Influenza A virus is an RNA virus that encodes up to eleven proteins and this small coding capacity demands that the virus utilize the host cellular machinery for many aspects of its life cycle1. Knowledge of these host cell requirements not only informs us of the molecular pathways exploited by the virus but also provides additional targets that could be pursued for antiviral drug development. Here, we employ an integrative systems approach, based upon genome-wide RNAi screening, to identify 295 cellular cofactors required for early-stage influenza virus replication. Within this group those involved in kinase-regulated signaling, ubiquitination and phosphatase activity are the most highly enriched and 181 factors assemble into a highly significant host-pathogen interaction network. Moreover, 219 of the 295 factors were confirmed to be required for efficient wild-type influenza virus growth and further analysis of a subset of genes revealed 23 factors necessary for viral entry, including members of the vacuolar ATPase (vATPase) and COPI-protein families, fibroblast growth factor receptor (FGFR) proteins, and glycogen synthase kinase 3 (GSK3)-beta. Additionally, 10 proteins were confirmed to be involved in post-entry steps of influenza virus replication. These include nuclear import components, proteases, and the calcium/calmodulin-dependent protein kinase (CaM kinase) II beta (CAMK2B). Importantly, growth of swine-origin H1N1 influenza virus is also dependent on the identified host factors and we show that small molecule inhibitors of several factors, including vATPase and CAMK2B, antagonize influenza virus replication.
Influenza viruses are a major cause of morbidity and mortality, and influenza A viruses in particular have the propensity to cause pandemic outbreaks such as occurred in 1918, 1957, 1968 and currently in 2009 with the swine-origin H1N1 influenza virus2. Two of the viral proteins, neuraminidase (NA) and the M2 ion channel protein are the targets for the FDA-approved influenza antiviral drugs; oseltamivir, zanamivir, amantadine and rimantadine 3. Unfortunately, there is now widespread resistance to both of these drug classes 4. Combined with the limited number of viral drug targets for influenza virus, this creates concern for the development of new influenza therapies.
An alternative therapeutic strategy that may greatly reduce the emergence of viral resistance is the pharmacological targeting of host factors required for viral replication. Genome-wide RNAi screens have enabled the identification of host factors required by a number of RNA viruses 5–7 8 9, 10 11, including an insect cell-based RNAi screen which implicated 110 Drosophila genes in influenza virus replication 12. In an effort to more comprehensively characterize the host machinery utilized by influenza virus in mammalian cells, we have performed a genome-wide siRNA screen with human lung epithelial (A549) cells. To facilitate the readout for the high-throughput screen, the coding region for the influenza A/WSN/33 virus hemagglutinin (HA) protein was replaced with that of Renilla luciferase (Figure 1a)13. As no HA is produced, this recombinant virus cannot complete its replication cycle. Thus our RNAi screen focuses on the cellular requirements for viral entry, uncoating, nuclear import, and viral RNA transcription/translation, but is not expected to identify factors involved in virus assembly, budding or release.
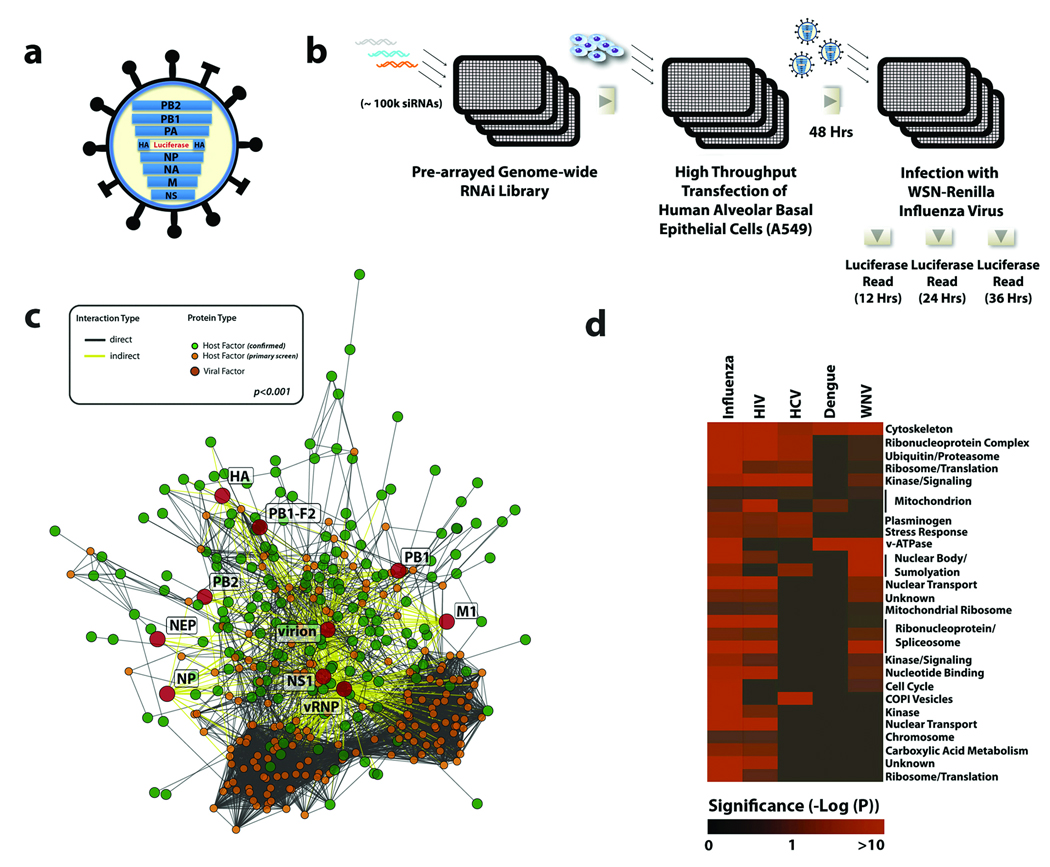
Figure 1. A Genome-wide RNAi Screen for Influenza Virus Host Cellular Factors.
