Summary
Membrane-anchored C-peptides (e.g. maC46) derived from human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein gp41 effectively inhibit HIV-1 entry in cell lines and primary human CD4+ cells in vitro. Here we evaluated this gene therapy approach in animal models of AIDS. We adapted the HIV gp41-derived maC46 vector construct for use in rhesus monkeys. Simian immunodeficiency virus (SIV and SHIV) sequence-adapted maC46 peptides, and the original HIV-1 derived maC46 expressed on the surface of established cell lines blocked entry of HIV-1, SIVmac251, and SHIV89.6P. Furthermore, primary rhesus monkey CD4+ T cells expressing HIV sequence-based maC46 peptides were also protected from SIV entry. Depletion of CD8+ T cells from PBMCs enhanced the yield of maC46-transduced CD4+ T cells. Supplementation with IL-2 increased transduction efficiency, whereas IL-7 and/or IL-15 provided no additional benefit. Phenotypic analysis showed that maC46-transduced and expanded cells were predominantly central memory CD4+ T cells that expressed low levels of CCR5 and slightly elevated levels of CD62L, β7-integrin, and CXCR4. These findings show that maC46-based cell surface-expressed peptides can efficiently inhibit primate immunodeficiency virus infection, and therefore serve as the basis for evaluation of this gene therapy approach in an animal model for AIDS.
Keywords: gene therapy, CD4+ T cells, rhesus monkey, AIDS, SIV
INTRODUCTION
Although combinations of antiretroviral drugs can control infection with human immunodeficiency virus (HIV) in many patients, mutations that confer drug resistance can arise. Furthermore, antiretroviral drugs can have serious toxicities, and they are costly. Gene therapy may offer an alternative to antiretroviral drugs as an effective treatment for HIV/AIDS. Genes can be introduced into cells ex vivo to advance a number of novel strategies that make cells more resistant to HIV. Such approaches target critical steps in the virus life cycle, from blocking virus entry into cells to interfering with the regulation of viral gene expression, as well as priming the immune system.1-3
Introducing genes that limit early steps in viral replication, like virus entry into CD4+ T cells, has the potential to select and expand HIV target cells that are resistant to infection.2,4 Peptide-based fusion inhibitors derived from HIV gp41 C-terminal hydrophobic alpha-helix inhibit virus fusion by interacting with the N-terminal hydrophobic alpha-helix. Thus, they block conformational changes essential for membrane fusion of the virus with the host cell.5 Peptides derived from heptad repeats (HR) such as T20, a 36-amino acid peptide (aa 638-673), disrupt the HIV gp41 conformational changes associated with membrane fusion. They are the basis of a new class of antiretroviral drugs.6 These peptides have poor oral bioavailability, a short serum half-life, and large amounts of these peptides are needed for an antiretroviral effect. Mutations in T20 emerge in the N- and C-terminal hydrophobic alpha-helixes that confer resistance to this class of antiretroviral drugs.7 In contrast, maC-peptide levels are not dependent on application intervals or restricted in length by the in vitro peptide synthesis process. The larger interacting surface of longer maC-peptides with the target domain of the HIV-1 gp41 envelope glycoprotein decreases the likelihood of resistance to this class of inhibitors.
Intracellular immunization of the PM-1 CD4+ T helper cell line with the M87 construct, a retroviral vector that expresses membrane-anchored T20, inhibited HIV replication at the level of virus entry.8 Optimization of the retroviral vector, the scaffold for presentation of the peptide on the cell surface and elongation of the C36- to a C46-peptide (M87o) have maximized expression of maC46 on PM-1 cells and primary lymphocytes. In addition antiretroviral activity against a broad range of viral isolates was enhanced.9 The improved M87o construct also minimized the emergence of HIV T20 escape mutants and inhibited their entry into human primary CD4+ T cells (9,10 and unpublished data). In a proof-of-concept study, M87o-transduced autologous CD8-depleted T lymphocytes were transfused into HIV-infected patients and could be detected in blood throughout the 1-year follow-up.11 Although transfer of the gene-modified T cells was safe, clinical efficacy was not established.
An animal model for AIDS, the simian immunodeficiency virus (SIV)-infected rhesus monkey, provides for pre-clinical testing of gene-therapy techniques. Here, we show that expression of maC46 on PM-1 cells blocks entry of the primate immunodeficiency viruses SHIV89.6P and SIVmac251. Furthermore, we demonstrate the potency of this gene therapy approach in blocking entry of SIVmac251 into maC46-expressing rhesus monkey CD4+ T cells. maC46-transduced primary T cells were predominantly central memory CD28+ CD95+ CD4+ T cells (CM) expressing low levels of CCR5. In addition, levels of CD62L, β7-integrin and CXCR4 were slightly elevated on the surface of these maC46-transduced cells. Our findings show that maC46-based cell surface-expressed peptides can efficiently inhibit primate immunodeficiency virus infection.
RESULTS
Sequence heterogeneity of the gp41 HR2 region in lentivirus infection
To evaluate the potential use of maC46 in an animal model for AIDS, we analyzed the deduced amino acid sequence homology among different C46 HIV gp41 C-terminal hydrophobic alpha-helix sequences in the HIV sequence database (http://www.hiv.lanl.gov/content/hiv-db/mainpage.html), including those from HIV-1 clade B (n = 176), SHIV (n = 11), and SIV (n = 22). We analyzed consensus sequences of the C46 region of the env gene obtained over time from 6 additional SIV-infected monkeys.12
Diversity of the C46 peptide region of gp41 was highest among HIV-1 clade B viruses (Figure 1A). The deduced amino acid changes predominated at the beginning of the C46 peptide. Nevertheless, the predicted contact sites with HR1 of the C46 peptide,5,13,14 the a and d positions, were conserved. In SHIV a similar pattern of changes in gp41 HR2 was observed (Figure 1B). Amino acid changes clustered at the N-terminus and the contact positions were conserved. In contrast, the C46 region of the relatively few SIV isolates was highly conserved (Figure 1C), possibly because sequence changes in SIV HR2 frequently disturb the six-helix bundle formation.15
Figure 1.
Sequence variability of HIV, SHIV and SIV envelope protein in the C46 region of the HR2 of gp41 Env. HIV sequences were derived from 176 HIV-1 clade B strains (A) and compared to the HXB2 envelope region (aa 117-163 of gp41). SHIV sequences were derived from 13 rhesus monkeys infected with SHIV (B) and compared to the SHIV89.6P gp41 region (aa 117-163). SIV sequences from 27 rhesus monkeys infected with SIVmac239 or SIVmac251 (C) were compared to gp41 of SIVmac239 (aa 112-157 of gp41). The bars represent the percent amino acid change at a specific position and are color coded for each amino acid.
