Abstract
The photochemical regulation of biological systems represents a very precise means of achieving high-resolution control over gene expression in both a spatial and a temporal fashion. DNAzymes are enzymatically active deoxyoligonucleotides that enable the site-specific cleavage of RNA, and have been used in a variety of in vitro applications. We have previously reported the photochemical activation of DNAzymes and antisense agents through the preparation of a caged DNA phosphoramidite and its site-specific incorporation into oligonucleotides. The presence of the caging group disrupts either DNA:RNA hybridization or catalytic activity, until removed via a brief irradiation with UV light. Here, we are expanding this concept by investigating the photochemical deactivation of DNAzymes and antisense agents. Moreover, we report the application of light-activated and light-deactivated antisense agents to the regulation of gene function in mammalian cells. This represents the first example of gene silencing antisense agents that can be turned on and turned off in mammalian tissue culture.
Introduction
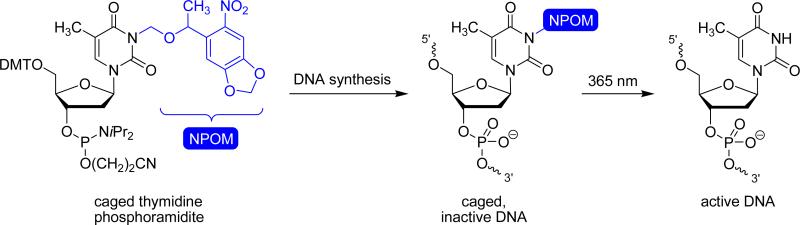
In order to achieve a detailed understanding of complex cellular and multicellular organisms, a precise external control over biological processes is required.1 Light is an ideal tool for the exogenous control of biological systems, e.g. at the gene transcription and translation level, as it possesses several advantages over traditional modulators of biological function. Perhaps the most beneficial feature is the ability to control light irradiation in both a spatial and a temporal fashion. Additionally, light irradiation is non-invasive, resulting in minimal secondary perturbations of cellular processes, and its amplitude can be regulated to enable tuning of the extent of biological activity. Light-induced activation of biological processes is most commonly achieved through the initial deactivation of a particular molecule via installation of a photo-protecting group at a critical functional motif required for biological activity. This renders the molecule inactive, in a practice known as “caging”.2-4 The photo-protecting group is removed upon irradiation with UV light, thus restoring the biological activity, in a practice known as “decaging” (Figure 1). Several very effective caging groups are known,4, 5 and ortho-nitrobenzyl (ONB) groups are by far the most common caging groups due to their facile synthesis, easy installation, and applicability to the caging of a wide range of functional groups. The photochemical properties of ONB groups can readily be tuned via electron donating groups (e.g. OCH3) to shift the absorption maximum to a longer wavelength, allowing efficient decaging with non-photodamaging UV light of 365 nm.6
Figure 1.
Caging and decaging of DNA. A caged monomeric building block (here, thymidine phosphoramidite) is incorporated into a deoxyoligonucleotide through standard DNA synthesis, rendering the oligomer inactive. Upon a brief irradiation with UV light, the caging group is removed, restoring the natural thymidine residue and thus the biological function of the DNA (e.g. the ability to undergo duplex formation, or catalytic activity). The 6-nitropiperonyloxymethylene (NPOM) caging group is shown in blue.
While the concept of caging biologically relevant molecules was introduced in 1978,7 only in the last 12 years have scientists investigated the synthesis of light-activatable oligonucleotides. Mostly, this was achieved through the introduction of light-cleavable groups on (or within) the phosphate backbone, the sugar, and the nucleotide base. However, examples of incorporating light-switchable motifs have been realized as well, and the several different approaches towards the light-regulation of oligonucleotide function possess distinct advantages and disadvantages.3
In our own development of light-activatable DNA,8-12 we found that a successful caging approach must address several criteria. Perhaps the most important one is that the installation of the caging group must completely abrogate the nascent function of the DNA oligomer, and afford a rapid restoration of activity upon a brief UV irradiation. Additionally, the caging group installation must be stable to both DNA synthesis and physiological conditions, as premature loss of the group under these conditions would nullify the value of the caging experiment. Moreover, it is advantageous if the number and location of caging groups installed on the DNA can be precisely controlled. Finally, a high yielding synthesis of the caging group and the caged DNA molecule under standard DNA synthesis conditions is favorable.
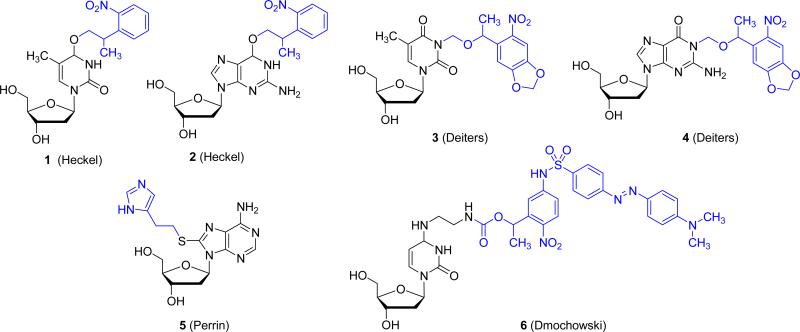
Three major approaches to DNA caging have been attempted. The first involves the statistical caging of the phosphodiester backbone with 1-(4,5-dimethoxy-2-nitrophenyl) diazoethane.13 However, due to the non-specific nature of this approach, the location and number of installed caging groups is difficult to control. Additionally, since much of the function of the DNA molecule is derived from base pairing interactions, the positioning of caging groups on the backbone is a less efficient means of disrupting function. A second approach, which has been increasingly applied in recent years, is the installation of a photocleavable linker into the backbone of the DNA oligomer. The fundamental basis of this approach was established by Taylor and Ordoukhanian by the simple incorporation of a 2-nitrobenzyl group between two DNA bases. Irradiation led to DNA scission, disrupting its hybridization.14 While a useful technology, due to the indirect caging of DNA, complete abrogation, followed by complete restoration of function requires substantial experimental design. The final approach to DNA caging involves the direct installation of a caging group on the nucleoside base. This approach is the most direct and has found substantial success in the disruption of nascent function of the oligomer due to the direct perturbation of hydrogen bonding and thus DNA hybridization. Several caged DNA nucleosides 1-6 have been prepared by us and others (Figure 2), and have been applied towards the regulation of various biological processes, including the photochemical activation of DNAzymes.10, 12, 15 Importantly, the direct caging of DNA-based antisense agents has enabled the photochemical control of gene silencing in mammalian cells.11 Antisense agents containing the caged nucleoside 3 at defined locations were unable to undergo hybridization to the mRNA until decaged through a brief UV irradiation. Moreover, locally restricted irradiation of a cellular monolayer of mouse fibroblast cells provided spatial control over gene silencing. Photochemical regulation of RNA interference was achieved as well, through the installation of a single caged nucleoside 1 at a crucial position of an siRNA reagent.16
Figure 2.
Structures of caged DNA nucleosides employed in the photochemical regulation of DNA function. The light-removable caging groups are shown in blue.
DNAzymes represent catalytically active oligonucletoides that have been evolved via in vitro selections to site-specifically cleave RNA substrates,17, 18 and recently DNA substrates.19 Unlike their ribozyme counterparts, DNAzymes are not naturally occurring; however, compared to ribozymes, they are more stable and less expensive to synthesize. The 10-23 DNAzyme was the first DNAzyme to be evolved by Joyce et al.17, 18, 20 and its RNA cleaving ability, catalytic activity, and mode of action been extensively studied.21 Since their initial discovery, several applications for DNAzymes have been developed both in vivo and in vitro, including roles as RNA cleavage and ligation catalysts, as molecular motors, and as sensors and detectors.21, 22 Moreover, they have been applied and proposed as gene silencing agents with therapeutic potential.23 Consequentially, the precise spatial and temporal control over DNAzyme activity with light has tremendous potential in the advancement of these technologies spanning both chemical and biological fields.