(a) A schematic of the recombinant WSN-Ren virus showing the HA segment modified to express Renilla luciferase but maintaining the HA packaging sequences. (b) An arrayed genome-wide RNAi library (100,000 siRNAs targeting over 19,000 human genes) was transfected into A549 cells. Cells were subsequently infected with WSN-Ren and virus replication was monitored by measuring luciferase activities at the indicated times. (c) A highly significant (p<0.001 based on permutation test) host-pathogen interaction map for influenza virus containing 4,266 interactions between 181 confirmed influenza virus-host cellular factors (green circles), 10 influenza virus-encoded proteins or complexes (red circles), and an additional 184 cellular proteins (orange circles). (d) Predicted complexes from the human interactome used by influenza virus, HIV, HCV, dengue virus, and West Nile virus (based upon results reported here and previous screens5–8, 10–12, 28). Intensity of red colour indicates the significance of enrichment (based on hypergeometric p-values) for proteins required by a virus within a complex.
An arrayed siRNA library targeting over 19,000 human genes was employed to transfect human A549 cells (Figure 1b and Supplementary Information). These cells were infected with the modified influenza virus (WSN-Ren), and luciferase readings were taken after 12, 24, and 36h. Data from two independent screens were analyzed using an integrative data analysis approach, which included Redundant siRNA Activity (RSA), as well as interactome and ontology-based analyses, (see Supplementary Information)6, 14. Using these methodologies, we were able to confirm 295 cellular genes for which at least 2 siRNAs reduced viral infection by 35% or greater (~2 standard deviations from mean of negative controls), without a concomitant induction of significant cellular toxicity (Supplementary Figure S1 and Supplementary Table S1). While some of these factors were previously known to be involved in influenza virus replication (confirming the robustness of our RNAi approach), the majority of the factors identified through this analysis represent host genes that have not previously been implicated in mediating influenza virus replication.
Analysis of over-represented biological annotations identified over 170 statistically enriched categories (Supplementary Table S2), which fell into 11 broadly related functional groups (Supplementary Figure S2, Supplementary Table S3). Signaling molecules, including those involved in the PI3K/AKT pathway, molecules that function to regulate cytoskeletal dynamics, and proteins involved in ubiquitination, phosphatase, and protease activities were overrepresented amongst the 295 factors, underscoring the importance of these cellular functions during influenza virus infection (Table 1, see also Supplementary Table S4 and S5). Consistent with these observations, we found that small molecule inhibition of two identified AKT pathway regulators, mTOR (FRAP1) and HSP90AA1, as well as microtubule assembly (TUBB), resulted in a dose-dependent inhibition of influenza virus replication (Supplementary Figure S3)15, 16.
Table 1.