Design of retroviral vectors
The M87o-based gammaretrovirus vectors (M87oHIV) were prepared using a strategy published previously.9 Briefly, all retroviral vectors had flanking long terminal repeats (LTRs), an MP71 leader sequence16 and an anti-HIV protein module with a signal peptide (SP), the C46 peptide fused to the hinge and linker domain and the membrane-spanning domain (MSD). We generated a set of maC46 low and high expressing vectors to determine whether inhibition of virus entry by membrane-anchored C46 (maC46) is dose-dependent. The low expressing vectors were generated by introducing a neomycin resistance gene under control of a polio internal ribosome entry site (IRES) behind the C46 module. The high expressing vectors carried two RNA elements in their 3' untranslated region (UTR), a rev responsive element (RRE) decoy17 as a second antiviral principle, and a woodchuck hepatitis post-transcriptional regulatory element (wPRE)18,19 to enhance transgene expression (Table 1). The amino acid sequences of C46 modules used in different vectors were derived from gp41 sequences obtained from HIV-1 HXB2, SHIV89.6P and SIVmac239 (Table 2A). Alanine-containing point mutations introduced into the C46 HIV sequence of M87oHIV generated the inactive C46 sequence. The targeted amino acid positions are essential for the interaction of HR2 C-domains with the N-domains of HR1 and formation of the entry-mediating six-helix bundle. The M87oSIV vector was adapted from the gp41 SIVmac239 wild-type sequence (KLNSWN) to the binding epitope of the broadly neutralizing anti-HIV antibody 2F5 (ELDKWA) permitting detection by flow cytometry using the 2F5 antibody.
Table 1.
Design of antiviral vectors derived from M87o and control vector.
| Vector | Leader regiona | C46 sequence | Hinge | Membrane spanning domain | Marker | Cis element | Expression PM-1/PBMC |
|---|---|---|---|---|---|---|---|
| M87oHIV | MP71 | HIV-1 HXB2 | Human IgG2 | Human CD34 | --- | wPRE/RRE | +++/+++ |
| M87oSHIV | MP71 | SHIV89.6P | Human IgG2 | Human CD34 | --- | wPRE/RRE | +++/+++ |
| M87oSIV | MP71 | SIVmac239 | Human IgG2 | Human CD34 | --- | wPRE/RRE | +++/+++ |
| M87oHIV low | MP71 | HIV-1 HXB2 | Human IgG2 | Human CD34 | IRES Neo | --- | +/n.d. |
| M87oSHIV low | MP71 | SHIV89.6P | Human IgG2 | Human CD34 | IRES Neo | --- | +/n.d. |
| M87oSIV low | MP71 | SIVmac239 | Human IgG2 | Human CD34 | IRES Neo | --- | +/n.d. |
| Neo control | MP1 | --- | --- | --- | IRES Neo | --- | n.a./n.a. |
| Inactive C46 | MP91 | mutated + 2F5 epitope | Human IgG2 | Human CD34 | --- | --- | +++/n.d. |
Table 2.
C46 sequences (A) and C46 sequence homology between HIV, SHIV, and SIV compared to M87o vectors (B)
| A | |
|---|---|
| M87oHIV | WMEWDREINNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWF |
| M87oSHIV | ----E---D---DY-YD-L-K--T------K---ELDKWA------ |
| M87oSIV | -Q--E-KVDFLEEN-TA-L--A-I-----MY---ELDKLWAVGF--- |
| SIVmac239 wt seq | -Q--E-KVDFLEEN-TA-L--A-I-----MY--QKLNSWDVFG--- |
| Inactive C46 | A--A---A--A--AA--AA---------------ELDKWA-AA-A- |
| Predicted aa contact sites between C4 6 and HR1 | abcdefgabcdefgabcdefgabcdefgabcdef |
| B | |||
|---|---|---|---|
| M87oHIV |
M87oSHIV |
M87oSIV |
|
| HIV-1 HXB2/NL4-3 | 100% | 91.1% | 65.8% |
| SHIV89.6P | 91.1% | 100% | 68.2% |
| SIVmac251 | 56.6% | 59.1% | 95.0% |
Inhibition of primate immunodeficiency virus entry into PM-1 cells by membrane-anchored C46
We have shown that cell surface-expressed maC46 can efficiently inhibit HIV entry in in vitro culture.9 Here we expanded the repertoire of HIV-based C46 vectors (M87oHIV) to vectors that express the C46 sequences adapted from SIVmac239 (M87oSIV) and SHIV89.6P (M87oSHIV), viruses that are commonly used in rhesus monkey experiments.
We first determined the efficacy of M87oHIV, M87oSIV, and M87oSHIV, in blocking the infection of transduced PM-1 cells with HIV-1 NL4-3, SIVmac251, and SHIV89.6P, respectively (Figure 2). The C46 region consensus sequence of the uncloned virus SIVmac251 is homologous to SIVmac239. As expected, PM-1 cells transduced with M87oHIV were completely resistant to infection with HIV (Figure 2A). Likewise, PM-1 cells transduced with M87oSHIV were resistant to infection with SHIV89.6P (Figure 2E), and PM-1 cells transduced with M87oSIV were resistant to infection with SIV (Figure 2I). Despite low sequence homology of HIV and SHIV C46 compared to SIVmac239 C46 (Table 2B), all vectors showed a similarly high efficiency of entry inhibition in this assay. Specifically, M87oHIV also inhibited entry of SHIV89.6P (Figure 2D) and SIVmac251 (Figure 2G), M87oSHIV also blocked entry of HIV (Figure 2B) and SIV (Figure 2H), and M87oSIV also impeded entry of HIV (Figure 2C) and SHIV89.6P (Figure 2F). The potential contribution of the RRE was not measurable with this assay since replication was almost completely inhibited. Also, in previous experiments we did not observe a different inhibitory capacity of RRE expressing and non-expressing vectors.9
Figure 2.
Inhibition of replication of HIV, SIV and SHIV in C46-expressing cell lines. PM-1 cells were transduced with retroviral vectors encoding for the antiviral maC46 sequence derived from HIV, SIV, or SHIV (A, D, and G: M87oHIV; B, E, and H: M87oSHIV; C, F, and I: M87oSIV). PM-1 cells transduced with the inactivate form of the C46 peptide were used as control cells. PM-1 cells were infected with HIV-1 NL4-3 (A-C), SIVmac251 (D-F), or SHIV89.6P (G-I) and the amount of p24 or p27 antigen in culture supernatants was determined by ELISA. The data shown are representative graphs from three experiments that were each performed in duplicate.
Given that the efficiency of virus entry inhibition is dependent on the level of maC46 cell surface expression (F.G. Hermann et al., unpublished observations), we next sought to determine whether low-level cell surface expression of maC46 could still cross-protect against infection with a heterologous virus. In contrast to maC46 high expressing vectors (M87oSIV, M87oSHIV and M87oHIV), cell surface expression of maC46 on PM-1 cells transduced with M87oSIV low, M87oSHIV low and M87oHIV low was significantly reduced (Figure 3A).
Figure 3.
