Abstract
The site on HIV-1 gp120 that binds to the CD4 receptor is vulnerable to antibodies. However, most antibodies that interact with this site cannot neutralize HIV-1. To understand the basis of this resistance, we determined co-crystal structures for two poorly neutralizing, CD4–binding site (CD4BS) antibodies, F105 and b13, in complexes with gp120. Both antibodies exhibited approach angles to gp120 similar to those of CD4 and a rare, broadly neutralizing CD4BS antibody, b12. Slight differences in recognition, however, resulted in substantial differences in F105- and b13-bound conformations relative to b12-bound gp120. Modeling and binding experiments revealed these conformations to be poorly compatible with the viral spike. This incompatibility, the consequence of slight differences in CD4BS recognition, renders HIV-1 resistant to all but the most accurately targeted antibodies.
More than 25 years after the discovery of HIV-1, an effective vaccine remains elusive. In this time, an estimated 60 million individuals have been infected by HIV-1, and ∼20 million have died (1). Identification of a site on the HIV-1 gp120 envelope glycoprotein that is vulnerable to neutralizing antibodies (2), along with the discovery that a major subpopulation of individuals infected with HIV-1 develops broadly effective antibodies against this site (3–7), has provided both a vaccine target and evidence that it is possible to induce broadly neutralizing antibodies in humans. The site of vulnerability corresponds to the initial site of attachment between gp120 on the viral spike and the CD4 receptor on the host cell. However, most CD4–binding site (CD4BS) antibodies do not effectively neutralize HIV-1 (8–10).
The HIV-1 gp120 envelope glycoprotein contains a number of features that help evade humoral immunity, including variable loops (11), N-linked glycosylation (12, 13), and conformational flexibility (14). How the initial site of CD4 attachment is protected, however, remains unclear. Of the variable loops, only one (V5, the least variable) is in close proximity, and the site itself is relatively conserved in sequence. N-linked glycosylation, meanwhile, surrounds half the site, though the site itself is free of glycosylation. In addition, though gp120 is conformationally flexible, the site of initial CD4 attachment is conformationally inert. In our experiment, we characterized neutralization breadth and potency for a panel of CD4BS antibodies, measured binding to gp120 variants with altered sites of CD4 binding, and determined co-crystal structures for two of these antibodies: (i) F105 (15) in complex with a YU2 gp120 core with V3 and (ii) b13 (16) in complex with an HXBc2 core stabilized to retain the CD4-bound conformation. The results show in atomic detail how the conformational flexibility of gp120 facilitates a decoy strategy that misdirects the humoral immune response.
To guide vaccine development, panels of reference HIV-1 isolates have recently been defined (17, 18). Tier 1 viruses are relatively easy to neutralize, whereas those in tier 2 are more difficult, and neutralization of tier 2 viruses is thought to be a necessary bar that an effective HIV-1 vaccine needs to surpass. To define the efficacy of CD4BS antibodies against these strains, we tested 10 CD4BS monoclonal antibodies, including the broadly neutralizing antibody b12 (19–21). Although CD4BS antibodies could neutralize diverse tier 1 viruses, except for antibody b12, they were largely ineffective against tier 2 isolates (table S1) (22).
To understand the inability of most CD4BS antibodies to neutralize tier 2 viruses, we employed robotic and variational techniques (23, 24) to crystallize a CD4BS antibody in complex with gp120. We assessed more than 10,000 crystallizations of 11 CD4BS antibody–gp120 complexes, including four variants of gp120, five CD4BS antibodies, and two V3-directed antibodies (table S2). Diffraction data to 2.9 Å resolution were collected from an R32 crystal of the antigen-binding fragment (Fab) of antibody F105 in complex with a YU2 gp120 core containing an intact V3. Structure solution of the two complexes in the asymmetric unit was accomplished by molecular replacement (25), and refinement to Rcryst 20.2% (Rfree 24.3%) defined the F105-gp120 structure, for all except gp120 residues 302 to 326 and 396 to 411, which correspond to the flexible V3 and V4 regions, respectively (Fig. 1A, fig. S1, and table S3).
Fig. 1.