Results and Discussion
Investigation and optimization of the light-activation of DNAzymes
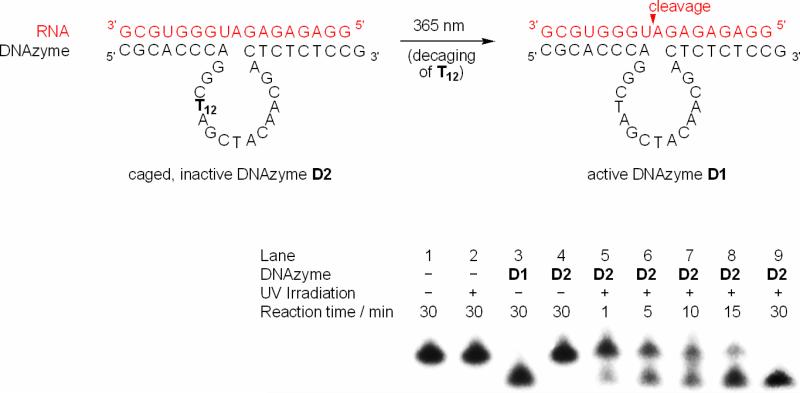
We have previously reported the preparation of a novel caging group (NPOM),24 and its implementation in the synthesis of a caged thymidine phosphoramidite of 3.12 The caged phosphoramidite was found to be stable under both physiological and standard DNA synthesis conditions, and was incorporated into a 10-23 DNAzyme at various positions to afford photoregulation of DNAzyme activity. DNAzyme activity was assessed by gel electrophoresis and imaging of the non-cleaved and cleaved 32P-labeled RNA substrate. We observed that complete DNAzyme deactivation was achieved through the installation of caging groups in either the hybridizing arms or at the essential T12 residue within the catalytic core of the 10-23 DNAzyme D2 (Figure 3).18, 20, 25 The DNAzyme mediated cleavage of the RNA substrate was restored after a brief UV irradiation (1 min, 365 nm, 25 W), thus removing the caging group from the inactive D2 and converting it into the active DNAzyme D1 (see Table 1 for all oligonucleotide sequences). Gel analysis of RNA cleavage (Figure 3) revealed that UV irradiation alone does not induce RNA degradation (lane 2). The non-caged DNAzyme D1 cleaves all RNA within 30 min (lane 3), while the caged DNAzyme D2 (lane 4) is completely inactive. However, a 1 min UV irradiation induced decaging of D2 leading to activation and RNA cleavage over 30 min (lanes 5-9).
Figure 3.
Light-activated DNAzyme. The caged 10-23 DNAzyme D2 (40 nM) binds to its complementary RNA (4 nM), but has no catalytic activity due to incorporation of the caged thymidine 3 at the crucial position T12 in the catalytic core. Irradiation at 365 nm removes the caging group, activates the DNAzyme, and induces RNA cleavage at a Mg2+ concentration of 100 mM (15 mM Tris buffer, pH 8.2). The RNA degradation was imaged by gel-separation of a 32P labeled RNA substrate.
Table 1.
Synthesized caged and non-caged deoxyoligonucleotide sequences.
| DNA | Sequence 5’ → 3’ |
|---|---|
| D1 | CGCACCCAGGCTAGCTACAACGACTCTCTCCG |
| D2 | CGCACCCAGGCT12AGCTACAACGACTCTCTCCG |
| D3 | CGCACCCAGGCTAGCT16ACAACGACTCTCTCCG |
| D4 | CGCACCCAGGCTAGCTACAACGACT25CTCTCCG |
| D5 | CGCACCCAGGCTAGCTACAACGACTCT27CTCCG |
| D6 | CGCACCCAGGCTAGCTACAACGACTCTCT29CCG |
| D7 | CGCACCCAGGCTAGCTACAACGACT25CT27CT29CCG |
| DD1 | GGAGAGAGATGGGTGCG |
| DD2 | GGAGAGAGAT10GGGT14GCG |
| DD3 | TCGTTGTAGCTAGCC |
| DD4 | T1CGT4TGTAGCT11AGCC |
| HP1 | CTAGCCTGGTGCGTTTTCGCACCCAGGCTAGCTACAACGACTCTCTCCG |
| HP2 | CCTGGTGCGTTTTCGCACCCAGGCTAGCTACAACGACTCTCTCCG |
| HP3 | CT2AGCCT7GGT10GCGTTTTCGCACCCAGGCTAGCTACAACGACTCTCTCCG |
| R1 | GCGCGCGAAACGCGCGCTACAACTCGGTGATGAGGCTAGCTACAACGAGTTCTCGGAGGAGCGGCGCGCGCAAAGCGCGCG |
| R2 | GCGCGCGAAACGCGCGCTACAACTCGGTGATGAGGCT37AGCTACAACGAGTTCTCGGAGGAGCGGCGCGCGCAAAGCGCGCG |
| R3 | GCGCGCGAAACGCGCGCTACAACTCGGTGATGAGGCA37AGCTACAACGAGTTCTCGGAGGAGCGGCGCGCGCAAAGCGCGCG |
| R4 | GCGCGCGAAACGCGCGCTACAACTCGGTGATGAGTTCTCGGAGGAGCGGCGCGCGCAAAGCGCGCG |
| R5 | GCGCGCGAAACGCGCGCTACAACT24CGGTGAT31GAGGCTAGCTACAACGAGTTCTCGGAGGAGCGGCGCGCGCAAAGCGCGCG |
| R6 | AT2CACCGAGT10TGT13AGCGCGCGAAACGCGCGCTACAACTCGGTGATGAGTTCTCGGAGGAGCGGCGCGCGCAAAGCGCGCG |
| R7 | GCGCGCGAAACGCGCGCCGCACCCAGGCTAGCTACAACGACTCTCTCCCGCGCGCGCAAAGCGCGCG |
Tnn = caged thymidine; underline = self-complementary region for hairpin formation; italics = protective hairpin formation for intracellular stabilization; A37 = mutation from thymidine to adenosine.
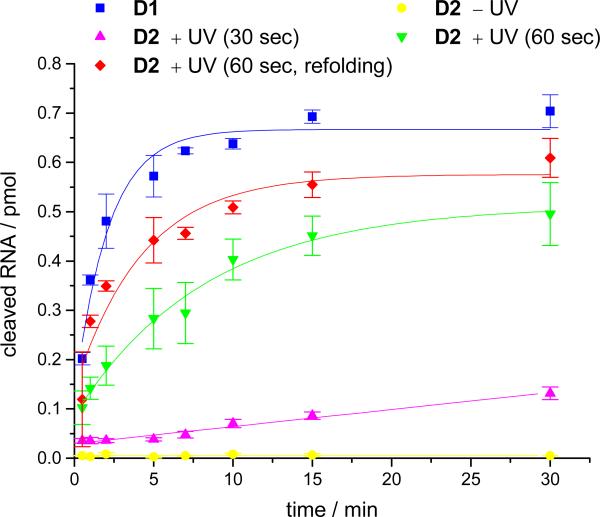
While the DNAzymes D2 and D7 could be activated with light, complete restoration of DNAzyme function was not achieved,12 affording a 53-54% restoration of the activity when compared to the non-caged analog D1. As a result we have further investigated the decaging process. In order to investigate DNAzyme activity, the Mg2+ concentration was reduced (10 mM) to decrease the reaction rate to a measurable level for the generation of a reaction time-course. Previously, the optimized conditions utilized a transilluminator (25 W) for 1 min at 365 nm (Figure 4). Increased irradiation times of up to 10 minutes did not lead to an increase in the catalytic activity of the DNAzyme D2. This led us to speculate that incomplete caging group removal was not the cause of the modest restoration of activity.
Figure 4.
Time course of RNA cleavage by the DNAzymes D1 (non-caged; 40 nM) and D2 (caged; 40 nM) under different irradiation and re-folding conditions. All reactions were performed with 4 nM 32P-labeled RNA substrate (10 mM MgCl2, pH 8.2, 15 mM Tris buffer). RNA cleavage was assessed via the removal of aliquots of the sample at given time points, followed by PAGE analysis (see Figure 3) and quantification of the radioactively labeled RNA substrate with ImageQuant. All experiments were conducted in triplicate and the error bars represent standard deviations.