Efficacy of entry inhibition of HIV, SIV and SHIV afforded by low and high maC46-expressing variants of retroviral vectors. PM-1 cell lines expressing high levels of maC46 were generated by fluorescence activated cell sorting. PM-1 cell lines with a low expression of maC46 were generated by selection with G418. Control PM-1 cells were transduced with a retroviral vector coding for IRES-Neo and selected with G418. (A) Expression level of maC46 was detected by 2F5 mAb staining. (B-D) Relative level of transduction compared to control PM-1 cells. Lentiviruses coding for eGFP were either pseudotyped with HIV-1 JRFL, SIVmac251 or SHIV89.6P envelope protein. Transduction efficacy was determined by measuring eGFP expression by flow cytometry.
For the challenge we used eGFP-expressing replication incompetent lentiviruses that were pseudotyped with HIV-1 JRFL, SHIV89.6P or SIVmac251 envelope proteins in single-round infection experiments. Single-round infection allowed for precise determination of the entry inhibitory potency of maC46 high and low expressing vectors. eGFP expression of these lentiviruses was RRE independent. As expected, the high expression vectors showed complete cross-inhibition of entry mediated through heterologous lentiviral envelopes (Figures 3B, 3C, and 3D). In contrast, PM-1 cell lines transduced with low expressing C46 vectors did not completely block virus entry. There was an association between the degree of protection against virus entry and the relative amount of cell surface expression of maC46 (Figures 3B and 3C). As shown in Figure 3A, the 2F5 binding was slightly different in the low expressing C46 variants of M87o (M87oHIV low: brightest 2F5 staining; M87oSHIV low: intermediate staining; and M87oSIV low: barely above background). Interestingly, almost no protection was seen in PM-1 cells expressing either one of the three low expressing maC46 variants (Figure 3D) when challenged with SIVmac251 pseudotyped lentivirus. Thus, adapting the C46 to the SIV sequence did not improve antiviral activity against homologous SIV strains.
Inhibition of SIV entry into primary rhesus monkey CD4+ T cells transduced with maC46
Primary human CD4+ T cells transduced with M87oHIV express maC46 at high levels, and therefore efficiently inhibit virus entry into the cell.9 Primary rhesus monkey peripheral blood CD4+ T cells transduced with M87oHIV express maC46 at high levels as well (Supplementary Figure 1). We then investigated whether rhesus monkey primary CD4+ T cells transduced with M87oHIV were, like PM-1 cells transduced with M87oHIV, protected against SIV infection (Figure 4).
Figure 4.
maC46 prevents infection of rhesus monkey CD4+ T cells with SIVmac251. Peripheral blood T lymphocytes from a naïve rhesus monkey were either transduced with the inactive C46 control vector or with M87oHIV and infected with SIVmac251. The expression of the maC46 peptide was determined by 2F5 mAb staining. The percentage of SIVmac251 infected cells was determined by anti-SIV p27 mAb staining. (A) Background p27 staining in non-transduced and C46 control vector-transduced, uninfected CD4+ T cells. (B) Detection of p27 antigen in non-transduced and C46 control vector transduced SIVmac251-infected CD4+ T cells. (C) Detection of p27 antigen in non-transduced and M87oHIV vector transduced, SIVmac251-infected CD4+ T cells. The percentage of p27 positive cells is indicated in each graph (top: transduced cells; bottom: non-transduced cells).
We observed low background SIV p27 Gag antigen staining in uninfected primary rhesus monkey CD4+ T cells whether the cells were or were not transduced with the inactive maC46 vector (Figure 4A). When these cells were infected with SIVmac251 a similar level of p27 antigen was detected in both non-transduced cells, and cells that were transduced with the inactive C46 vector (Figure 4B). Primary rhesus monkey CD4+ T cells transduced with M87oHIV expressed maC46 at high levels, and were protected from infection with SIVmac251 (Figure 4C). In contrast, non-transduced cells in the same culture had relatively high p27 staining and were not protected against SIV entry (Figure 4C). High maC46 expression completely abolished SIV entry in the single round infection assay. We, therefore, assume that the RRE element did not contribute to the absent virus replication as determined by p27 staining. Since M87oHIV was capable of blocking entry and replication of SIVmac251 in primary rhesus monkey CD4+ T cells and PM-1 cells, we decided to use this construct in the experiments described below.
Optimized transduction of primary rhesus monkey CD4+ T cells
Our eventual goal is to test the potential benefit of adoptive transfer of autologous, M87oHIV-transduced CD4+ T cells into SHIV- and SIV-infected animals to modulate infection and prevent disease progression. The large number of gene-modified T cells that will be needed for these experiments necessitated optimization of the in vitro transduction and culture conditions. Primary human PBMCs (both CD4+ and CD8+ T cells) stimulated with anti-CD3/anti-CD28 coated paramagnetic beads in the presence of an M87o-based vector are transduced at a relatively high efficiency (>50%) and expand rapidly in culture. Primary rhesus monkey CD4+ T cells were less efficiently stimulated, however, and were therefore at a relatively low efficiency (<20%, Figure 5B).
Figure 5.
Expansion, M87oHIV transduction and phenotype of maC46-transduced CD4+ T cells. CD8+ lymphocyte-depleted and total lymphocytes obtained from peripheral blood were transduced with the M87oHIV retroviral vector and cultured with anti-CD3 and anti-CD28 coated beads for 7 days. (A) Rate of expansion of CD4+ T cells 7 days after initiation of in vitro culture. Cell counts were determined by hemacytometer. The black bar represents the bulk culture, the white bar the CD8+ cell-depleted culture. (B) Transduction efficacy of CD4+ T cells with the retroviral vector M87oHIV. The cells were transduced twice with a multiplicity of infection (MOI) of 1.3-2 or more than a MOI of 3.3. M87oHIV expression was determined by staining with a C46 specific mAb. The black bar represents the bulk culture, the white bar the CD8+ cell-depleted culture. (C) and (D) Phenotype of expanded CD4+ T cells. On day 7 after culture, cells were stained with anti-CD28 and anti-CD95 mAbs to determine T cell subsets and with 2F5 to determine C46 expression. The black bars represent M87oHIV-expressing cells, the white bars M87oHIV negative cells. The bars in all graphs represent median values, error bars the interquartile range (n = 4-10).
To optimize the transduction efficiency of primary rhesus monkey CD4+ T cells, we depleted peripheral blood mononuclear cells of CD8+ lymphocytes with anti-CD8 coupled paramagnetic beads prior to stimulation. Following 7 days of culture, we observed reduced numbers of expanded CD4+ T cells in the CD8+ lymphocyte-depleted cultures relative to the bulk cultures (Figure 5A). With CD8+ T cell depletion however, the efficiency of transduction with an M87o-based vector increased from 20% to 35% (MOI of more than 3.3) and from 7% to 25% (MOI of 1.3-2); Figure 5B).