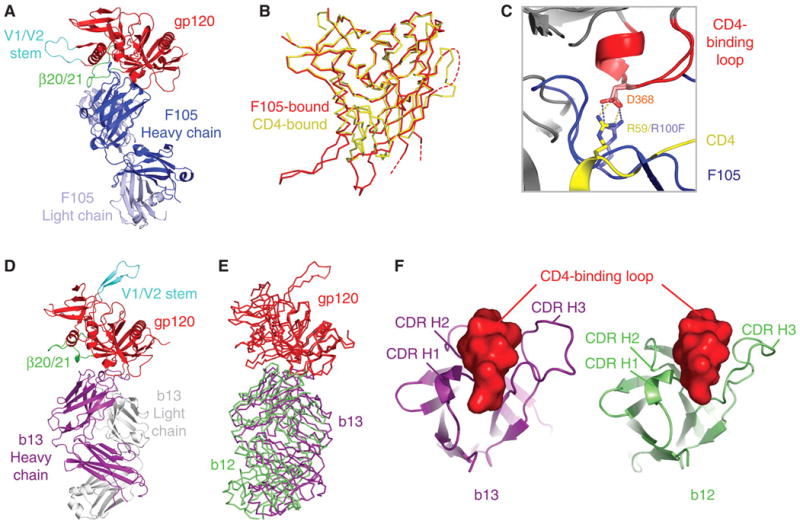
Co-crystal structure of antibodies F105 and b13 in complex with HIV-1 gp120. (A) Fab F105 in complex with a YU2 gp120 core with intact V3. Polypeptide chains are depicted in ribbon representation, with F105 in dark and light blue for heavy and light chain, respectively, and gp120 in red (with β20/β21 and V1/V2 stem highlighted in green and cyan, respectively). (B) Cα-backbone traces for F105- and CD4-bound conformations of gp120. The F105-bound structure corresponds to the core with V3 determined here, whereas the YU2 structure corresponds to the original core, with V3 truncation (46). Dashed lines correspond to the disordered V3 (fig. S1) and V4 regions. (C) Similarities in recognition of Asp368 of gp120 by Arg100F of F105 (blue) and Arg59 of CD4 (yellow) (47). (D) Fab b13 in complex with an HXBc2 gp120 core restrained to be in the CD4-bound state. Polypeptide chains are depicted in ribbon representation with b13–heavy chain in purple, light chain in gray, and gp120 colored as in (A). (E) Cα-backbone traces for heavy chain of antibody b12 (green) and antibody b13 (purple) in complexes with gp120 (red) after gp120-outer domain superposition. (F) Heavy-chain complementarity-determining regions (CDRs) for b13 (purple) and b12 (green) binding the CD4-binding loop (red) of gp120.
Overall, F105 binding to gp120 occurred primarily through heavy-chain interactions, which were similar in many respects to those of CD4. Instances of precise mimicry were observed, for example, between Arg100F of F105 and Arg59 of CD4, both of which make analogous hydrogen bonds to Asp368 of gp120 (Fig. 1C) (26). The approach angle of F105 was also comparable to that of CD4 (fig. S2); the F105-heavy chain resided 18% more within the CD4 envelope of approach than that of b12 (27). Moreover, F105 recognized a conformation in gp120 similar to that induced by CD4 (Fig. 1B); superposition of the inner and outer domains of core gp120 in F105 and CD4-bound conformations showed a root mean square deviation (RMSD) in Cα-positions of 1.97 Å, only about twice the RMSD of 0.78 Å between HIV-1 gp120 cores from strains HXBc2 and YU2, when both are bound to CD4. One critical part of gp120, however, the four-stranded “bridging sheet” (Fig. 2), was considerably altered from the CD4-bound state. All four strands of the sheet were displaced to uncover a hydrophobic surface (Fig. 2A), which served as a focus of F105 binding.
Fig. 2.