Virtually complete removal of the caging group from the thymidine T12 of D2 after a 1 min irradiation (365 nm, 25 W) was confirmed by HPLC (see Supporting Information Figure S1). Under these conditions 87% of the oligomers are decaged, however, only 54% of the enzymatic activity was restored. Thus, it may be possible that the incorporation of a caging group elicits a conformational perturbation in the tertiary structure of a sub-population of the DNAzymes, requiring re-folding after photochemical removal for catalytic activation. To investigate this possibility, the DNAzyme D2 was decaged for 1 minute (365 nm, 25 W), followed by a brief heating to 90 °C for 1 minute and cooling to room temperature to afford proper re-folding prior to the addition of RNA substrate. This led to an enhanced restoration of activity, as shown in Figure 4. Conditions which employ this re-folding step led to higher cleavage activities than their corresponding conditions without re-folding.
Having successfully demonstrated the efficient photochemical activation of DNAzyme function, we also wanted to devise a means of deactivating DNAzymes with light. This will be particularly useful in the spatio-temporal activation of gene expression through a light-deactivatable DNAzyme as an antisense agent (see below). If the DNAzyme is constitutively active, gene expression will be suppressed via mRNA cleavage; however, upon light irradiation, deactivation of DNAzyme activity will lead to intact mRNA and thus expression of the gene of interest. Thus, we investigated several approaches towards this photochemical deactivation of DNAzymes.
Light-deactivation of DNAzyme activity using caged DNA decoys
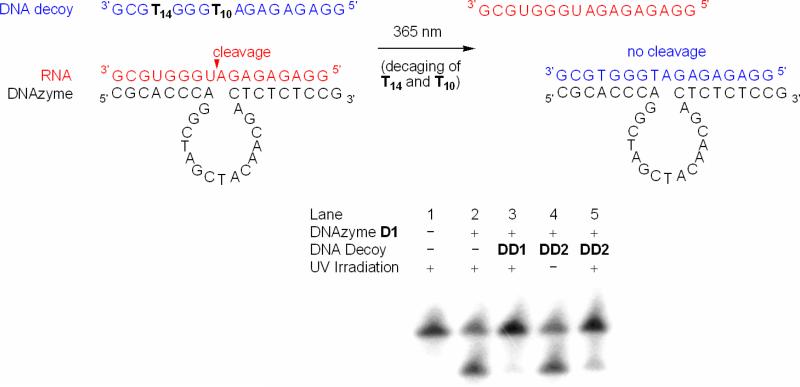
We first hypothesized that by incubating the DNAzyme with a DNA strand complementary to the DNAzyme binding arms should lead to a competition between the DNA and RNA substrate, since the DNAzyme is inactive towards cleavage of a deoxyoligonucleotide. If used in excess, DNA:DNA hybridization should be most prevalent, efficiently prohibiting binding and cleavage of the RNA substrate. If the DNA decoy is caged, it would be incapable of hybridization and inhibition of the DNAzyme until the photolabile protecting groups are removed through UV irradiation. In the absence of DNA:DNA hybridization the DNAzyme catalysis would function normally, cleaving the RNA substrate.
To validate this approach, we first attempted an experiment with a non-caged DNA decoy to determine optimal conditions for DNAzyme deactivation (see Supporting Information Figure S2). As with previous experiments, the RNA substrate was labeled with γ-32P ATP and employed in the experiments along with the 10-23 DNAzyme and the decoy DNA complement DD1 (see Table 1 for sequence information). Several experimental parameters were varied to optimize DNAzyme deactivation, including Mg2+ concentration, DNA substrate to RNA substrate ratio, and temperature. Ultimately, it was found that a 10:1 ratio of DNA inhibitor to RNA substrate, at 10 mM Mg2+ for 30 minutes at 25 °C, was optimal for suppressing DNAzyme activity. Based on these results we investigated the regulation of this process in a photochemical fashion, by employing a DNA decoy DD2 (see Table 1 for sequence information) containing two caged thymidine residues 3. Based on previous discoveries,9, 11, 12 2-3 caged thymidine nucleotides are sufficient to effectively inhibit DNA:DNA hybridization of a 17-mer deoxyoligonucleotide to its complement. Employing the optimized conditions, we evaluated the photoregulation of the caged decoy, as shown in Figure 5.
Figure 5.
Photochemical deactivation of the DNAzyme D1 (40 nM) with the caged complementary DNA decoy DD2 (40 nM) in the presence of RNA substrate (4 nM; 10 mM MgCl2, pH 8.2, 15 mM Tris buffer). Prior to irradiation the decoy is inactive and does not undergo hybridization to the DNAzyme; however, upon a brief irradiation (1 min, 365 nm, 25 W), the caging groups are removed enabling the hybridization of the decoy to the DNAzyme and inhibiting RNA cleavage.
The irradiated DNA decoy efficiently prevented RNA cleavage. The non-irradiated DNA decoy DD2 remained completely inactive towards DNAzyme inhibition (Figure 5; lane 4), as identical levels of RNA cleavage were observed as when no competing DNA inhibitor was added (lane 2). In contrast, the irradiated decoy DD2 (lane 5) induced a virtually complete deactivation of the DNAzyme, comparable to the non-caged decoy DD1 (lane 3). Overall, these experiments demonstrated the photochemical deactivation of DNAzyme activity with an excellent on/off ratio.
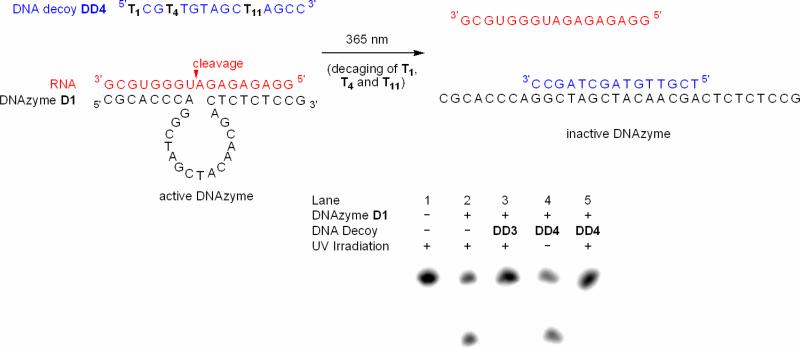
In order to avoid a competition between the DNA decoy and the RNA substrate for binding to the DNAzyme, we designed the DNA decoy DD3 (see Table 1 for sequence information) which is complementary to the catalytic core of the DNAzyme. Duplex formation of this decoy with the DNAzyme should significantly change the DNAzyme secondary structure, thus inhibiting catalytic activity and RNA:DNAzyme hybridization. Again, several variables were altered to ascertain the ideal reaction conditions for DNAzyme inactivation. Based on the previous experiment, we altered magnesium concentrations and DNA inhibitor to RNA substrate ratios, as described in the Supporting Information (Figure S3). As expected, employing DD3 in the DNAzyme inhibition was a much more efficient strategy than using a DNA decoy (DD1) complementary to the binding arms of the DNAzyme. Virtually no DNAzyme cleavage of the RNA substrate is observed under any condition using the active site inhibitor DD3. Here, even at high magnesium concentrations where the 10-23 DNAzyme has been demonstrated to be highly active, its catalytic ability is suppressed at low decoy/RNA substrate ratios (5:1; see Supporting Information). This is not surprising, as the two sequences (RNA and DNA decoy) are now not competing for binding, but rather have different sites for hybridization. Based on these results, we prepared the caged DNAzyme catalytic core inhibitor DD4 (see Table 1 for sequence information), containing three caged thymidines 3. We used the same reaction conditions as in Figure 5, but lowered our caged decoy/RNA ratio to 1:1. The reaction mixture was either kept in the dark or irradiated for 1 minute at 365 nm. Analysis by SDS PAGE after a 30 minute incubation at 25 °C revealed that in the absence of any DNAzyme the RNA remains uncleaved (Figure 6; lane 1); however, standard cleavage can be detected in the presence of the DNAzyme without the decoy (lane 2). Introduction of the non-caged decoy DD3 completely inactivated the DNAzyme (lane 3). However, the DNAzyme was fully active in the presence of the non-irradiated caged decoy DD4, leading to the same level of RNA cleavage as in the absence of an inhibitor (lane 4). As expected, UV irradiation of the caged decoy DD4 completely deactivated the DNAzyme effectively inhibiting catalysis (lane 5).