The phenotype of primary rhesus monkey CD4+ T cells that express membrane-anchored C46
To evaluate if the CD8+ cell depletion would potentially alter the phenotype of expanded and transduced CD4+ T cells, we utilized anti-CD28 and anti-CD95 mAbs to determine the relative percentage of maturation-associated T cell subsets. Specifically the percentage of naïve CD4+ T cells (CD28+ CD95-), central memory (CM) CD4+ T cells (CD28+ CD95+) and effector memory (EM) CD4+ T cells (CD28- CD95+).20 Because the anti-CD3/anti-CD28 coated beads were removed before staining, there was no inhibition of the anti-CD28 antibody. Moreover, the anti-CD3/anti-CD28 coated beads do not shed their anti-CD28 antibodies.21 CD8+ lymphocyte depletion prior to expansion and transduction did not alter the CD4+ T cell phenotype (Figures 5C and 5D). As expected, CD4+ T cells under both culture conditions consisted mainly of CM T cells and up to 13% EM T cells. No naïve CD4+ T cells were found. M87oHIV-transduced cells did not show a different phenotype compared to non-transduced cells.
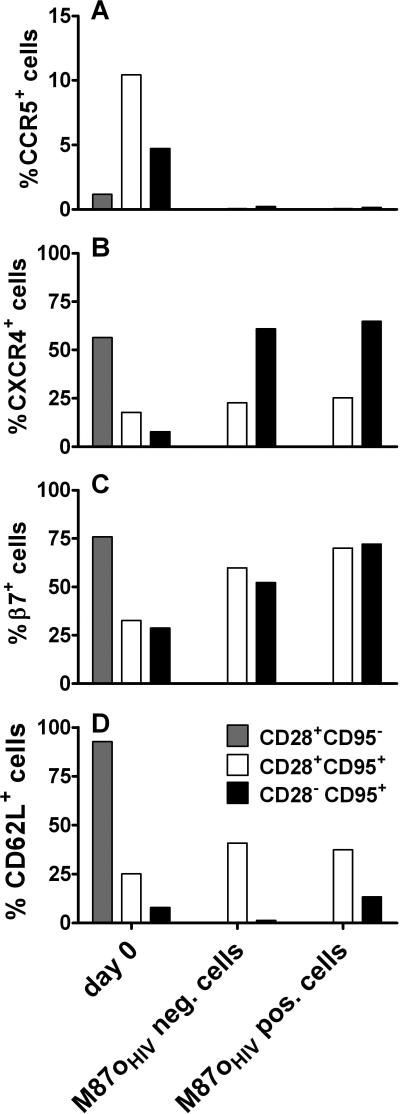
We next divided the CD4+ T cells (both transduced and not) into maturation-associated cell subsets (i.e., naïve, CM, and EM) and determined the percentage of cells that expressed CCR5, CXCR4, β7, and CD62L in each of these cell subsets. Like others,22 we observed that stimulation of CD4+ T cells with anti-CD3/anti-CD28 coated beads and expansion in culture markedly reduced expression of CCR5 on the cell surface (Figure 6A). The percentage of CCR5+ cells was marginally higher for CM CD4+ T cells transduced with M87oHIV (median: 0.06 (range: 0.01-2.47)) than for non-transduced cells (median: 0.05 (range: 0.02-3.57)).
Figure 6.
Phenotype of M87oHIV-transduced CD4+ T cells. Peripheral blood from 4 or 5 naïve rhesus monkeys was used to determine the expression of CCR5 (A), CXCR4 (B), β7-integrin (C) and CD62L (D) on CD4+ T cell subsets prior to transduction and on day 7 after transduction and in vitro culture. The CD28 and CD95 paradigm was used to determine the lymphocyte maturation-associated T cell subsets. PBMC separated by Ficoll gradient were used to determine cell surface expression of β7-integrin, CXCR4 and CD62L on day 0. The cell surface expression of CCR5 on day 0 was determined by whole blood staining. The bars in all graphs represent median values.
Unlike CCR5, CXCR4 was highly expressed on the surface of freshly isolated primary naïve CD4+ T cells. Levels of CXCR4 were lower on CM and EM CD4+ T cells (Figure 6B). Expression of CXCR4 on the surface of CM CD4+ T cells did not significantly change following transduction and 7 days in culture. Interestingly, the level of CXCR4 expression on expanded EM CD4+
T cells after 7 days of culture was the same as observed on freshly isolated naïve CD4+ T cells. Expression of maC46 did not affect CXCR4 cell surface expression.
Adhesion-associated molecules, such as the integrin molecule β7, mediate lymphocyte homing to gut-associated lymphoid tissue (GALT). Because GALT is a principal site for primate immunodeficiency virus replication, retention of β7 integrin on CD4+ T cells transduced with M87o constructs to sustain trafficking to the gut is critical. For freshly isolated lymphocytes, the highest level of β7 expression was found on naïve cells; CM and EM CD4+ T cells had lower levels of β7 expression (Figure 6C). Following culture, β7 expression on CM and EM T cells was increased regardless of maC46 peptide expression.
Finally, we analyzed cell surface expression of the homing receptor CD62L. This receptor enables lymphocytes to traverse high endothelial venules and enter lymph nodes. As expected, freshly isolated naïve CD4+ T cells nearly uniformly expressed high levels of CD62L, whereas CM cells expressed an intermediate level and EM CD4+ T cells expressed very low levels of CD62L (Figure 6D). Stimulation of cells for 7 days in culture led to increased CD62L surface expression on CM CD4+ T cells. No difference in expression of the CD62L molecule was observed between cells transduced with M87oHIV and non-transduced cells after stimulation. In contrast, non-transduced stimulated EM CD4+ T cells showed decreased CD62L expression. Compared to freshly isolated EM CD4+ T cells, cells transduced with M87oHIV and stimulated in culture had marginally elevated CD62L expression.
Primary rhesus monkey CD4+ T cells propagated and transduced with cellular growth factors
To further optimize stimulation and transduction of autologous rhesus monkey CD4+ T cells ex vivo, we investigated the potential of cytokines to enhance the stimulation and transduction. IL-2, IL-7 and IL-15 play an essential role in T cell homeostasis in vivo.23 IL-2 can potently induce T cell expansion in vitro. This growth factor can activate, stimulate, and sustain growth of T cells, independent of antigen. IL-7 promotes survival and homeostatic proliferation of memory T cells; when used in combination with IL-15 this activity increases.22,24 Accordingly, we tested these cytokines alone and in combination to achieve the maximum effect on primary lymphocyte transduction and expansion.
Primary rhesus monkey lymphocytes cultured in media supplemented with human recombinant IL-2 (40 U/ml), rhesus recombinant IL-7 (100 U/ml), and rhesus recombinant IL-15 (100 U/ml), alone and in combination. A control culture without any of these cytokines was also included. Cell culture supernatants supplemented with fresh cytokines were partially exchanged daily during days 4 through 6 of culture. Expansion of CD4+ T cells was measured after day 7 (Figure 7A).