Selected functional categories in early steps of influenza virus replication
| Functional Category |
Gene Names a | Cellular function | Proposed/known role in influenza virus replication b |
Replication block |
|---|---|---|---|---|
| IP3-PKC pathway | ROCK1, CDC42BPA, KSR2, ARAF, PRKCI, CDC42BPB, CIT, AKAP13, RACGAP1, EIF2AK2, CDK4, ACVR2A, MAPK1, PRKCD, MAP2K2, GRK5, ARAF, HIPK1, PAK3, MAP2K3, PTPMT1, LIMK1, PIP5K1C, PAK2, GRK6, SGK1 |
Signaling | Viral entry, promoting apoptosis; inhibitors of PKC block viral RNA replication and PKC activators enhance virus replication |
Entry (MAP2K3) |
| COPI vesicles | ARCN1, COPA, COPB2, COPG, USE1 | early endosome maturation, retrograde golgi to ER transport |
entry (endosomal trafficking) | Entry (ARCN1) |
|
Endosomal uptake, maturation, acidification and fusion |
ATP6V1A, ATP6V1B2, ATP6V0B, ATP6V0C, ATP6AP1, ATP6V0D1, ATP6AP2, RABEP1, PIP5K1C, VPS16, TRPV2, MARCH2, EPHB2 |
vATPase complex: acidification of intracellular organelles, including endosomes |
Low pH-dependent entry, ubiquitin/vacuolar protein sorting pathway required for entry |
Entry (ATP6V1A, ATP6V1B2, ATP6V0B, ATP6V0C, ATP6AP1) |
|
Actin organization and function |
GAK, APC2, CIT, PAK2, CDC42BPB, PAK3, CDC42BPA, SGCA, FSCN1, OXSR1, CD81, FGFR2, FGFR4, ITGA3, AKAP13, ACTC1, FGFR1, LIMK1, ROCK1, PIK3R4 |
Actin organization and function |
Intracellular transport, Particle movement at cell periphery prior to virus fusion; Actin also implicated in NP functions such as transcription, replication, and genome- trafficking |
Entry (CD81, FGFR2 FGFR4, ITGA3, AKAP13) |
|
PI3K–AKT pathway |
AKT1, BCL3, FRAP1, GSK3B, HRAS, HSP90AA1, IKBKE, ITGA3, JAK2, MAP2K2, MAPK1, MDM2, PIK3R4, PIK3R4 |
Signaling | important for viral protein yields and nuclear export of vRNPs. Involved in inhibition of apoptosis. |
Entry (GSK3B) |
|
Endosomal recycling pathway |
RAB11B, RAB17 | Vesicle trafficking | entry (endosomal trafficking) | Entry (RAB11B) |
| MAPK pathway | MAP2K3, DUSP3, MAP3K12, MAPK1/ERK, MAP2K2/MEK, ARAF, CAD, CREB1, EPHB2, FGFR4, HRAS, JUN, NTRK2, PAK2, PRKACA, PRKCD, MAP2K5, MAP4K4, HIPK3, ATP6AP2, STK39, MINK1, PBK, TAOK1, ZNF436, CANT1, KSR2 |
Signaling | important for viral protein yields and nuclear export of vRNPs. |
Entry (MAP2K3, DUSP3) |
| Proteases | CTSW, PCSK7, KLK9, ANPEP, PRSS35 | Post-translational processing |
HA cleavage | Post-entry (PRSS35) |
|
Calcium/ Calmodulin Proteins |
CAMK2B, PRKACA, DAPK2, ADRA1B, CREB1, PRKAG2, DCLK2, GRK5, GRK6, KCNJ3, PKD1, PRKCD, STX5, CACGN4, AGTRAP |
Calcium regulation and signaling |
Transcriptional regulation | Post-entry (CAMK2B) |
|
Nuclear trafficking |
CSE1L, KPNB1, NUP214, NUP153, TNPO3 |
Nuclear trafficking | Nuclear import of vRNA or viral/host proteins | Post-entry (KPNB1, CSE1L) |
| Trafficking | STX10, STX5, GOPC, CLL1, NRBP1 | Membrane and Receptor trafficking |
Glycoprotein trafficking | ND |
| Sumoylation | SUMO2, SUMO1, SAE1, SUMO4 | Post-translational modification |
unknown | ND |
|
Microtubule organization and function |
MID1IP1, TUBB, PRKCI, PLK4, MARK2, DCLK1, NUDCD3, RACGAP1, MAP1LC3C |
Cytoskeletal organization |
Microtubules implicated in intracellular viral transport. |
ND |
| Autophagy | PRKAG2, MAP1LC3C, FRAP1, HRAS | Stress response | post-entry, blocks viral protein production | ND |
| Ubiquitination | MDM2, UBQLN4, HECTD1, CBLL1, DTX2, EPS8L3, FBXO44 |
Post-translational modification |
unknown | ND |
References are contained in Supplemental Table 5. ND = No Data
To understand the network of host-pathogen interactions that govern the early steps of influenza virus replication, we employed several protein interaction datasets to construct a host-pathogen interaction map depicting associations between the identified host factors, viral-encoded proteins, and other cellular proteins (Figure 1c, Supplementary Figures S4 and S5). The integration of RNAi and interactome datasets produced a network containing 181 confirmed host cellular factors that mediate 4,266 interactions between viral or cellular proteins, and act as central rate-limiting “hubs” in cellular pathways or processes required for influenza virus replication (Supplementary Figure S6, Supplementary Table S6). Although the coverage and quality of currently available protein interaction databases remains difficult to assess17, the influenza interaction map was found to be highly significant (p<0.001), indicating that this network topology is not randomly derived, and likely reflects a unique cellular sub-network.