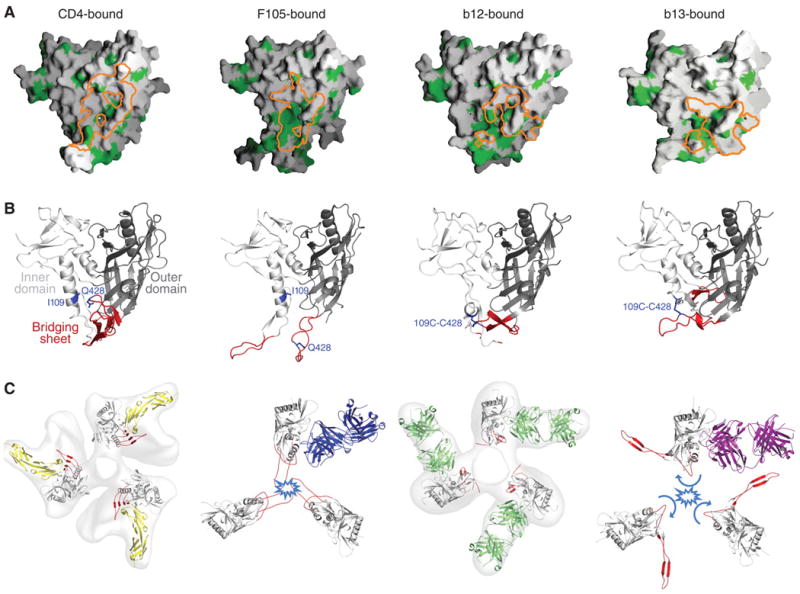
Epitopes, bound conformations, and trimer modeling. (A) Epitope hydrophobicity. The surface of gp120 is shown in gray, with hydrophobic residues highlighted in green. Binding surfaces for CD4, F105, b12, and b13 are outlined in orange. (B) Ligand-bound conformation of gp120. Polypeptides of gp120 are depicted in ribbon representation with inner domains shown in light gray, outer domains in dark gray, and regions that in the CD4-bound state correspond to the bridging sheet shown in red. Residues 109 and 428 are highlighted in blue and shown in stick representation. (C) Viral spike compatibility. Density maps derived from the cryo–electron tomography of HIV-1 BaL isolate spike are shown in gray for CD4 and 17b- and b12-bound states (first and third from left, respectively), along with optimal fits of atomic-level models (30). To model F105- and b13-bound forms of gp120 into likely viral spike orientations, the invariant β-sandwich of the gp120 inner domain was superimposed. Likely clashes of V1/V2 in the superimposed conformation with neighboring protomers close to the trimer axis are highlighted in light blue.
To assess the necessity and generality of access to this hydrophobic surface for CD4BS antibodies, we mutationally tethered the loop between strands β20 and β21 of the bridging sheet to the α1 helix by creating a disulfide bond between residues 109 and 428 (28). A monomeric 109-428 disulfide variant was recognized at close to wild-type levels by CD4 and CD4-induced antibodies, 17b and m6. In contrast, binding was ablated to eight of the CD4BS antibodies, and only b12 and b13 were able to recognize the 109-428 variant (tables S4 and S5). The results suggest that most CD4BS antibodies rely on access to the hydrophobic surface under the bridging sheet (29). Such access necessitates movement of bridging-sheet strands β20 and β21, which abut the initial site of CD4 attachment, as well as of the neighboring strands, β2 and β3, from which the V1/V2 loops emanate. With F105 bound, the tip of the V1/V2 stem shifts up to 40 Å, with respect to the equivalent region in the CD4-bound state (fig. S3).
To model the consequences of the F105-recognized alteration of the bridging sheet in the context of the oligomeric viral spike, we used the cryo–electron microscopy (EM) tomograms of the viral spike from the BaL isolate of HIV-1 (Fig. 2C) (30). In the BaL spike tomograms of CD4- and b12-bound states, ligand positions were used to orient placement of gp120 atomic-level models. We used these atomic-level models as guides for placement of the F105-bound conformation of gp120. Major clashes between equivalent protomers around the trimer threefold were predicted for expected positions of V1/V2 stem (Fig. 2C) (31). Although the precise displacement of the bridging sheet probably depends on antibody particulars, because of the constrained location of this sheet and especially of the V1/V2 stem and its proximity to neighboring protomers at the trimer interface, displacement from the CD4-proximal face of gp120 probably results in a clash in the oligomeric context. Thus, CD4BS antibodies that access the hydrophobic region under the bridging sheet recognize or induce conformations in gp120 that are poorly compatible with the functional viral spike (32).