Figure 6.
Photochemical DNAzyme inactivation using the caged DNA decoy DD4 (4 nM) complementary to the catalytic core of the DNAzyme (40 nM) in the presence of RNA substrate (4 nM; 10 mM MgCl2, pH 8.2, 15 mM Tris buffer). Prior to UV irradiation normal DNAzyme function is observed. However, upon decaging (1 min, 365 nm, 25 W), the DNA decoy is capable of hybridizing to the catalytic core, completely inhibiting DNAzyme catalyzed cleavage of RNA.
Light-deactivation of DNAzyme activity via phototriggered DNA hairpin formation
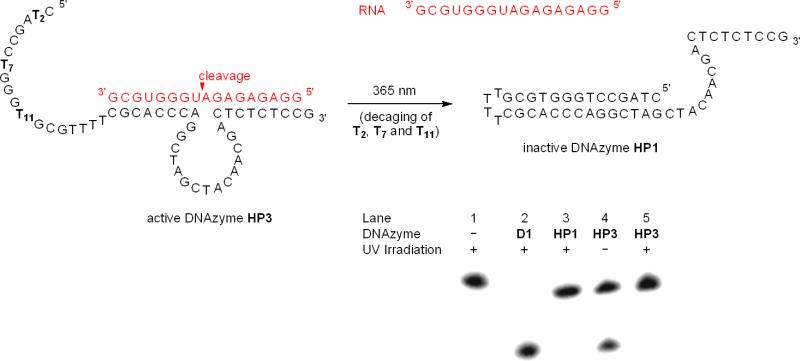
The approaches for the light-triggered deactivation of DNAzyme function described in Figures 5 and 6 rely on the co-localization of the decaged DNA decoy and the DNAzyme. An alternative approach is the covalent attachment of the caged decoy to the DNAzyme for a cis-acting DNAzyme inhibition. This can be achieved via the synthesis of a DNAzyme with a self-complementary binding arm. When caged, the DNAzyme would remain active; however, upon photochemical removal of the caging groups an intramolecular hybridization event will occur, forming a hairpin and suppressing RNA hybridzation and cleavage. Advantages of this strategy are a faster and more stable intramolecular hybridization and the use of only a single oligonucleotide, facilitating its synthesis and application. Two hairpin DNAzymes, HP1 and HP2 (see Table 1 for sequence information), with different degrees of self-complementarity were prepared, in order to test how many nucleotides need to be engaged in hairpin formation to achieve complete inhibition of catalytic activity. The two DNAzymes only differ in the length of the self-complementary region, with the hairpin of HP1 extending into the catalytic core of the DNAzyme, while HP2 has a shorter hairpin, only blocking one recognition arm. We conducted test reactions at 10-100 mM Mg2+ using the previously described radioactively labeled RNA substrate. These studies revealed that only the DNAzyme HP1 was capable of suppressing RNA cleavage, whereas the DNAzyme HP2 was constitutively active. In order to photochemically control deactivation of the DNAzyme HP1 we installed caging groups on three thymidine bases of the hairpin portion of HP1, yielding the caged DNAzyme HP3 (see Table 1 for sequence information). DNAzyme light-deactivation was assayed under the same reaction conditions as before (10 mM Mg2+, 25 °C, 30 min reaction time) (Figure 7).
Figure 7.
Photochemical DNAzyme inactivation using a caged hairpin strategy. Prior to UV irradiation normal DNAzyme function of HP3 (40 nM) is observed. However, upon decaging (1 min, 365 nm, 25 W), the complementary sequence is capable of hairpin formation, thus disrupting RNA-binding and the catalytic core and thereby inhibiting DNAzyme HP1 catalyzed cleavage of RNA substrate (4 nM; 10mM MgCl2, pH 8.2, 15 mM Tris buffer).
In the absence of any DNAzyme, the RNA substrate remains uncleaved (Figure 7; lane 1), even when irradiated with UV light of 365 nm. Complete cleavage occurs with the natural 10-23 DNAzyme D1 (lane 2). The non-caged hairpin DNAzyme HP1 is completely inactive leading to no degradation of the RNA substrate (lane 3). However, the caged hairpin DNAzyme HP3 remains active in the absence of UV irradiation, but displays slightly less RNA cleavage than the natural 10-23 DNAzyme D1 after a short 30 min reaction time (lane 4). Gratifyingly, irradiation of the caged hairpin for 1 minute with 365 nm UV light photochemically converts the DNAzyme HP3 into HP1 thus deactivating RNA cleavage (lane 5).
In summary, we have demonstrated three different approaches to the DNAzyme deactivation using UV light irradiation. These results complement our previously developed photochemical activation of DNAzymes.12
Light-regulation of gene function in mammalian cells using caged antisense agents
Having successfully developed approaches for the photochemical activation and deactivation of DNAzymes we next tested this methodology in the photochemical regulation of gene function in mammalian cell culture. The therapeutic application of DNAzymes as antisense agents has been proposed, as the enzymes can target gene transcripts for degradation in a sequence specific fashion.26 However, unmodified single-stranded DNA is intrinsically unstable in an intracellular environment.27 A recent report demonstrated that the creation of double-stranded hairpins on the ends of a single-stranded DNAzyme showed enhanced stability against exonuclease degradation and allowed for the silencing of reporter gene activity in mammalian cell cuture.28
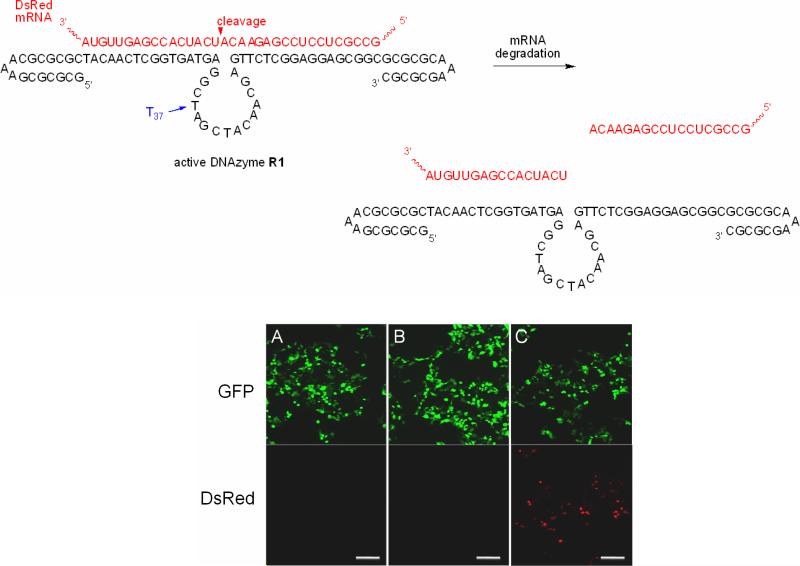
In order to test that photocaged DNAzymes can be used as light-activated gene silencing agents in eukaryotic cells, we targeted the DsRed reporter gene using a specifically designed DNAzyme R1 and its caged analog R2 (see Table 1 for sequence information). The thymidine at position 37 was selected for the introduction of a single caging group in R2 based on our in vitro data, which indicated that this residue in the catalytic core is essential for DNAzyme activity (see D2 in Figure 3).12 Both DNAzymes, R1 and R2 (500 pmol each), were co-transfected (X-TremGENE) with a plasmid bearing a CMV-driven DsRed reporter gene (CreStoplight,29 1 □g) and a CMV-driven eGFP control plasmid (C117,30 1 □g) as a transfection control into human embryonic kidney cells (HEK293T). After 4 hours of incubation the cells were either irradiated for 2 minutes at 365 nm (25 W) or kept in the dark. Cells were subsequently incubated for 48 hours to afford expression and maturation of the fluorescent proteins, and then imaged by fluorescence microscopy (Figure 8). Interestingly, no DsRed was detected in either sample (Figure 8A and 8B), indicating that the DNAzyme R2 retained its gene silencing activity despite the loss of RNA cleavage activity through caging of the residue T37. DsRed expression was clearly visible in cells transfected with a DNAzyme control R7 (Figure 8C). Based on these results we suspected that the DNAzyme was not necessarily silencing the DsRed transcript based on its intrinsic enzymatic RNA cleavage activity, but was rather acting as a classical DNA antisense agent leading to suppression of gene function via an established RNase H mediated mechanism.31
Figure 8.