Of the 295 identified host factors required for influenza virus replication, 53 were previously identified in RNAi screens for different RNA viruses (Supplementary Figure S7, Supplementary Table S7), including 9 mammalian orthologues of host proteins required for influenza virus infection of Drosophila cells 12. It is currently not clear if this statistically significant (p=3.1 × 10−9), but modest, overlap reflects false-negative activities in the current or the aforementioned screen, or the differential host cell requirements between insect and mammalian cells for influenza virus replication. However, functional classification and protein interaction analysis of these shared factors revealed that, collectively, these viruses rely on common host cellular mechanisms to promote discrete stages of their life cycles (Figure 1d, Supplementary Figure S8; Supplementary Table S8).
To verify that the genes identified through the use of the reporter virus reflect the requirements in the context of a wild-type (WT) virus infection, 219 of 295 identified genes were confirmed to inhibit multi-cycle replication of WT WSN virus with at least two siRNAs per gene. Furthermore, 76% of the remaining genes had one siRNA that inhibited WT influenza replication, indicating a high confirmation rate (Figure 2a, Supplementary Table S9). For a subset of these genes additional assays were undertaken to confirm that depletion of these genes resulted in reduced viral gene expression (Figure 2a, Supplementary Table S9), and also to ensure that inhibition of viral replication was not being triggered by a non-specific siRNA-mediated induction of an antiviral state (Supplementary Table S10).
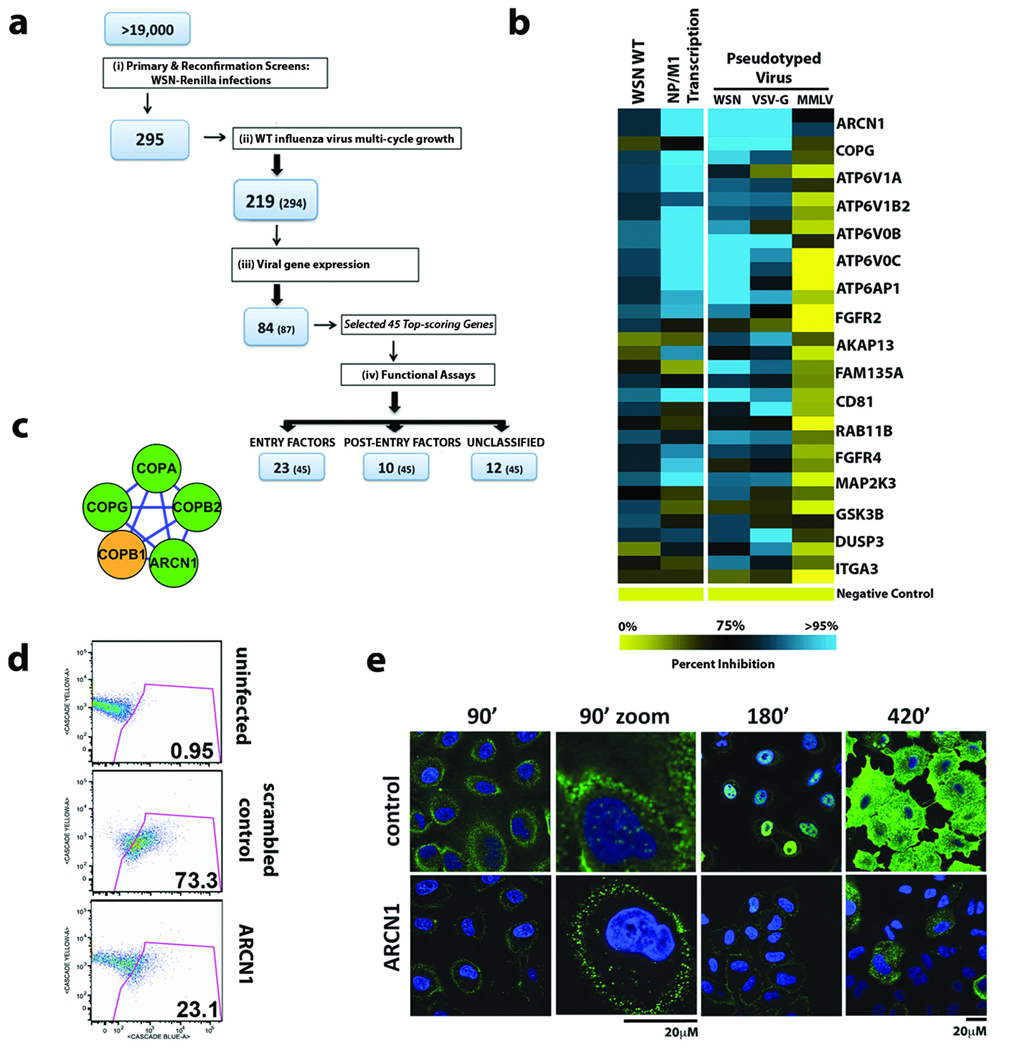
Figure 2. Identification of Host Factors Involved in Influenza Virus Entry.