Unlike other CD4BS antibodies, antibody b13 showed substantial, though reduced, binding to the 109-428 variant. In addition, b13, like b12, was able to recognize an outer domain–only variant of gp120 (tables S4 and S5). To understand how b13 recognizes both 109-428 and outer domain–only variants of gp120 (yet still cannot effectively neutralize HIV-1), we again turned to crystallography. In light of our difficulty in obtaining CD4BS antibody–complex crystals with unconstrained versions of gp120, we crystallized the Fab of b13 with a 2-disulfide variant of gp120, which we had previously crystallized with b12 (crystals were also obtained with an outer domain–only variant, but these were not suitable for analysis) (2). The crystals formed in two space groups, C222 and C2221, which diffracted to minimum Bragg spacings of 2.5 and 3.2 Å, respectively. We determined the structures of both by molecular replacement and refined to Rcryst values of 17.8 and 19.6% (Rfree values of 23.9 and 23.7%), respectively (table S3). We describe the higher-resolution C222 structure, which is depicted in Fig. 1D.
Antibody b13 bound gp120 similarly to antibody b12 (Fig. 1E). A rotation of only 17° would superimpose the variable portions of these two antibodies precisely (table S6). With b13, as with b12, heavy chain–only recognition was employed, with the central focus of the b13–complementarity determining loops on the CD4-binding loop of gp120 (Fig. 1F) (33). Despite this overall similarity in antibody recognition, the 17° difference in recognition moved the b13 epitope 6 Å toward the bridging-sheet region. This lead to a 13 Å movement in strands β20 and β21 of gp120. The β20–β21 movement was amplified by a more dramatic alteration in the neighboring β2–β3 strands, which twisted 152° from the CD4-facing side to the “back”-side of gp120. As the connection between strands β2 and β3 extends into V1/V2, we would expect such a change to be even more pronounced in the context of a full-length V1/V2 loop. Modeling the consequences of the ligand-induced alteration on the EM tomograms of the HIV-1 BaL isolate revealed substantial clashes between equivalent protomers around the trimer threefold to achieve the expected positions of V1/V2 in the b13-bound conformation (Fig. 2C) (34). Because the precise conformation observed in the monomeric crystal structure containing the flexible V1/V2 stem may not reflect solution or oligomeric behavior, we tested binding for the panel of CD4BS antibodies to an uncleaved form of the ectodomain (gp140) appended to a trimerization domain to confirm clash predictions (35). We did this for two primary isolate-derived variants, though because of avidity effects, we only measured on-rates. CD4 and b12 showed on-rates that were reduced four- to sixfold from that of gp120, whereas all other CD4BS antibodies showed more substantially reduced on rates (tables S4 and S7). We also measured binding to cell-surface viral spikes from the primary HIV-1 isolate JR-FL, in cleaved and uncleaved states, as the cell-surface cleaved state more accurately mimics the functional viral spike (36). CD4 and antibody b12 recognized cleaved and uncleaved spikes with similar affinity, whereas all other CD4BS antibodies showed markedly reduced recognition of the cleaved state (fig. S4). Thus, despite similarities in epitopes recognized by F105 and b13 and those recognized by CD4 and b12 (Fig. 3 and fig. S3), small differences in recognition induced [or selected (32)] more substantial differences in gp120 conformation. In particular, both F105 and b13 recognized conformational shifts in the position of the V1/V2 stem, resulting in a V1/V2 orientation poorly compatible with the functional viral spike. As functional viral spikes, which contain the properly formed attachment site, are an immunogenic minority—with monomeric gp120 and viral debris in vast abundance—such conformational diversity begins to explain why CD4BS antibodies are frequently elicited but do not neutralize.
Fig. 3.