Terminal hairpins introduced on the DNAzyme R1 increase intracellular stability in mammalian tissue culture and allow for mRNA cleavage. Fluorescence image of HEK293T cells co-transfected with DsRed and GFP expressing plasmids and the DNAzymes R1 (non-caged) and R2 (caged at T37). A) Transfection of the non-caged DNAzyme R1 leading to the silencing of DsRed expression. B) Transfection of the DNAzyme R2 caged at the essential residue T37 in the catalytic core, previously shown to abrogate DNAzyme activity; however, in this case DNAzyme complete silencing of DsRed is still observed. C) Control DNAzyme R7 transfection leading to the expression of both DsRed and GFP. Scale bar = 200 μm.
To further probe the mechanism of gene silencing by the DNAzymes/antisense oligonucleotides, we obtained two additional non-caged oligomers; R3 which had the essential thymidine T37 in the catalytic core mutated to an adenosine (T37 → A37) inhibiting catalytic activity,25 and R4 where the entire catalytic core was removed from the DNAzyme (see Table 1 for sequence information). If silencing is observed in with these two constructs it will confirm that the DNAzyme is not functioning via its intrinsic RNA cleavage activity, but rather by a classical antisense mechanism.31 Thus, the DNAzyme constructs R3 and R4 (500 pmol each) were co-transfected with the two plasmids expressing DsRed and GFP plasmid (1 μg each), and fluorescence was imaged after 48 h (see Supporting Information, Figure S4). Unexpectedly, the DsRed gene is efficiently silenced with all of the DNAzyme constructs. The DNAzyme R3, bearing a mutated, inactive catalytic core, and the oligonucleotide R4, without any catalytic core, had identical silencing effects as the normal DNAzyme R1. This suggests, in contrast to a previous report,28 that the RNA cleaving activity of the DNAzymes is not necessary for highly efficient gene silencing. As a result, these oligonucleotides will subsequentially be referred to as antisense agents rather than DNAzymes.
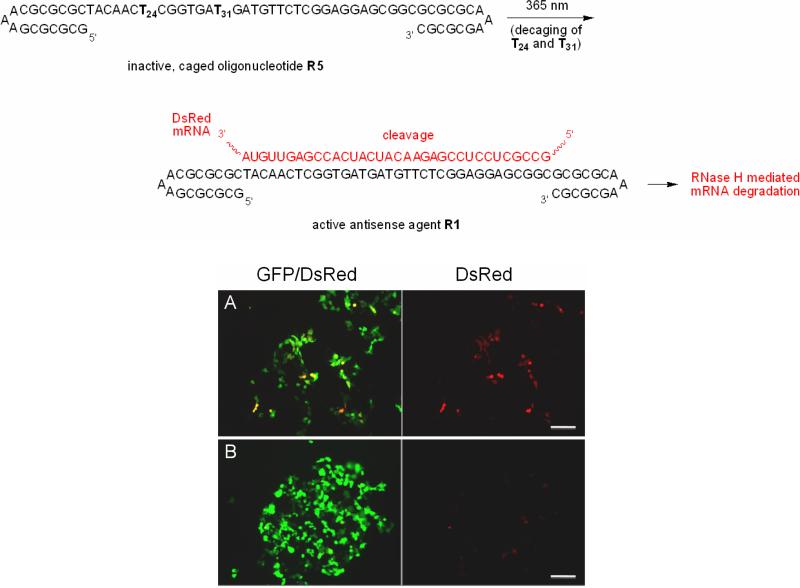
With this information, a new strategy was developed for the photochemical activation and deactivation of a strictly DNA-based antisense agent in mammalian cells: we employed the stability enhancing effects of the terminal hairpins but removed the redundant catalytic core from the DNAzyme R1, thus generating the oligonucleotide R4 (see Table 1 for sequence information). In order to achieve photochemical control of antisense activity we selected two thymidines in the mRNA hybridizing sequence of R4 and installed caged thymidine residues 3, generating the caged oligonucleotide R5 (see Table 1 for sequence information). In the absence of UV irradiation, the two caging groups will prevent hybridization9 of the antisense oligonucleotide to the DsRed mRNA transcript and allow for DsRed expression; however, upon a brief UV irradiation, the caging groups will be removed, the antisense agent will become active and DsRed will be silenced via RNase H mediated mRNA degradation. After transfection of HEK293T cells with the DsRed and GFP reporter constructs and the caged DNA-based antisense agent R5, one set of cells was irradiated for 2 minutes (365 nm, 25 W), while the other set of cells remained non-irradiated. After 48 hours, the cells were imaged with a fluorescence microscope (Figure 9).
Figure 9.
Photochemical activation of antisense oligonucleotide function in mammalian tissue culture. A) Normal DsRed expression is observed prior to irradiation with light, since the caging groups prevent mRNA:DNA hybridization. B) Irradiated cells (365 nm, 2 min, 25 W) no longer express DsRed due to light-activation of the DNA leading to DsRed silencing. GFP/DsRed (left) corresponds to an overlay of the GFP and DsRed images, while only the DsRed channel is shown on the right. Scale bar = 150 μm.
The cells transfected with the non-irradiated caged antisense agent R5 exhibited DsRed expression (Figure 9A), because the caged antisense agent is inactive. Conversely, the irradiated cells display no DsRed expression, indicating that the activity of the antisense agent R5 has been restored via the irradiation and decaging event (Figure 9B). Thus, we demonstrated that light-activation of gene silencing in mammalian cell culture can be achieved using a caged antisense agent comprised purely of DNA without the necessity of using any of the typically employed backbone and sugar modifications (as seen in antisense agents based on phosphorothioate nucleic acids, locked nucleic acids, peptide nucleic acids, morpholinos, etc.23).