(a) Illustration of the screen progression from primary genome-wide analysis to the identification of factors involved in entry and post-entry steps in the virus life cycle. The number of confirmed genes and number of genes tested at each stage (in parentheses) are indicated. (b) The relative effects of gene depletion (2 siRNAs/gene) on infection of luciferase-encoding HIV particles pseudotyped with WSN, VSV or MMLV envelopes (right panel). Effects of RNAi upon wild-type WSN virus replication and transcription of viral NP and M1 genes are also shown (left panel). Inhibition in each assay is shown as a continuum of blue (high inhibition) to yellow (low inhibition). (c) The endosomal coat protein complex (COPI) was identified as one of seventeen biochemical modules (MCODE clusters) likely to be important for influenza virus entry (see Figure 1c for legend; also Supplementary Figure S5)29. (d) Infection of siRNA-transfected A549 cells with influenza virus VLPs carrying a beta-lactamase (Bla-M1) fusion protein. The percentages of cells containing detectable cytoplasmic beta-lactamase activity are indicated. (e) Cells depleted of ARCN1 and infected with wild-type WSN virus were fixed and stained for NP (green) and nuclei (blue) at the indicated times and analyzed by confocal microscopy. The enlarged images at 90min post-infection indicate the lack of incoming RNP complexes in the nucleus in cells depleted of ARCN1.
Next, to identify potential factors specifically involved in virus entry steps, 45 of the top-scoring genes in the WT WSN assay were selected to be tested in a pseudotyped particle (PP) entry assay, designed to identify host factors that impede low-pH-dependent entry mediated specifically by influenza virus HA (WSN) and vesicular stomatitis virus (VSV)-G protein, while not affecting pH-independent entry promoted by the murine leukemia virus (MMLV) envelope (Env) 18, 19. WSN-PP infection was reduced in the presence of siRNAs targeting 23 of these genes, including CD81, FGFR4, GSK3B, MAP2K3 and the v-ATPase subunit ATP6V0C (Figure 2a, 2b, Supplementary Table S11, Supplementary Figure S9). These genes were also required for efficient VSV-G-PP (but not MMLV-PP) infection, suggesting a role in low-pH-dependent virus entry. Importantly, small molecule inhibitors of FGFR4, GSK3B, and v-ATPase activities attenuated replication of WSN virus, further highlighting their importance in influenza virus infection (Supplementary Figures S3 and S10).
The COPI coat complex is made up of seven subunits, four of which (ARCN1, COPA, COPB2, COPG) were among the confirmed factors in the protein interaction network (Figures 1c and 2c). COPI association with endosomes is pH-dependent and coatomer complex is required for the formation of intermediate transport vesicles between the early and late endosomes 20, 21. Consistent with this role, depletion of COPG and ARCN1 both blocked WSN-PP infection (Figure 2b). The requirement for ARCN1 during the influenza virus entry step was further demonstrated using a more direct virus-like particle (VLP) assay (Figure 2d)22, as well as immunolocalization studies (Figure 2e).
