only those which had a length >10 bases. Additionally, deletions were required to have a ≥10-base long gapless aligned region at both ends. With this constraint, the hope was to select deletions that are biological while weeding out those that could be generated as alignment artifacts through gapped matching of a few error-prone bases. Two deletion-related quantities were defined for each sequence: (i) Total deletion length, representing the cumulative length of all qualifying deletions, and (ii) maximum deletion length, representing the length of the longest qualifying deletion. Then the spectrum of total deletion lengths that can be assayed by our study ranges from 11 to ~900 bases, the upper limit being imposed by the longest amplicon. For each possible deletion length in this range, sequences with cumulative deletions of at least that size were selected and tested for excess in vector-treated mice (un-pooled analysis). Statistical significance was inferred by Mann–Whitney test and displayed as –log10 of the P value (Figure 6b,c). Repeating analysis with maximum deletion length did not alter results significantly.
SUPPLEMENTARY MATERIALTable S1. Samples studied and numbers of pyrosequence reads.Table S2. Sequences of oligonucleotides and barcodes used in this study.
Supplementary Material
Samples studied and numbers of pyrosequence reads.
Sequences of oligonucleotides and barcodes used in this study.
Acknowledgments
We are grateful to members of the Bushman and Riley laboratories for help and suggestions, Carl June (Department of Pathology and Laboratory Medicine, University of Pennsylvania) and Gwen Binder (Translational Research Program University of Pennsylvania) for assistance in establishing this collaboration, VIRxSYS (Gaithersburg, MD) for providing the lentiviral vector used in this study, Jake Yalley-Ogunro, Gary Thomas, and Mark Lewis (all from Bioqual, Rockville, MD) for manipulating the HIV-1-infected mice, and Gwenn Danet-Desnoyers and Anthony Secreto (both from the Stem Cell and Xenograft Core, University of Pennsylvania) for maintaining the animals before HIV-1 infection. This work was supported by NIH grants RO1AI0802020, U19AI082628, U19AI066290, Penn Genome Frontiers Institute, and a grant with the Pennsylvania Department of Health. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. We do not have any conflicts of interest to declare.
REFERENCES
- Reyes-Darias JA, Sánchez-Luque FJ., and , Berzal-Herranz A. Inhibition of HIV-1 replication by RNA-based strategies. Curr HIV Res. 2008;6:500–514. doi: 10.2174/157016208786501454. [DOI] [PubMed] [Google Scholar]
- Rossi JJ, June CH., and , Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer L, Rossi JJ., and , Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- Lu X, Yu Q, Binder GK, Chen Z, Slepushkina T, Rossi J, et al. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. J Virol. 2004;78:7079–7088. doi: 10.1128/JVI.78.13.7079-7088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine BL, Humeau LM, Boyer J, MacGregor RR, Rebello T, Lu X, et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc Natl Acad Sci USA. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G, et al. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther. 2009;17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Schaffer DV., and , Arkin AP. Theoretical design of a gene therapy to prevent AIDS but not human immunodeficiency virus type 1 infection. J Virol. 2003;77:10028–10036. doi: 10.1128/JVI.77.18.10028-10036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chain(null) mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ., and , Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE. 2007;2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Minkah N, Leipzig J, Wang G, Arens MQ, Tebas P, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM., and , Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML., and , Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky LM., and , Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YN, Malim MH., and , Bieniasz PD. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol. 2008;82:8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD., and , Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Polson AG., and , Bass BL. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., and , Carmichael GG. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro RM., and , Bonhoeffer S. Production of resistant HIV mutants during antiretroviral therapy. Proc Natl Acad Sci USA. 2000;97:7681–7686. doi: 10.1073/pnas.97.14.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JL, Schlienger K, Blair PJ, Carreno B, Craighead N, Kim D, et al. Modulation of susceptibility to HIV-1 infection by the cytotoxic T lymphocyte antigen 4 costimulatory molecule. J Exp Med. 2000;191:1987–1997. doi: 10.1084/jem.191.11.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, et al. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol Ther. 2004;9:902–913. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples studied and numbers of pyrosequence reads.
Sequences of oligonucleotides and barcodes used in this study.