Figure 7.
Effect of different cytokines on efficacy of expansion and level of transduction of CD4+ T cells. PBMC were depleted of CD8+ cells and expanded for 7 days with anti-CD3 and anti-CD28-coated paramagnetic beads. The different cytokines or combinations of cytokines were added on day 0. Cell culture supernatants were partially replaced with fresh media containing the different cytokines on days 4, 5 and 6. On days 4 and 5 cells were transduced with M87oHIV retroviral vectors. (A) Fold expansion of CD4+ T cells expanded in different cytokines or cytokine combinations relative to IL-2 treated cells that were set to an expansion factor of 1. (B) M87oHIV transduction efficacy of CD4+ T cells expanded in different cytokines or cytokine combinations. The bars represent median values from 5 animals. (C-F) Side and Forward Scatter of one representative dot plot of CD4+ T cells expanded in media without cytokines (C), supplemented with IL-2 (D), IL-7 (E), and IL-7 + IL-15 (F).
Compared to IL-2 alone, all cytokines and cytokine combinations produced comparable expansion and proliferation, with the exception of IL-7 alone, which was lower. As shown in Figure 7B, primary rhesus monkey lymphocytes propagated in IL-7 containing media and media without cytokines had dramatically reduced proliferation and transduction efficacy (i.e., a median of 10% compared to 38% for IL-2 stimulated cultures). IL-15 when used alone or in combination with IL-7 supported only a relatively low transduction level (median: 17% and 24%, respectively). Cultures containing IL-2 had the highest maC46 transduction efficiency, reaching 38%.
Lymphocyte activation was monitored using a FSC/SSC dot plot. Cells cultured in the presence of IL-2 were larger than cells cultured in the presence of IL-7 or in the absence of any cytokine (Figure 7D, 7E and 7C). Regardless of which cytokines were used in culture, the memory phenotype was preserved (Supplementary Figure 2).
DISCUSSION
In this study, we show that gene-modified primary rhesus monkey CD4+ T cells that express maC46 peptides inhibit primate immunodeficiency virus entry and confer protection against infection. Adaptation of the HIV-based maC46-peptide to the corresponding sequence of the SIV or SHIV C-terminal heptad repeat in gp41, however, did not improve antiviral activity against these monkey immunodeficiency viruses. Therefore, we optimized the conditions for the transduction of primary rhesus monkey CD4+ T cells with the original construct M87oHIV, with respect to high transduction levels, and expansion ex vivo. These data support a proof-of-concept study for engineering rhesus monkey lymphocytes that cannot be infected by SIV.
Although the HIV C46 peptide shares 91.1% identity with the corresponding SHIV C46 peptide and 56.6% with the SIV C46 peptide, each of the three C46 sequence variants cross-inhibited entry of the other two viruses. Primate immunodeficiency viruses share the same envelope protein structure25,26 including the N- and C-terminal hydrophobic alpha-helixes27 that form a six-helix bundle during virus fusion with the cell membrane. Despite limited deduced amino acid sequence homology among different primate immunodeficiency viruses (Table 2B), the principle alpha-helical structures are preserved.28 Furthermore, the essential amino acid contact residues between the N-heptad repeat and the C46 sequence within the C-heptad repeat29 are conserved amongst different clades of HIV.30 Thus, the cross-reactivity among the three vectors in inhibiting entry of primate immunodeficiency viruses is likely determined by the ability of the peptide to intercalate into the three dimensional structure of the peptide, and not the primary amino acid sequence per se.
Single-round infection of PM-1 cells that express low-level membrane-anchored C-peptide showed distinct differences in the extent of inhibition of different primate immunodeficiency viruses. Specifically, SIV pseudotyped virions were inhibited to a far lesser extent by all three maC46 when expressed at low levels. In contrast, SHIV and HIV pseudotyped virions were inhibited by the different maC46 peptides in a concentration-dependent manner. Thus, the threshold for entry inhibition seems to be higher for SIV envelope-mediated entry than for the other two viral envelopes. Others have reported weak inhibition of SIV by soluble HIV C34 peptides as well.28,31 After binding of soluble CD4, exposure of the pre-hairpin loop is longer for HIV than for SIV.31 Accordingly, the shorter pre-hairpin exposure time for SIV envelope gives the C-peptides a shorter window of time in which they can bind to the N-heptad repeat to prevent formation of the six-helix bundle. HIV-2 derived C-peptides have a greatly enhanced SIV inhibition in soluble form.28 These soluble peptides might overcome the inferior inhibition of SIV as membrane-anchored peptides although membrane-anchored C-peptides have a slightly different inhibition kinetics than soluble C-peptides.32
Despite these findings, we observed a nearly complete inhibition of SIV entry by HIV maC46 expressed on the surface of primary rhesus monkey CD4+ T cells (Figure 4). This inhibition was likely due to the relatively high expression of maC46 on rhesus monkey CD4+ T cells (Supplementary Figure 1), which was comparable to the amount of maC46 expression on human cells. The reduced CCR5 expression that we detected in maC46-transduced cells might also enhance the relative resistance against viral entry. However, low CCR5 expression on transduced and expanded cells alone was not sufficient to prevent viral entry in vitro since inactive vector-transduced cells could readily be infected. Previous reports indicate that primate immunodeficiency virus-infected humans or non-human primates may benefit from adoptive CD4+ T cell transfer of expanded CCR5 negative non gene-modified cells.39,40 Further in vivo benefit should be gained when these cells also express maC46 HIV on their surface, and are therefore protected from AIDS virus entry.
The original HIV maC46 is sufficient to inhibit SIV entry into rhesus monkey CD4+ T cells, provided transgene expression reaches a high level. However, integrated retroviral vectors can be silenced in long term cultures and in vivo.33-36 So far, we have not observed this effect in in vitro cultures of HIV C46-expressing CD4+ T cells (data not shown). Also, murine hematopoietic stem cells transduced with M87 HIV and transplanted into conditioned mice, showed no silencing of HIV C46 expression within 12 months.37 Nevertheless, further in vivo studies are warranted to show if sufficiently high maC46 expression levels on CD4+ T cells can be achieved to efficiently block SIV entry into cells in SIV-infected rhesus monkeys.
For future in vivo studies, large quantities of M87oHIV-transduced rhesus monkey CD4+ T cells will be necessary. Accordingly, we optimized the protocol for obtaining a high yield of M87oHIV gene-modified cells by varying the culture conditions. We did not culture the cells for more than 7 days in vitro to prevent exhaustion and ensure better engraftment of gene-modified cells in future in vivo studies. Prior CD8+ cell depletion increased the number of CD4+ T cells transduced by M87oHIV, probably by higher virus to CD4+ T cell ratio. The addition of IL-7 and/or IL-15 to the culture did not improve transduction efficacy. Nevertheless, at this time we do not completely rule out that other cytokines, in addition to IL-2 (e.g. IL-7 and IL-15), might have a potential benefit for the functionality of expanded lymphocytes used for in vivo experiments in rhesus monkeys. Although the transduction efficiency in IL-7- and/or IL-15-stimulated cultures was lower as compared to IL-2, the relative cell size of these transduced cells was smaller, indicating a relatively lower level of activation (Figure 7F). The smaller cell size however, could potentially be an advantage, since relatively quiescent cells may be less likely to be retained in small vessels or undergo apoptosis in vivo.