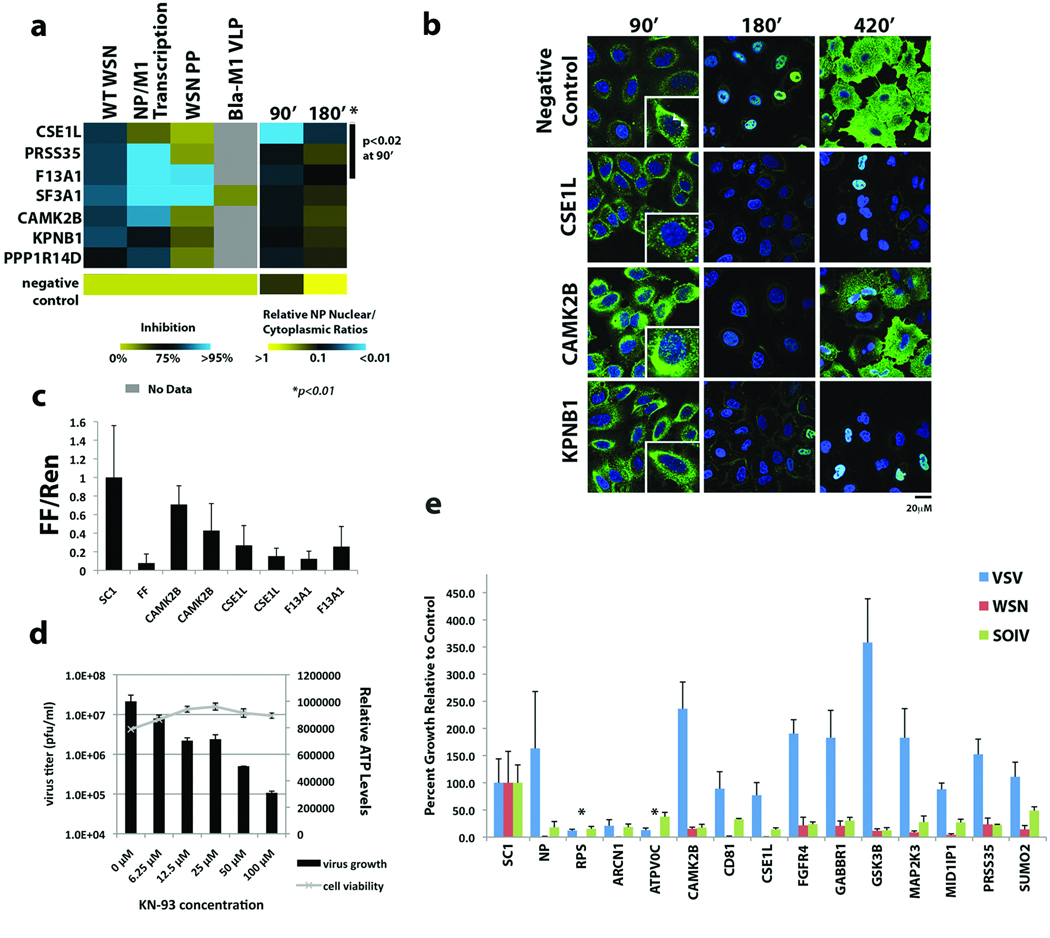
To evaluate those factors that affect virus replication but not influenza virus entry, we monitored the localization of the influenza virus nucleoprotein (NP) in siRNA-depleted cells after infection with influenza A/WSN/33 virus (Figure 3a; Supplementary Figure S11, see Supplementary Information). In comparison to controls, cells depleted of CSE1L, PRSS35, F13A1, SF3A1, CAMK2B, KPNB1, and PPP1R14D show a significant decrease (p<0.01) of nuclear to cytoplasmic ratios of NP protein at 180 min. With the exception of F13A1, depleting these factors did not inhibit entry by WSN pseudotyped virus or β-lactamase (Bla)-M1 VLPs (Figure 3a), confirming their role in post-entry steps of influenza virus infection. Depletion of CSE1L, PRSS35, and F13A1 also led to a statistically significant (p<0.02) reduction of nuclear to cytoplasmic NP ratios at 90 minutes post-infection, suggesting that they may play specific roles in early post-entry steps, such as viral uncoating or nuclear import of viral ribonucleoproteins (vRNPs; see also Supplementary Figure S12). Consistent with a role in nuclear trafficking, imaging at higher resolution confirmed that RNAi-mediated inhibition of CSE1L, but not CAMK2B or KPNB1, results in a decrease in nuclear vRNPs typically seen 90 min after infection with influenza virus (Figure 3b)23. Furthermore, CSE1L specifically inhibited influenza virus gene expression in a mini-genome replicon assay, suggesting that CSE1L activity is required for the nuclear import of vRNPs as well as newly synthesized viral proteins (Figure 3c, Supplementary Table S12).
Figure 3. Characterization of Factors in Post-Entry Replication Events and Conserved Requirement by Different Influenza Viruses.