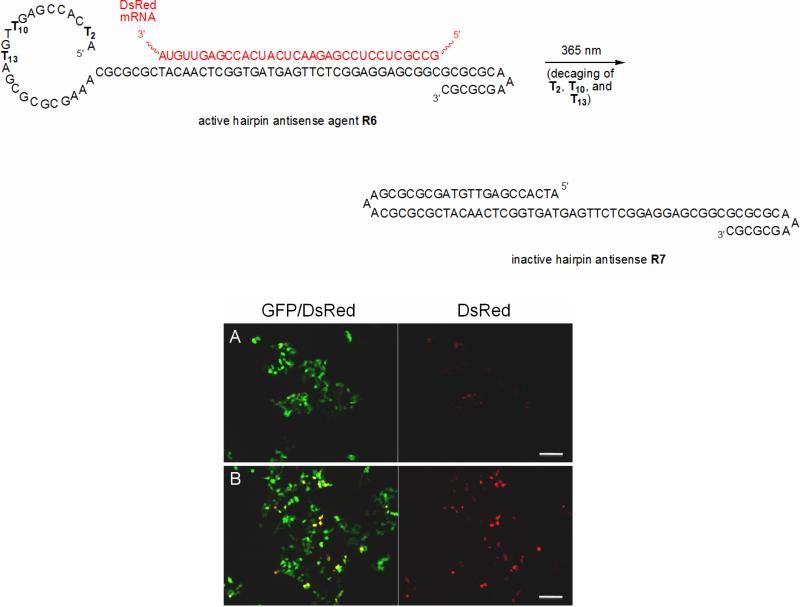
Next, we wanted to achieve the first photochemical deactivation of an antisense agent. Although the light-activation of antisense agents (including phosphorothioate DNA, peptide nucleic acids, and morpholinos) has been reported before using various approaches,11, 32 the photochemical deactivation of antisense activity has not been demonstrated. Having shown that we can deactivate antisense function through light-induced hairpin formation (see Figure 7), we assembled a caged construct in which we removed the catalytic core of the DNAzyme (since it is not necessary for gene silencing) and extended the hairpin into the binding arms as a means of disrupting hybridization to the mRNA transcript. The oligonucleotide R6 was designed to possess three caged thymidines which prevent hairpin formation until decaging through UV irradiation. This should result in initial activity of the construct, silencing DsRed expression, until photolysis removes the caging groups and the intramolecular hybridization forming a hairpin displaces mRNA binding, deactivating transcript degradation and enabling DsRed expression. HEK293T cells were transfected with the reporter plasmids (as described above) and R6, followed by a 2 minute irradiation (365 nm, 25 W) of one set of cells. The cells were incubated for 48 hours, and then imaged by fluorescence microscopy (Figure 10). In the absence of UV light, the antisense agent remains active, effectively suppressing DsRed expression (Figure 10A). However, upon irradiation the caging groups are removed leading to hairpin formation and deactivation of the antisense agent, followed by DsRed expression (Figure 10B).
Figure 10.
Deactivation of antisense oligonucleotide function in mammalian tissue culture. A) The hairpin antisense agent R6 is active in the absence of light irradiation, silencing the DsRed gene. B) Irradiation of cells removes the caging groups, enabling the formation of a hairpin that blocks the antisense agent from recognizing the mRNA transcript. Inactivation of the antisense agent then allows for DsRed expression. GFP/DsRed (left) corresponds to an overlay of the GFP and DsRed images, while only the DsRed channel is shown on the right. Scale bar = 200 μm.
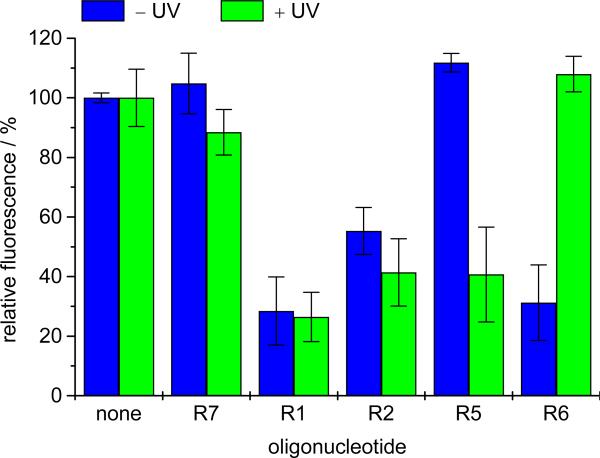
In addition to the fluorescence imaging, DsRed silencing was quantified for each condition by fluorescence measurement of GFP (488/509 nm) and DsRed (560/585 nm) after cell lysis. The GFP signal was used to normalize the relative fluorescence units (RFU) for differences in transfection efficiency and cell confluency (Figure 11). The obtained data confirmed the previous imaging results shown in Figures 8-10. No change in DsRed fluorescence is observed in the absence of antisense agents or with a scrambled control sequence (R7). The previously reported antisense agent R128 led to an approximate 70% reduction in DsRed fluorescence irrespective of light irradiation. A similar level of silencing was observed with R2 caged at the catalytic core in the presence or absence of irradiation, albeit a slightly higher DsRed signal is observe in the absence of irradiation. The antisense agent R5 caged on the binding arms showed normal DsRed expression in the absence of UV irradiation and a substantial decrease in fluorescence upon UV irradiation. A similar silencing activity as in case of the wild type antisense agent R1 was obtained, demonstrating an excellent off/on ratio before/after UV irradiation. Conversely, the caged hairpin antisense agent R6 is initially active in its caged form, as hairpin formation is inhibited. Upon irradiation, however, caging group removal occurs and intramolecular hairpin formation dominates, leading to the restoration of DsRed expression. Together with the fluorescent images presented in Figures 9 and 10, these results demonstrate the ability to both photochemically activate and deactivate antisense agents in mammalian cell culture.
Figure 11.
Measurement of DsRed and GFP fluorescence to quantify intracellular antisense activity. Transfections were performed as previously described, and the cells were either irradiated (2 min, 365 nm, 25 W), or not exposed to UV irradiation. After 48 hours fluorescence was measured on a Molecular Devices Gemini EM microplate spectrofluorimeter. Relative fluorescence (DsRed/GFP) normalized to a transfection in the absence of DNA is shown. Error bars represent standard deviations from three independent experiments. The mRNA translational suppression of ~30% with the non-caged analog R1 is in accordance with previous reports.28
Conclusion
In summary, we have demonstrated both the photochemical activation and deactivation of DNAzyme function and antisense oligonucleotide function. The light-activation of DNAzymes was achieved through the site-specific installation of caging groups on thymidine residues located either in the catalytic domain of the DNAzyme or the sequences binding to the RNA target. A brief UV irradiation at 365 nm removes those caging group and restores >80% of DNAzyme activity. Two different approaches towards the light-deactivation of DNAzyme activity were investigated, including 1) caged, trans-acting DNA decoys targeting either the binding arms or the catalytic core of the DNAzyme, and 2) caged hairpin motifs enabling the cis-regulation of DNAzyme activity. The latter provides the advantage of only requiring one deoxyoligonucleotide for an experiment, and ensures co-localization of the light-activatable inhibitor and the DNAzyme. These developments have substantial applications towards the regulation of gene function, as DNAzymes are capable of site-specifically recognizing mRNA, suppressing its translation, and inducing its degradation. Towards this application we discovered that previously described DNAzymes, stabilized for cellular function, in fact achieve the majority of their silencing not through their mRNA cleavage activity, but rather via a typical antisense mechanism. We were able to employ our previously described light-regulation approaches to the photochemical activation and deactivation of these antisense agents enabling gene deactivation and activation in mammalian tissue culture. These findings indicate a potentially improved means of antisense inhibition, as the hairpin single-stranded DNA displays a sufficient cellular stability. Previous studies have involved the utilization of caged siRNA,33, 34 caged phosphorothioates,11 and morpholinos and peptide nucleic acids possessing a complementary sequence fused by a photocleavable tether.32, 35, 36 In the case of siRNA the photoregulation by non-specific caging of the backbone was comparatively leaky33 (i.e., gene silencing was observed in case of the caged oligonucleotide) compared to the antisense approaches, which was later remedied via specific caging of the 5’ phosphate34 or the application of fluorinated analogs (siFNAs).37 The application of tethered antisense decoys has the disadvantages that two oligomers are released intracellularly after photochemical tether cleaved, and that the selection of a suitable decoy sequence is not trivial.36 All of these technologies provide the ability to spatio-temporally modulate gene function; however, unlike the caged DNA antisense agents reported in this study, previous reagents are more synthetically difficult to prepare than the simple DNA phosphoroamidites employed here. In addition, none of the previous reports of light-activated antisense agents allow for the photochemical deactivation of gene silencing. This methodology can rapidly be extended to the photochemical regulation of endogenous genes with both high spatial and temporal resolution via the tuning of the DNA sequence. Thus, critical experiments to be performed, elucidating the complexities of gene expression during the development of cells and organisms where genes are regulated with high spatio-temporal resolution.