We predict that a substantial percentage of expanded and gene-modified CD4+ T cells will home to lymphatic organs and the intestine, since a significant percentage of these cells expressed CD62L (40%) and β7 integrin (60%). Future in vivo studies will have to demonstrate whether cells that express maC46 can replenish virus-depleted CD4+ T cells and reconstitute the local lymphatic structures in an animal model for AIDS.
MATERIALS AND METHODS
Animals
We obtained EDTA anti-coagulated blood samples from 12 virus naïve rhesus monkeys (Macaca mulatta). All animals were maintained in accordance with the guidelines of the Committee on Animals for the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals.41
Viruses
Primate immunodeficiency viruses used in this study included SIVmac25143, SHIV89.6P44,45, and HIV-1 NL4-3.42 SIVmac251 and SHIV89.6P were propagated in PM-1 cells. HIV-1 NL4-3 was expanded in human PBMC cultures. Virus titers in culture supernatants of SIVmac251 and SHIV89.6P, and HIV-1 NL4-3 were measured by a p27 enzyme-linked immunosorbent assay (ELISA) and an HIV p24 focus-forming assay on U87 cells,46 respectively. The identity of the viruses was verified by analysis of coreceptor usage and sequencing of the env gene.
Cell lines
The PM-1 cell line, kindly provided by Buchacher and coworkers,47 was cultured in RPMI-1640 medium supplemented with 10% FCS and 2% Glutamine. The Phoenix packaging cells, generously provided by G. P. Nolan,48 and the 293T cell line (ATCC) were cultured in DMEM medium supplemented with 10% FCS and 2% Glutamine.
Generation of high expressing retroviral vectors
Construction of the vector M87o-RRE and the Neo control have been described previously.8,9 M87o-RRE vector was engineered to contain the HIV C46 sequence (M87oHIV). Generation of the homologous M87oSIV and M87oSHIV as well as the low expressing vectors M87oHIV low, M87oSIV low, and M87oSHIV low is described in detail in the supplementary materials and methods.
Generation of cell lines expressing membrane-anchored peptides
Retroviral particles were produced by transfection of Phoenix packaging cells as described previously.48 Low multiplicities of infection (MOI) were used for transduction of PM-1 cells to obviate multiple vector integrations. Cells stained with the human monoclonal antibody (mAb) 2F5 (kindly provided by H. Katinger) directed against a motif in the C-peptide of gp418 and a monoclonal goat anti-human IgG antibody coupled to phycoerythrin (PE) (Jackson ImmunoResearch Laboratories, West Grove, PA) were analyzed by flow cytometry to assess transduction efficiency. PM-1 cells, transduced with the vectors that contain a neomycin resistance gene, were selected with G418 (0.8 mg/ml) for 10 days. Transduced cells without the neomycin resistance gene were enriched by fluorescence-activated cell sorting for C-peptide expression to more than 95% purity using a FACSCalibur (Becton-Dickinson, Heidelberg, Germany).
Inhibition of HIV, SIV and SHIV by membrane-anchored peptides expressed on PM-1 cells
The different PM-1 cell lines were plated in 24 well plates with a seeding density of 2×105 cells per well and subsequently infected with HIV-1 NL4-3 (MOI = 0.001), SIVmac251 (1.25 focus forming units (FFU)), and SHIV89.6P (92 FFU). About 24h after infection, cells were washed once with PBS. HIV p24 or SIV, SHIV p27 antigen was measured in the culture supernatants collected at different time points by ELISA (Innogenetics, Heiden, Germany).
Generation of lentiviral vector supernatants
The different primate immunodeficiency virus Env expression plasmids were generated from viral RNA or synthetic HIV-1 89.6 (SynGP160) in pcDNA3.1 kindly provided by R. Wagner (Regensburg, Germany). The env gene of SIVmac251 was amplified from viral RNA in the supernatants of infected PM-1 cells by reverse transcription-PCR using Superscript II (Invitrogen, Karlsruhe, Germany) with a specific primer SIV9319oR. The nested PCR with the Expand HiFidelity Plus PCR System (Roche, Mannheim, Germany) was performed using SIV9319oR and SIV6508oF for the first and SIVkozak6601iF and SIV9246iR for the second step. The PCR product DNA was sequenced and cloned into the pHCMV vector.50
The vector particles, pseudotyped with HIV-1 JRFL, SIVmac251 or SHIV89.6 Env were then generated by transient transfection of 293T cells as described51 with minor modifications. Specifically, we used the lentiviral transfer plasmid pHR9SIN-SEW,52 which has an enhanced green fluorescence protein (eGFP) marker gene (kindly provided by Dr. M. Grez, Georg-Speyer-Haus), for packaging. Virus particles in the culture supernatant were concentrated by ultracentrifugation (25,000 rpm, 2h, 4°C) and stored at -80°C.
Lentiviral single-round infection assay
The lentiviral transduction was carried out as described previously.51 Briefly, a 96-well plate was coated with 10 ng/μl fibronectin (Invitrogen, Karlsruhe, Germany) and then seeded with PM-1 cells transduced with the different C-peptide-encoding retroviral vectors or the control vector in triplicate (1×104 cells per well). The cells were transduced with replication-incompetent lentiviral particles (MOI 0.1 – 0.2) with HIV-1 JRFL, SIVmac251 or HIV-1 89.6 Env. After 5 days, the cells were stained for C46 expression using a biotinylated mAb 2F5 and Streptavidin-APC (Becton-Dickinson). About 7×104 viable cells per sample were analyzed by flow cytometry to determine the percentage of cells positive for expression of C46 and GFP.
Primary lymphocyte cultures and transduction of primary rhesus monkey T lymphocytes
Peripheral blood mononuclear cells (PBMC) were obtained from naïve rhesus monkeys by Ficoll gradient separation (PAA, Cölbe, Germany). In the case of CD8+ cell depletion, 107 PBMC were incubated for 30 min with anti-CD8 (cM-T807) coupled to epoxy activated M-450 beads (Invitrogen, Carlsbad, CA) in 1 ml PBS/2% HS at 4°C. The tube was placed in a magnet for 2 min to remove bead-bound CD8+ cells and the supernatant was used in subsequent steps. 1.5×106 cells were stimulated in 3 ml X-Vivo (containing 5% HS / 2% Glutamine / IL-2 at 40U/ml; IL-2 was provided by Hoffman-La Roche, Nutley, NJ) for 4 days with anti-CD3 (SP34; Becton Dickinson) and anti-CD28 (L293; Becton-Dickinson) coated epoxy activated M-450 beads at a cell-to-bead ratio of 1:3 in a 6-well plate. Cells were transduced 2 times on days 4 and 5 after isolation by transferring them onto retronectin (Takara; Japan) coated non-tissue culture plates preloaded with either M87oHIV or inactive C46 retroviral vector supernatants. Cells were further cultivated in X-Vivo (containing 5% HS / 2% Glutamine / IL-2 at 40 U/ml) at 37°C for 2 days and subsequently used for SIV infection or phenotypic characterization by flow cytometry. In some experiments, recombinant rhesus monkey IL-7 and/or IL-15 cytokines (100 U/ml each; Resource for Nonhuman Primate Immune Reagents, Atlanta, GA) were used instead of, or in combination with, recombinant human IL-2 (40 U/ml).