(a) The impact of host factor depletion on the nuclear localization of viral NP protein at 90 and 180 minutes after A/WSN/33 virus infection is shown (right panel). Significant effects (p<0.01 based on Welch T-test) are seen at 180min with all genes and with CSE1L, PRSS35, F13A1 (p<0.02) at 90min. Levels of virus replication (WT WSN), viral gene (NP/M1) transcription and entry of WSN pseudotyped particles or Bla-M1 VLPs in cells lacking these factors are shown in the left panel. Values relative to negative controls (bottom row) are depicted in a continuum of blue (>95% inhibition) to yellow (little or no inhibition). (b) Confocal imaging of influenza virus NP protein localization at the indicated times following A/WSN/33 virus infection in cells depleted of CSE1L, CAMK2B and KPNB1. Arrows in the 90’ inset indicate nuclear RNPs. (c) The effects of host factor depletion on replication of an influenza virus mini-genome firefly reporter. The normalized fold reduction of firefly luciferase for each gene is shown relative to the scrambled siRNA control (SC1) +/− standard deviation. All reductions are significant (p<0.05) by Student’s T-test. (FF=firefly luciferase siRNA). (d) KN-93, a selective inhibitor of CAMK2B, inhibits A/WSN/33 influenza viral replication in a dose-dependent manner in MDCK cells, without affecting cell viability (ATP levels). Mean titers +/− standard deviation of triplicate samples are shown. (e) A549 cells were transfected with siRNAs targeting the indicated genes and subsequently infected with influenza A/WSN/33 virus, swine-origin influenza A/Netherlands/602/2009 (H1N1) virus (SOIV) or VSV. Virus growth is shown as the average percent relative to the scrambled siRNA control (SC1) +/− standard deviation. *below level of detection (1 × 104 pfu/ml). NP=siRNA for influenza A virus NP, RPS=siRNA for RPS27A.
Calcium/calmodulin-dependent protein kinase (CaM kinase) II beta (CAMK2B) is a ubiquitously expressed calcium sensor that regulates diverse cellular functions, including actin cytoskeletal regulation and CREB-dependent transcription 24. Our data implicate this kinase in the regulation of viral RNA transcription as RNAi-knockdown of the kinase had a moderate effect on expression of an influenza mini-genome (Figure 3c), but did not delay nuclear accumulation of vRNPs at 90 min post-infection (Figure 3b). We also show that a specific inhibitor of CAMK2B, KN-93, inhibits influenza virus growth (Figure 3d, Supplementary Figure S13), suggesting that pharmacological targeting of this kinase may be an effective strategy for the development of host factor-directed antivirals25.
Finally, we assessed the requirements for twelve identified host cellular factors in the replication of a swine-origin influenza virus (SOIV) isolate from the 2009 pandemic (A/Netherlands/602/2009 (H1N1)) in comparison with influenza A/WSN/33 virus and VSV. Viral growth in siRNA-treated A549 cells revealed that these proteins are all required for both SOIV and WSN replication but none of these factors, with the exception of the vATPase and COPI factors, inhibited VSV replication (Figure 3e, Supplementary Table S13, Supplementary Figure S14). These results indicate that factors identified here are likely important for the replication of multiple influenza virus strains.
This genome-wide analysis of influenza virus host factor requirements described here has revealed a large number of cellular proteins and biological pathways previously unknown to be involved in the influenza virus life-cycle. These include the identification of COPI complex, FGFR, GSK3B, CAMK2B, PRSS35, and others. Since this study focused on host factors that regulate the early steps of influenza virus replication, additional analyses will likely help to elucidate the full complement of cellular proteins required during the complete replication cycle. Further understanding of the roles for these proteins in influenza virus infection will provide new insight into the host-pathogen interactions that orchestrate the viral replication cycle and novel opportunities for the development of host factor-directed antiviral therapies.
Methods
Renilla luciferase influenza virus
The coding region for the viral hemagglutinin (HA) protein was replaced with that of Renilla luciferase and the packaging signals for the HA segment were incorporated, as previously described 13. The recombinant WSN-Ren virus was generated by reverse genetics in the presence of complementing HA and amplified in HA-expressing MDCK cells 13.
Genome-wide RNAi screen
Genome-wide libraries comprising 98,737 synthetic siRNAs targeting 19,628 unique human genes were arrayed in 384-well plates (7ng/siRNA) such that each well contained either two (47,560 wells) or one (3617 wells) unique and identifiable siRNA per gene. Although use of low screening concentrations of RNAi may help to minimize off-target activities, this may also contribute to false-negative activities. The library matrix was introduced into A549 cells through a high throughput transfection process 26, 27 and after 48h the cells were infected with WSN-Ren virus at a multiplicity of infection (MOI) of 0.5. EnduRen Live Cell substrate (Promega) was added after 5 hours and relative luminescence for each well was analyzed on a plate reader (Viewlux) at 12h, 24h and 36h after infection. For the toxicity screen Cell-titer-glo (Promega) reagent was added 72h after siRNA transfection. The screens were run minimally in duplicate and analyzed using a scaling methodology that sets the positive control siRNA at an arbitrary value of 0.1, and the negative control siRNAs at 1. siRNAs targeting host factors were assigned a score based on the distribution of these values. For more details see Supplementary Information.