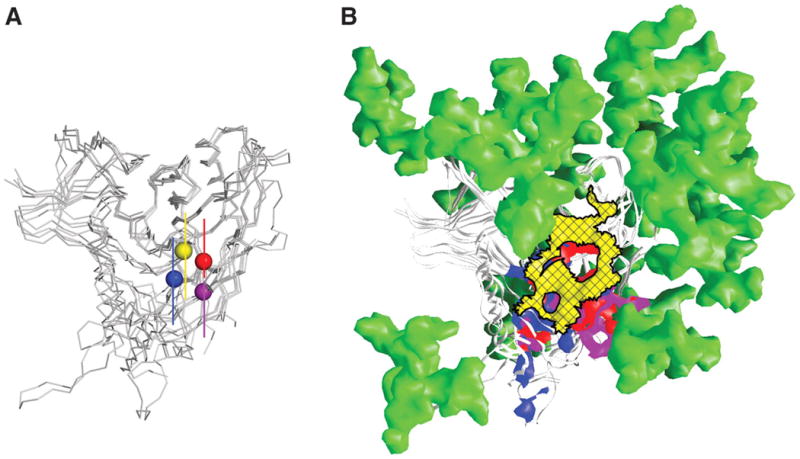
Immune evasion at the site of initial CD4 attachment. (A) Recognition similarity. Centers of recognition for CD4, F105, b12, and b13. After superposition of gp120 outer domains, the centers of the recognition surface of each ligand on gp120 is denoted by balls for CD4 (yellow), F105 (blue), b12 (red), and b13 (purple). (B) Immune evasion. The initial site of CD4 attachment (cross-hatched yellow surface) is circumscribed by a combination of glycan (green) and conformational constraints. The surface on gp120 recognized by F105, b12, and b13 (that strays beyond the site of CD4 attachment) is shown in blue, red, and purple, respectively. Glycosylation sterically crowds the immune response toward the bridging-sheet region (blue surface that F105 recognizes) or toward the V3 region (purple surface that b13 recognizes) (48). In either case, recognition of these regions of gp120 results in antibody-bound conformations of gp120 that are poorly compatible with the functional spikes of HIV-1 virions from tier 2 primary isolates.
Our results reveal how induced conformation can modify the recognition of a site that is itself conformationally invariant. This mechanism represents a twist on that of conformational masking (37), whereby the virus uses quaternary constraints to resist conformational changes required for exposure or formation of a particular antibody epitope (and thereby resists binding of and neutralization by that particular antibody). Conformational masking was previously shown for epitopes like the CD4- and co-receptor–binding sites on HIV-1, neither of which is fully formed in the nascent viral spike, or the V3, which is not accessible on most primary isolates. With the initial site of CD4 attachment, however, the target site is fully formed, reasonably accessible, and conformationally inert. However, the bridging sheet and V1/V2 loops detect and amplify any recognition that strays outside the target site (Fig. 3 and fig. S3). Should an antibody stray, even by just a few angstroms, then gp120 conformational changes that are poorly compatible with the functional viral spike become a constraining factor for binding and neutralization.
Supplementary Material
Acknowledgments
L. Chen produced and assessed for crystallization CD4BS antibodies with unconstrained gp120, crystallized the F105-gp120 complex, assisted with F105-gp120 data collection and structure solution, and carried out mutagenesis and SPR binding experiments. Y.D.K. assisted with F105-gp120 crystallization, data collection, and structure solution and refined and analyzed the F105-gp120 structure. T.Z. purified, crystallized, solved, and analyzed the b13-gp120 complex. X.W., S.O.'D. and J.R.M. assessed neutralization potency and breadth of CD4, patient sera, and CD4BS antibodies. L. Chen and M.P. carried out cell-surface JR-FL binding experiments. M.T. and R.W. provided YU2 core gp120; L.X. and G.J.N. provided stabilized-core gp120; Z.-Y.Y. and G.J.N. converted b13 from Fab to IgG format and provided b13 IgG; L. Cavacini and M.R.P. provided F105; M.-Y.Z. and D.S.D. provided m6, m14, and m18; A.J.H. and D.R.B. provided b3, b6, b11, b12; and J.A. provided dodecameric CD4. J.A., D.R.B., D.S.D., G.J.N., M.R.P., J.S., R.W., and J.R.M. assisted with analysis and writing, and P.D.K. assisted with crystallography and experimental planning and wrote the first draft. Figures, tables, and supporting online material were produced by L. Chen, Y.D.K., M.P., T.Z. and X.W. We thank L. Shapiro and members of the Structural Biology Section, Vaccine Research Center (VRC), for discussions and comments on the manuscript, M. Connors for patient serum, M. Fung for antibodies G3-42 and G3-299, J. Robinson for antibodies 1.5e and F91, J. Stuckey for assistance with figures and tables, S. Subramaniam for EM tomograms, C. Winter and C. Huang for S2 production of core YU2 gp120, X. Yang for preparation of JR-FL gp120, and the Flow Cytometry Core, VRC, for assistance with antibody binding to cell-surface–expressed HIV-1 spikes. Support for this work was provided by the Intramural Research Program of NIH, the International AIDS Vaccine Initiative, a grant from the Bill and Melinda Gates Foundation Grand Challenges in Global Heath Initiative, and grants from NIH. The use of insertion device 22 (Southeast Region Collaborative Access Team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38. Coordinates and structure factors for the F105-gp120 complex (accession code 3HI1) and the b13-gp120 complexes (accession codes 3IDX and 3IDY for C222 and C2221 forms, respectively) have been deposited with the Protein Data Bank.