Experimental
DNA synthesis protocol
DNA synthesis was performed using an Applied Biosystems (Foster City, CA) Model 394 automated DNA/RNA Synthesizer using standard β-cyanoethyl phosphoramidite chemistry at the Wake Forest University Nucleotide Core Facility. All caged DNAzymes were synthesized using 40 nmole scale, low volume solid phase supports obtained from Glen Research (Sterling, VA). Reagents for automated DNA synthesis were also obtained from Glen Research. Standard synthesis cycles provided by Applied Biosystems were used for all normal bases using 2 minute coupling times. The coupling time was increased to 10 minutes for the positions at which the caged-T modified phosphoramidites were incorporated. Each synthesis cycle was monitored by following the release of dimethoxy trityl (DMT) cations after each deprotection step. No significant loss of DMT was noted following the addition of the caged-T for any of the DNAzymes, so 10 minutes was sufficient to allow maximal coupling of the caged-T. Yields of all DNAzymes were close to theoretical values routinely obtained.
Decaging and DNAzyme activation through UV irradiation
RNA substrate (5’ GGAGAGAGAUGGGUGCG 3’) was purchased from IDT and end-labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P] ATP (37°C, 30min). The labeled substrate was purified by Microcon 3 centrifugation and resuspended in DEPC water. DNAzyme assays were performed under single turnover conditions with the DNAzyme (40 nM) and the RNA substrate (4 nM) in standard reaction buffer (100 mM MgCl2, pH 8.2, 15 mM Tris base). The DNAzyme was equilibrated at 37 °C in the reaction buffer for approximately 15 minutes, followed by the addition of RNA substrate to initiate the reaction. In the case of photochemical activation of caged DNAzyme, the substrate was irradiated in a disposable cuvette with a UVP transilluminator (25 W) prior to equilibration at 37 °C and RNA substrate addition. Following the irradiation, the DNAzyme reaction was heated to 90 °C for 1 minute and cooled to room temperature to afford proper re-folding, and was then incubated at 37 °C for 15 minutes prior to RNA substrate addition. Aliquots of the reaction were removed at time points between 0-30 min and the reaction was stopped via addition of 6× stop/loading dye (10 mM Tris-HCl (pH 7.6), 0.03% bromophenol blue, 0.03% xylene cyanol FF, 60% glycerol, 60 mM EDTA)The samples were analyzed by 15% denaturing PAGE (160 V, 40 min). Acrylamide gels were visualized using a Storm 840 Phosphorimager, and radioactive band intensities were quantified using Image Quant 5.2 and correlated to RNA concentrations.
HPLC assessment of decaging
To demonstrate effective decaging, samples were analyzed on a Hamilton reverse phase HPLC column (10 μM, 250 × 4.1 cm, PRP-1) using an Agilent 1100 HPLC. A 10 μM sample of non-caged DNAzyme was initially analyzed to establish optimal conditions (90% H2O/10% acetonitrile isocratic for 5 min, ramp to 35% acetonitrile within 15 min; each solvent contained 0.1% TFA). See the Supporting Information for chromatograms (Figure S1).
Deactivation of DNAzyme function with DNA decoys
DNAzyme assays were performed as described above with the DNAzyme (40 nM) and the RNA substrate (4 nM) in standard reaction buffer (100 mM or 10 mM MgCl2, pH 8.2, 15 mM Tris base). The DNAzyme was equilibrated at 37 °C or 25 °C in the reaction buffer for approximately 15 minutes in the presence or absence of DNA decoy (0, 4, 20, or 40 nM), followed by the addition of RNA substrate to initiate the reaction. In the case of photochemical activation of the caged DNA decoy, the reaction was irradiated in a PCR tube with a UVP transilluminator (365 nm, 25 W) prior to equilibration at either 25 °C, or 37 °C (see Supporting Information) and RNA substrate addition. The reaction was incubated for 30 minutes, and stopped via addition of 6× stop/loading dye. The samples were analyzed by 15% denaturing PAGE (160 V, 40 min). Acrylamide gels were visualized using a Storm 840 Phosphorimager. The effects of different magnesium concentrations and different temperatures on DNAzyme catalysis are shown in the Supporting Information (Figures S2 and S3).
Deactivation of DNAzymes via hairpin formation
DNAzyme assays were performed as described above with the DNAzyme (40 nM) and the RNA substrate (4 nM) in standard reaction buffer (100 mM or 10 mM MgCl2, pH 8.2, 15 mM Tris base). The DNAzyme was then equilibrated at either 37 °C or 25 °C for 15 min prior to the addition of RNA substrate. In the case of photochemical activation of caged hairpin DNAzymes, the reaction was irradiated in a PCR tube with a UVP transilluminator (365 nm, 25 W) prior to equilibration at either 25 °C or 37 °C and RNA substrate addition. The reaction was incubated for 30 minutes, and stopped via addition of 6× stop/loading dye. The samples were analyzed by 15% denaturing PAGE (160 V, 40 min). Acrylamide gels were visualized using a Storm 840 Phosphorimager. The effects of different magnesium concentrations and different temperatures on DNAzyme catalysis are shown in the Supporting Information.
Photochemical regulation of antisense agents mammalian cell culture
Human embryonic kidney cells (HEK293T) were grown at 37 °C and 5% CO2 in Dulbecco's modified Eagle's media (DMEM; Hyclone); supplemented with 10% Fetal Bovine serum (FBS; Hyclone) and 10% streptomycin/ampicillin (MP Biomedicals). Cells were passaged into chamber slides (1000 μL per well; ~1×105 cells) and grown to ~70% confluence within 24 hours. The media was changed to OPTIMEM (Invitrogen), and the cells were co-transfected with CreStoplight plasmid (1 μg), C117eGFP plasmid (1 μg), and the DNAzyme (500 pmol) using X-TremeGENE (3:2 reagent/DNA ratio; Invitrogen). The following conditions were used: no DNAzyme construct, a control DNA oligomer with no sequence homology to DsRed, and the caged DNAzyme constructs. All transfections were performed in triplicate. Cells were incubated at 37 °C for 6 hours and the transfection media was removed. One of two 96 well plates was briefly irradiated with a hand-held UV lamp (365 nm, 23 W) for 2 min. The media was then replaced with standard growth media and the cells were incubated for an additional 48 hours. The cells were observed and no changes in growth or morphology were visible when comparing the irradiated cells with the non-irradiated cells. The cells were then imaged on a Lecia DM5000B microscope to assess DsRed and GFP expression. Cells were then lysed (Lysis Buffer, Promega) for 10 min and the lysate was measured on a Molecular Devices Gemini EM microplate spectrofluorimeter at both 488/509 nm (GFP) and 560/585 nm (DsRed). All experiments were conducted in triplicate, the ratio of DsRed expression to GFP expression was calculated for each of the triplicates, the data was averaged, and standard deviations were calculated using Microsoft Excel.
Supplementary Material
Acknowledgement
This research was supported in part by the National Institutes of Health (R01GM079114), the Beckman Foundation (Beckman Young Investigator Award for AD), and the Research Corporation (Cottrell Scholar Award for AD). DDY acknowledges a graduate research fellowship from the ACS Medicinal Chemistry Division.
Footnotes
Supporting Information Available. HPLC analysis of DNA decaging, optimization of DNA decoy and hairpin deactivation experiments, and validation of DsRed DNAzyme antisense activity. This information is available free of charge via the World Wide Web at http://pubs.acs.org.