Inhibition of virus growth
siRNA-transfected A549 cells were infected with either influenza A/WSN/33 virus or VSV (MOI of 0.01) or swine origin influenza A/Netherlands/602/2009 virus (SOIV) (MOI of 1) at 48h post siRNA transfection. At 36h post infection supernatants were harvested and virus titers were determined by plaque assay on MDCK cells (for A/WSN/33 and A/Netherlands/602/2009) or on Vero cells for VSV.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by National Institutes of Health grants U01 AI1074539, U54 AI057158 (Northeast Biodefense Center), HHSN272200900032C, HHSN266200700010C, 1 PO1 AI058113 and 1R21AI083673. S.S. is supported by a fellowship from the German Research Foundation and D.M.T is supported by NIH fellowship 1F32AI081428. M.B.O is supported by NIH training grant 1 T32 AI07647 and Mount Sinai Medical Scientist Training Program T32 GM007280. AI is supported by a Japan Society for the Promotion of Science Fellowship. The small molecule screen was performed at the National Screening Laboratory for the Regional Centers of Excellence in Biodefense (NSRB), Harvard Medical School, Boston and was supported by NIH grant U54 AI057159. Confocal laser scanning microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported with funding from NIH-NCI shared resources grant (5R24 CA095823-04), NSF Major Research Instrumentation grant (DBI-9724504) and NIH shared instrumentation grant (1 S10 RR0 9145-01). We thank Ron Fouchier (Erasmus Medical Center, Rotterdam, The Netherlands) for providing influenza A/Netherlands/602/2009 (H1N1) virus.
Footnotes
Further methods used in this study can be found in the Supplementary Information.
AUTHOR CONTRIBUTIONS
RK, SS, AGS, JATY, PP, MLS, SKC designed research; RK, SS, AI, HHH, SuB, SEA, JGA, DMT, MBO, YL, QG, PD, LP, CS, performed research; RK, CS, PD, BPT, LM, GB performed the screens; AO provided siRNAs; RK, SS, BPT, MLS, SKC, PD analyzed data; YZ, GB, SoB, TI performed bioinformatics analysis; RK, SS, AGS JATY, PP, MLS, SKC co-wrote the manuscript.
AUTHOR INFORMATION
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
The authors declare no competing financial interests.
REFERENCES
- 1.Palese P, Shaw ML. Orthomyxoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Edition. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1647–1689. [Google Scholar]
- 2.Wang TT, Palese P. Unraveling the mystery of swine influenza virus. Cell. 2009;137:983–985. doi: 10.1016/j.cell.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 3.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Layne SP, Monto AS, Taubenberger JK. Pandemic influenza: an inconvenient mutation. Science. 2009;323:1560–1561. doi: 10.1126/science.323.5921.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 6.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao L, et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 15.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A. 2000;97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beer C, Andersen DS, Rojek A, Pedersen L. Caveola-dependent endocytic entry of amphotropic murine leukemia virus. J Virol. 2005;79:10776–10787. doi: 10.1128/JVI.79.16.10776-10787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClure MO, Sommerfelt MA, Marsh M, Weiss RA. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71(Pt 4):767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 20.Whitney JA, Gomez M, Sheff D, Kreis TE, Mellman I. Cytoplasmic coat proteins involved in endosome function. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 21.Aniento F, Gu F, Parton RG, Gruenberg J. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol. 1996;133:29–41. doi: 10.1083/jcb.133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tscherne DM, Manicassamy B, Garcia-Sastre A. An Enzymatic Virus-like Particle Assay For Sensitive Detection of Virus Entry. J of Virological Methods. doi: 10.1016/j.jviromet.2009.10.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 24.Colbran RJ. Targeting of calcium/calmodulin-dependent protein kinase II. Biochem J. 2004;378:1–16. doi: 10.1042/BJ20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sumi M, et al. The newly synthesized selective Ca2+/calmodulin dependent protein kinase II inhibitor KN-93 reduces dopamine contents in PC12h cells. Biochem Biophys Res Commun. 1991;181:968–975. doi: 10.1016/0006-291x(91)92031-e. [DOI] [PubMed] [Google Scholar]
- 26.Chanda SK, et al. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12153–12158. doi: 10.1073/pnas.1934839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aza-Blanc P, et al. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Molecular cell. 2003;12:627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 28.Sui B, et al. The use of Random Homozygous Gene Perturbation to identify novel host-oriented targets for influenza. Virology. 2009;387:473–481. doi: 10.1016/j.virol.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.