Footnotes
References and Notes
- 1.UNAIDS. Joint United Nations Programme on HIV/AIDS; 2006. 2006 Report on the Global AIDS Epidemic. www.unaids.org/en/knowledgeCentre/HIVData/GlobalReport/2006/ [Google Scholar]
- 2.Zhou T, et al. Nature. 2007;445:732. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sather DN, et al. J Virol. 2009;83:757. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley JM, et al. J Virol. 2008;82:11651. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, et al. Nat Med. 2007;13:1032. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, et al. J Virol. 2009;83:1045. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhillon AK, et al. J Virol. 2007;81:6548. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roben P, et al. J Virol. 1994;68:4821. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid JF, et al. Nature. 2009;458:636. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 10.Pantophlet R, et al. J Virol. 2003;77:642. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starcich BR, et al. Cell. 1986;45:637. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt R, et al. Nature. 1998;393:705. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 13.Wei X, et al. Nature. 2003;422:307. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 14.Myszka DG, et al. Proc Natl Acad Sci USA. 2000;97:9026. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posner MR, et al. J Immunol. 1991;146:4325. [PubMed] [Google Scholar]
- 16.Barbas CF, III, et al. J Mol Biol. 1993;230:812. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 17.Mascola JR, et al. J Virol. 2005;79:10103. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweighardt B, et al. J Acquired Immune Defic Syndr. 2007;46:1. doi: 10.1097/QAI.0b013e318074eb5a. [DOI] [PubMed] [Google Scholar]
- 19.Burton DR, et al. Science. 1994;266:1024. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 20.Two of the CD4BS antibodies were tested as full-length immunoglobulin Gs (IgGs), four as Fabs, and four in both IgG and Fab format. Neutralization by IgG and Fab for b12, b13, m18, and F105 showed similar breadths and potencies (table S1).
- 21.Materials and methods are available as supporting material on Science Online.
- 22.We also tested soluble versions of the CD4 receptor, including a monomeric four-domain version (sCD4, 1mer) (38), a dimeric immunoglobulin chimera (CD4-IgG, 2mer) (39), and a dodecameric version (CD4 dodecamer, 12mer) (40), the latter designed to mimic oligomeric membrane-associated CD4 at the cell surface (table S1). Certain HIV-1 isolates and primary viral swarms are ineffectively neutralized by monomeric versions of soluble CD4 (37, 41). Nonetheless, against tier 2 viruses, monomeric sCD4 was quite effective [94% breadth, 7.6 μg/ml average median inhibitory concentration (IC50)], though not quite as potent as oligomeric versions (0.9 μg/ml average IC50 for the CD4 dodecamer) (table S1) (42).
- 23.Morris DW, Kim CY, McPherson A. Biotechniques. 1989;7:522. [PubMed] [Google Scholar]
- 24.Kwong PD, et al. J Biol Chem. 1999;274:4115. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- 25.The F105-gp120 structure described here was solved before release of the coordinates for the unbound F105 structure (43); bound- and unbound-Fab structures, however, do closely resemble each other.
- 26.Residue positions for antibodies are referred by Kabat numbering (44); thus, residue “100F” corresponds to insertion “F” at position “100” in Kabat nomenclature.