References
- 1.a Banaszynski LA, Wandless TJ. Chem Biol. 2006;13:11. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]; b Buskirk AR, Liu DR. Chem. Biol. 2005;12:151. doi: 10.1016/j.chembiol.2004.11.012. [DOI] [PubMed] [Google Scholar]; c Lewandoski M. Nat. Rev. Genet. 2001;2:743. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]; d Padidam M. Curr. Opin. Plant Biol. 2003;6:169. doi: 10.1016/s1369-5266(03)00005-0. [DOI] [PubMed] [Google Scholar]
- 2.a Deiters A. Chembiochem. 2010;11:47. doi: 10.1002/cbic.200900529. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Deiters A. Curr. Opin. Chem. Biol. 2009;13:678. doi: 10.1016/j.cbpa.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Lee HM, Larson DR, Lawrence DS. ACS Chem. Biol. 2009;4:409. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Young DD, Deiters A. Org. Biomol. Chem. 2007;5:999. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]; e Curley K, Lawrence DS. Curr. Opin. Chem. Biol. 1999;3:84. doi: 10.1016/s1367-5931(99)80015-5. [DOI] [PubMed] [Google Scholar]
- 3.a Casey JP, Blidner RA, Monroe WT. Mol. Pharm. 2009;6:669. doi: 10.1021/mp900082q. [DOI] [PubMed] [Google Scholar]; b Tang X, Dmochowski IJ. Mol. Biosyst. 2007;3:100. doi: 10.1039/b614349k. [DOI] [PubMed] [Google Scholar]; c Mayer G, Heckel A. Angew. Chem. Int. Ed. 2006;45:4900. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 4.Adams SR, Tsien RY. Annu. Rev. Physiol. 1993;55:755. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 5.Pelliccioli AP, Wirz J. Photochem. Photobiol. Sci. 2002;1:441. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
- 6.a Dong Q, Svoboda K, Tiersch TR, Monroe WT. J. Photochem. Photobiol. B. 2007;88:137. doi: 10.1016/j.jphotobiol.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Robert C, Muel B, Benoit A, Dubertret L, Sarasin A, Stary A. J. Invest. Dermatol. 1996;106:721. doi: 10.1111/1523-1747.ep12345616. [DOI] [PubMed] [Google Scholar]; c Schindl A, Klosner G, Honigsmann H, Jori G, Calzavara-Pinton PC, Trautinger F. J. Photochem. Photobiol. B. 1998;44:97. doi: 10.1016/s1011-1344(98)00127-4. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan JH, Forbush B, Hoffman JF. Biochemistry. 1978;17:1929. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 8.a Young DD, Lusic H, Lively MO, Deiters A. Nucleic Acids Res. 2009;37:e58. doi: 10.1093/nar/gkp150. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Young DD, Govan JM, Lively MO, Deiters A. Chembiochem. 2009;10:1612. doi: 10.1002/cbic.200900090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young DD, Edwards WF, Lusic H, Lively MO, Deiters A. Chem. Commun. 2008:462. doi: 10.1039/b715152g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lusic H, Lively MO, Deiters A. Mol. Biosyst. 2008;4:508. doi: 10.1039/b800166a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young DD, Lusic H, Lively MO, Yoder JA, Deiters A. Chembiochem. 2008;9:2937. doi: 10.1002/cbic.200800627. [DOI] [PubMed] [Google Scholar]
- 12.Lusic H, Young DD, Lively MO, Deiters A. Org. Lett. 2007;9:1903. doi: 10.1021/ol070455u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a Ghosn B, Haselton FR, Gee KR, Monroe WT. Photochem. Photobiol. 2005;81:953. doi: 10.1562/2004-11-15-RA-373. [DOI] [PubMed] [Google Scholar]; b Monroe WT, McQuain MM, Chang MS, Alexander JS, Haselton FR. J. Biol. Chem. 1999;274:20895. doi: 10.1074/jbc.274.30.20895. [DOI] [PubMed] [Google Scholar]
- 14.Ordoukhanian P, Joyce GF. J. Am. Chem. Soc. 2002;124:12499. doi: 10.1021/ja027467p. [DOI] [PubMed] [Google Scholar]
- 15.a Krock L, Heckel A. Angew. Chem. Int. Ed. 2005;44:471. doi: 10.1002/anie.200461779. [DOI] [PubMed] [Google Scholar]; b Cordero D, Marcucio R, Hu D, Gaffield W, Tapadia M, Helms JA. J. Clin. Invest. 2004;114:485. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Tang X, Dmochowski IJ. Org. Lett. 2005;7:279. doi: 10.1021/ol047729n. [DOI] [PubMed] [Google Scholar]
- 16.Mikat V, Heckel A. Rna. 2007;13:2341. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 18.Santoro SW, Joyce GF. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4262. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandra M, Sachdeva A, Silverman SK. Nat. Chem. Biol. 2009;5:718. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santoro SW, Joyce GF. Biochemistry. 1998;37:13330. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- 21.a Silverman SK. Chem. Commun. 2008:3467. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]; b Baum DA, Silverman SK. Cell. Mol. Life Sci. 2008;65:2156. doi: 10.1007/s00018-008-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a Schlosser K, Li YF. Chem. Biol. 2009;16:311. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]; b Hobartner C, Silverman SK. Biopolymers. 2007;87:279. doi: 10.1002/bip.20813. [DOI] [PubMed] [Google Scholar]
- 23.a Chan CW, Khachigian LM. Intern. Med. J. 2009;39:249. doi: 10.1111/j.1445-5994.2009.01799.x. [DOI] [PubMed] [Google Scholar]; b Flintoft L. Nature Rev. Gen. 2008;9:6. [Google Scholar]; c Isaka Y. Curr. Opin. Mol. Ther. 2007;9:132. [PubMed] [Google Scholar]; d Dass CR. Trends Pharmacol. Sci. 2004;25:395. doi: 10.1016/j.tips.2004.06.001. [DOI] [PubMed] [Google Scholar]; e Eckstein F. Expert Opin. Biol. Ther. 2007;7:1021. doi: 10.1517/14712598.7.7.1021. [DOI] [PubMed] [Google Scholar]; f Scherer LJ, Rossi JJ. Nat. Biotechnol. 2003;21:1457. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 24.a Lusic H, Deiters A. Synthesis. 2006:2147. [Google Scholar]; b Young DD, Deiters A. Bioorg. Med. Chem. Lett. 2006;16:2658. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Zaborowska Z, Furste JP, Erdmann VA, Kurreck J. J. Biol. Chem. 2002;277:40617. doi: 10.1074/jbc.M207094200. [DOI] [PubMed] [Google Scholar]
- 26.a Fahmy RG, Waldman A, Zhang G, Mitchell A, Tedla N, Cai H, Geczy CR, Chesterman CN, Perry M, Khachigian LM. Nat. Biotechnol. 2006;24:856. doi: 10.1038/nbt1225. [DOI] [PubMed] [Google Scholar]; b Trepanier J, Tanner JE, Momparler RL, Le ON, Alvarez F, Alfieri C. J. Viral. Hepat. 2006;13:131. doi: 10.1111/j.1365-2893.2005.00684.x. [DOI] [PubMed] [Google Scholar]
- 27.Eder PS, DeVine RJ, Dagle JM, Walder JA. Antisense Res. Dev. 1991;1:141. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- 28.Abdelgany A, Wood M, Beeson D. J. Gene Med. 2007;9:727. doi: 10.1002/jgm.1061. [DOI] [PubMed] [Google Scholar]
- 29.Yang YS, Hughes TE. BioTechniques. 2001;31:1036. doi: 10.2144/01315st03. [DOI] [PubMed] [Google Scholar]
- 30.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D'Amato RJ, Mulligan RC. Nature. 2004;431:471. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 31.Dash P, Lotan I, Knapp M, Kandel ER, Goelet P. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7896. doi: 10.1073/pnas.84.22.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.a Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Nucleic Acids Res. 2008;36:559. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shestopalov IA, Sinha S, Chen JK. Nat. Chem. Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]; c Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. J. Am. Chem. Soc. 2007;129:11000. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 33.Shah S, Rangarajan S, Friedman SH. Angew. Chem. Int. Ed. 2005;44:1328. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 34.a Shah S, Friedman SH. Oligonucleotides. 2007;17:35. doi: 10.1089/oli.2006.0067. [DOI] [PubMed] [Google Scholar]; b Shah S, Jain PK, Kala A, Karunakaran D, Friedman SH. Nucleic Acids Res. 2009;37:4508. doi: 10.1093/nar/gkp415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK. J. Am. Chem. Soc. 2009;131:13255. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards JL, Tang X, Turetsky A, Dmochowski IJ. Bioorg. Med. Chem. Lett. 2008;18:6255. doi: 10.1016/j.bmcl.2008.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blidner RA, Svoboda KR, Hammer RP, Monroe WT. Mol. Biosyst. 2008;4:431. doi: 10.1039/b801532e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.