Inhibition of SIVmac251 in membrane-anchored peptide-expressing rhesus monkey T cells
4×106 prestimulated and transduced T cells were infected with 60 FFU SIVmac251 in 4 ml X-Vivo (containing 5 % HS / 2% Glutamine / IL-2 (40 U/ml)). Cells were washed with PBS 1 day after infection and re-stimulated with freshly isolated primary lymphocytes from another donor animal on day 4 post infection. The cells were stained 7 days after infection with anti-p27 (Tebu-bio, Offenbach, Germany), anti-CD3 (SP34, BD Biosciences, San Jose, CA), anti-CD4 (L200; BD Biosciences, San Jose, CA) and 2F5 antibody (as described above) and the expression was quantified by flow cytometry.
Phenotypic characterization of membrane-anchored peptide-expressing rhesus monkey T cells
To determine the phenotypic changes that may occur through the different culture conditions, we performed polychromatic flow cytometric assays on whole blood, freshly isolated lymphocytes prior to culture, and cultured lymphocytes 7 days after transduction. We determined the cell surface expression of the Phycoerytherin-conjugated monoclonal antibodies against CCR5 (3A9; Pharmingen, San Diego, CA), CD62L (SK11; Pharmingen, San Diego, CA), β7 (FIB504; Pharmingen, San Diego, CA), and CXCR4 (12G5; Pharmingen, San Diego, CA) using the following antibody panel: anti-CD3-AlexaFlour700 (SP34.2; BD Biosciences, San Jose, CA), anti-CD4-AmCyan (L200; BD Biosciences, San Jose, CA), anti-CD8-Allophycocyanin-Cy7 (SK1; BD Biosciences, San Jose, CA) anti-CD28-PerCP-Cy5.5 (CD28.2; Coulter, Miami, FL), and anti-CD95-Fluorescein isothiocyanate (DX2; Pharmingen, San Diego, CA). Transduction of cells was determined by staining with 2F5 (as described above). Cell surface expression of the different antigens was measured with a LSR II flow cytometer (Becton Dickinson, San Jose, CA).
DNA Sequences
One hundred seventy six HIV-1 clade B sequences were downloaded from the HIV sequence database (http://www.hiv.lanl.gov/content/hiv-db/mainpage.html; accession numbers are described in Supplementary Table 1). The SHIV and SIV sequences used for this study were from publicly available isolates in the HIV sequence Database (http://www.hiv.lanl.gov/content/hiv-db/mainpage.html), and for intra-host SIV sequence analysis, SIVmac samples were animal isolates from a recombination study which was published recently.12
Sequence analysis
C46 sequence analysis was performed using ClustalW.53 Assessment of degree of divergence and percent identity was performed. Divergence was calculated by comparing sequence pairs in relation to the phylogeny reconstructed by MegAlign (v7.1.0, DNASTAR, WI). Percent identity compares sequences directly, without accounting for phylogenetic relationships.
Supplementary Material
ACKNOWLEGMENTS
This work was supported by the German Government grant: BMBF TreatID and by the National Institute of Health grants AI060354, AI061797, CA73473 and RR00168.
Footnotes
Supplementary information is available at Gene Therapy's website.
REFFERENCES
- 1.Puls RL, Emery S. Therapeutic vaccination against HIV: current progress and future possibilities. Clin Sci. 2006;110:59–71. doi: 10.1042/CS20050157. [DOI] [PubMed] [Google Scholar]
- 2.von Laer D, Hasselmann S, Hasselmann K. Gene therapy for HIV infection: what does it need to make it work? J Gene Med. 2006;8:658–667. doi: 10.1002/jgm.908. [DOI] [PubMed] [Google Scholar]
- 3.Strayer DS, Akkina R, Bunnell BA, Dropulic B, Planelles V, Pomerantz RJ, et al. Current status of gene therapy strategies to treat HIV/AIDS. Mol Ther. 2005;11:823–842. doi: 10.1016/j.ymthe.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 4.von Laer D, Hasselmann S, Hasselmann K. Impact of gene-modified T cells on HIV infection dynamics. J Theor Biol. 2006;238:60–77. doi: 10.1016/j.jtbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Gallo SA, Finnegan CM, Viard M, Raviv Y, Dimitrov A, Rawat SS, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 6.Poveda E, Briz V, Soriano V. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 2005;7:139–147. [PubMed] [Google Scholar]
- 7.Baldwin C, Berkhout B. HIV-1 drug-resistance and drug-dependence. Retrovirology. 2007;4:78. doi: 10.1186/1742-4690-4-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildinger M, Dittmar MT, Schult-Dietrich P, Fehse B, Schnierle BS, Thaler S, et al. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J Virol. 2001;75:3038–3042. doi: 10.1128/JVI.75.6.3038-3042.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egelhofer M, Brandenburg G, Martinius H, Schult-Dietrich P, Melikyan G, Kunert R, et al. Inhibition of human immunodeficiency virus type 1 entry in cells expressing gp41-derived peptides. J Virol. 2004;78:568–575. doi: 10.1128/JVI.78.2.568-575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohrengel S, Hermann F, Hagmann I, Oberwinkler H, Scrivano L, Hoffmann C, et al. Determinants of human immunodeficiency virus type 1 resistance to membrane-anchored gp41-derived peptides. J Virol. 2005;79:10237–10246. doi: 10.1128/JVI.79.16.10237-10246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Lunzen J, Glaunsinger T, Stahmer I, von Baehr V, Baum C, Schilz A, et al. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- 12.Kim EY, Busch M, Abel K, Fritts L, Bustamante P, Stanton J, et al. Retroviral recombination in vivo: viral replication patterns and genetic structure of simian immunodeficiency virus (SIV) populations in rhesus macaques after simultaneous or sequential intravaginal inoculation with SIVmac239Deltavpx/Deltavpr and SIVmac239Deltanef. J Virol. 2005;79:4886–4895. doi: 10.1128/JVI.79.8.4886-4895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 14.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 15.Marti DN, Bjelic S, Lu M, Bosshard HR, Jelesarov I. Fast folding of the HIV-1 and SIV gp41 six-helix bundles. J Mol Biol. 2004;336:1–8. doi: 10.1016/j.jmb.2003.11.058. [DOI] [PubMed] [Google Scholar]
- 16.Hildinger M, Abel KL, Ostertag W, Baum C. Design of 5' untranslated sequences in retroviral vectors developed for medical use. J Virol. 1999;73:4083–4089. doi: 10.1128/jvi.73.5.4083-4089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer G, Valdez P, Kearns K, Bahner I, Wen SF, Zaia JA, et al. Inhibition of Human Immunodeficiency Virus-1 (HIV-1) Replication After Transduction of Granulocyte Colony-Stimulating Factor-Mobilized CD34+ Cells From HIV-1-Infected Donors Using Retroviral Vectors Containing Anti-HIV-1 Genes. Blood. 1997;89:2259–2267. [PubMed] [Google Scholar]