- 27.“Envelopes of approach” were assessed by comparing occluded volumes of binding ligands.
- 28.In addition to covering the hydrophobic site under the bridging sheet, tethering residues 109 and 428 was previously shown to impair binding of CD4BS antibodies (2); however, this previous study was carried out in the context of other mutations (e.g., Ser375 → Trp375), which by themselves may ablate F105 binding, and so the effect of the 109-428 linkage on CD4BS antibodies had not been clearly assessed previously.
- 29.The percentage of gp120-hydrophobic side chains contacted in the epitopes for antibodies b12, b13, and F105 is 27.5, 23.4, and 39.2%, respectively.
- 30.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Nature. 2008;455:109. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Many tier 1 HIV-1 isolates, including the BaL isolate, are neutralized by CD4BS antibodies (table S1). So whereas the F105-bound conformation of gp120 may not be favored in the functional viral spike, in tier 1 isolates, there appears to be sufficient spike flexibility to permit binding.
- 32.CD4BS antibodies probably use a mixture of induced fit and conformational selection to bind gp120 (45). We use the term “induced” to describe the antibody-bound conformation of gp120, though “selected” might also be appropriate.
- 33.Antibodies b12 and b13 are phage display–derived constructs. The binding of these two antibodies by their heavy chains is likely to be a direct function of this derivation, and these antibodies may not represent the expressed B cell repertoire in humans.
- 34.A lattice interaction of convergent stems influences the position of the V1/V2 stem in the b13-gp120 crystal. Nonetheless, it is unlikely that these crystals could form, unless the structural alterations described here caused the V1/V2 stem to twist so drastically from the CD4-bound state. Also, though the particular gp120 conformation recognized by b13 is a consequence in part of the gp120 alterations used in the crystallization, such alterations constrain the conformation of gp120 to be in the CD4-bound state. So the extent that these alterations affect conformation is likely to be similar to the extent that they decrease the differences between b13- and CD4-bound states.
- 35.Yang X, et al. J Virol. 2002;76:4634. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pancera M, Wyatt R. Virology. 2005;332:145. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 37.Kwong PD, et al. Nature. 2002;420:678. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 38.Deen KC, et al. Nature. 1988;331:82. doi: 10.1038/331082a0. [DOI] [PubMed] [Google Scholar]
- 39.Traunecker A, Schneider J, Kiefer H, Karjalainen K. Nature. 1989;339:68. doi: 10.1038/339068a0. [DOI] [PubMed] [Google Scholar]
- 40.Arthos J, et al. J Biol Chem. 2002;277:11456. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- 41.Moore JP, McKeating JA, Huang YX, Ashkenazi A, Ho DD. J Virol. 1992;66:235. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whereas CD4 effectiveness provides partial validation of the CD4BS as a vaccine target, additional validation can be found in patient sera. Here we demonstrate one patient serum to be effective against 30 of 31 tier 2 clade B and C viruses (table S1). Against a subset of these isolates, we further analyzed the effect of adsorption by gp120 and by a gp120 variant with an Asp-to-Arg substitution at position 368, a critical site in the CD4-binding loop. In two-thirds of the cases tested, the wild-type gp120 adsorption reduced neutralization by more than a factor of 2 over the Asp368 → Arg368 mutant (table S8). These results, along with others that have recently been published (3–7), suggest that naturally elicited CD4BS antibodies can occur at titers sufficient to neutralize tier 2 viruses.
- 43.Wilkinson RA, et al. J Virol. 2005;79:13060. doi: 10.1128/JVI.79.20.13060-13069.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5. U.S. Department of Health and Human Services, National Institutes of Health; Bethesda, MD: 1991. [Google Scholar]
- 45.Hammes GG, Chang YC, Oas TG. Proc Natl Acad Sci USA. 2009;106:13737. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwong PD, et al. Structure. 2000;8:1329. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 47.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 48.Calculations of distances between gp120 glycans and binding surfaces of CD4 and antibodies (table S9) show that for distance cutoffs of 10 to 15 Å, the surfaces recognized by b12 and CD4 are closer to more glycans than surfaces recognized by b13 and F105.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