- 18.Schambach A, Wodrich H, Hildinger M, Bohne J, Krausslich HG, Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- 19.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element Enhances Expression of Transgenes Delivered by Retroviral Vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 21.Pene J, Rahmoun M, Temmerman S, Yssel H. Use of anti-CD3/CD28 mAb coupled magnetic beads permitting subsequent phenotypic analysis of activated human T cells by indirect immunofluorescence. J Immunol Methods. 2003;283:59–66. doi: 10.1016/j.jim.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Onlamoon N, Hudson K, Bryan P, Mayne AE, Bonyhadi M, Berenson R, et al. Optimization of in vitro expansion of macaque CD4 T cells using anti-CD3 and co-stimulation for autotransfusion therapy. J Med Primatol. 2006;35:178–193. doi: 10.1111/j.1600-0684.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- 23.Ma A, Koka R, Burkett P. Diverse Functions of IL-2, IL-15, and IL-7 in Lymphoid Homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 24.Geginat J, Sallusto F, Lanzavecchia A. Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4(+) T cells. J Exp Med. 2001;194:1711–1719. doi: 10.1084/jem.194.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu P, Chertova E, Bess J, Jr., Lifson JD, Arthur LO, Liu J, et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu P, Liu J, Bess J, Jr., Chertova E, Lifson JD, Grise H, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 27.Malashkevich VN, Chan DC, Chutkowski CT, Kim PS. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc Natl Acad Sci USA. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustchina E, Hummer G, Bewley CA, Clore GM. Differential inhibition of HIV-1 and SIV envelope-mediated cell fusion by C34 peptides derived from the C-terminal heptad repeat of gp41 from diverse strains of HIV-1, HIV-2, and SIV. J Med Chem. 2005;48:3036–3044. doi: 10.1021/jm049026h. [DOI] [PubMed] [Google Scholar]
- 29.Mo H, Konstantinidis AK, Stewart KD, Dekhtyar T, Ng T, Swift K, et al. Conserved residues in the coiled-coil pocket of human immunodeficiency virus type 1 gp41 are essential for viral replication and interhelical interaction. Virology. 2004;329:319–327. doi: 10.1016/j.virol.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Dong XN, Xiao Y, Dierich MP, Chen YH. N- and C-domains of HIV-1 gp41: mutation, structure and functions. Immunol Lett. 2001;75:215–220. doi: 10.1016/s0165-2478(00)00302-3. [DOI] [PubMed] [Google Scholar]
- 31.Gallo SA, Sackett K, Rawat SS, Shai Y, Blumenthal R. The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors. J Mol Biol. 2004;340:9–14. doi: 10.1016/j.jmb.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 32.Melikyan GB, Egelhofer M, von Laer D. Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J Virol. 2006;80:3249–3258. doi: 10.1128/JVI.80.7.3249-3258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aker M, Tubb J, Groth AC, Bukovsky AA, Bell AC, Felsenfeld G, et al. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum Gene Ther. 2007;18:333–343. doi: 10.1089/hum.2007.021. [DOI] [PubMed] [Google Scholar]
- 34.Yannaki E, Tubb J, Aker M, Stamatoyannopoulos G, Emery DW. Topological constraints governing the use of the chicken HS4 chromatin insulator in oncoretrovirus vectors. Mol Ther. 2002;5:589–598. doi: 10.1006/mthe.2002.0582. [DOI] [PubMed] [Google Scholar]
- 35.Emery DW, Yannaki E, Tubb J, Stamatoyannopoulos G. A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc Natl Acad Sci USA. 2000;97:9150–9155. doi: 10.1073/pnas.160159597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mok HP, Javed S, Lever A. Stable gene expression occurs from a minority of integrated HIV-1-based vectors: transcriptional silencing is present in the majority. Gene Ther. 2007;14:741–751. doi: 10.1038/sj.gt.3302923. [DOI] [PubMed] [Google Scholar]
- 37.Schambach A, Schiedlmeier B, Kuhlcke K, Verstegen M, Margison GP, Li Z, et al. Towards hematopoietic stem cell-mediated protection against infection with human immunodeficiency virus. Gene Ther. 2006;13:1037–1047. doi: 10.1038/sj.gt.3302755. [DOI] [PubMed] [Google Scholar]
- 38.Brice GT, Riley JL, Villinger F, Mayne A, Hillyer CD, June CH, et al. Development of an animal model for autotransfusion therapy: in vitro characterization and analysis of anti-CD3/CD28 expanded cells. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:210–220. doi: 10.1097/00042560-199811010-00002. [DOI] [PubMed] [Google Scholar]
- 39.Levine BL, Bernstein WB, Aronson NE, Schlienger K, Cotte J, Perfetto S, et al. Adoptive transfer of costimulated CD4+ T cells induces expansion of peripheral T cells and decreased CCR5 expression in HIV infection. Nat Med. 2002;8:47–53. doi: 10.1038/nm0102-47. [DOI] [PubMed] [Google Scholar]
- 40.Villinger F, Brice GT, Mayne AE, Bostik P, Mori K, June CH, et al. Adoptive transfer of simian immunodeficiency virus (SIV) naive autologous CD4+ cells to macaques chronically infected with SIV is sufficient to induce long-term nonprogressor status. Blood. 2002;99:590–599. doi: 10.1182/blood.v99.2.590. [DOI] [PubMed] [Google Scholar]
- 41.Institute of Laboratory Animal Resources (U.S.) Guide for the care and use of laboratory animals. National Academy Press; Washington, D.C.: 1996. p. xii. 125pp. [Google Scholar]
- 42.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniel MD, Letvin NL, King NW, Kannagi M, Sehgal PK, Hunt RD, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 44.Reimann KA, Li JT, Voss G, Lekutis C, Tenner-Racz K, Racz P, et al. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collman R, Balliet JW, Gregory SA, Friedman H, Kolson DL, Nathanson N, et al. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clapham PR, McKnight A, Weiss RA. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lusso P, Cocchi F, Balotta C, Markham PD, Louie A, Farci P, et al. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, et al. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 49.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Pt A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 51.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, et al. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 52.Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.