Abstract
UAP56, an ATP dependent RNA helicase that also has ATPase activity, is a DExD/H box protein that is phylogenetically grouped with the eukaryotic initiation factor eIF4A, the prototypical member of the DExD/H box family of helicases. UAP56, also known as BAT1, is an essential RNA splicing factor required for spliceosome assembly and mRNA export but its role in protein synthesis is not known. Here we demonstrate that UAP56 regulates protein synthesis and growth in cardiomyocytes. We found that wild-type (WT) UAP56 increased serum induced protein synthesis in HeLa cells. UAP56 mutants lacking ATPase and/or helicase activity inhibited protein synthesis compared with WT UAP56, suggesting that the ATPase and RNA helicase activity of UAP56 is important for protein synthesis. UAP56 siRNA inhibited phenylephrine (PE) induced protein synthesis in cardiomyocytes and inhibited PE induced cardiomyocyte hypertrophy. Our data demonstrate that UAP56 is an important regulator of protein synthesis and plays an important role in the regulation of cardiomyocyte growth.
Keywords: cardiac hypertrophy, protein synthesis, growth, UAP56, helicase
INTRODUCTION
Cardiac hypertrophy occurs in response to various factors including mechanical, hemodynamic and pathological stimuli [1]. While this response is initially adaptive, it can result in pathologic hypertrophy and subsequent heart failure. Regression or inhibition of cardiac hypertrophy may lead to improved outcomes as was the case in the HOPE trial in which the angiotensin converting enzyme (ACE) inhibitor ramipril was shown to decrease the development of and cause regression of electrocardiographic markers of left ventricular hypertrophy (LVH) [2]. These changes were independent of blood pressure reduction and were associated with reduced risk of death, myocardial infarction, stroke and congestive heart failure. Enhanced protein synthesis is a critical component of cardiac hypertrophy. Cardiac hypertrophy in vivo, and cardiac myocyte hypertrophy in vitro are characterized by increases in total protein content, increased cell size and up-regulation of fetal genes including atrial natriuretic peptide and B-type natriuretic peptide [3]. Initiation of translation, peptide chain elongation and translation termination are the main stages of protein synthesis. Increased rates of protein synthesis in cardiomyocytes correlate with an increase of activity of translation initiation factors [3].
UAP56 is an ATP dependent RNA helicase that also has ATPase activity [4, 5], utilizing the energy derived from ATP hydrolysis to unwind double stranded RNA. UAP56 is a DExD/H box protein and is phylogenetically grouped with eukaryotic initiation factor eIF4A, the prototypical member of the DExD/H box family of helicases [5–7]. UAP56 was initially identified in an analysis of genes centromeric to HLA-B in the human major histocompatibility complex and was named BAT1 (HLA B associated transcript 1) [8]. BAT1 was rediscovered as an essential RNA splicing factor and renamed UAP56 [9]. UAP56 is required for spliceosome assembly and mRNA export from the nucleus to the cytoplasm [10–12]. Binding of ATP to lysine 95 (K95) of UAP56 is required for mRNA export [11]. Depletion of HEL, the Drosophila equivalent of UAP56 has been shown to inhibit protein synthesis [10]. Given the role of UAP56 in RNA splicing and mRNA export, we hypothesized that UAP56 plays an important role in protein synthesis and subsequent hypertrophy. In the present study we found that UAP56 regulates protein synthesis and cardiomyocyte growth an effect that is blocked by helicase dead mutant UAP56.
METHODS
Cell culture
Neonatal rat cardiomyocytes were isolated from the cardiac ventricles of 2–3 day old Sprague-Dawley rat neonates as described previously [13]. Briefly, the ventricular tissue parts were excised and rinsed in Hank’s balanced salt solution (HBSS) prior to digestion with multiple rounds of collagenase type II (Worthington). Cells were collected by centrifugation, resuspended in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 μg/ml streptomycin. Non-myocytes were removed by two rounds of pre-plating on culture dishes at 37 °C for 1 hour. Cells were then cultured in DMEM containing 20 μM cytosine β-D- arabinofuranoside (Ara-C) (Sigma) for 24 hours on 0.2% gelatin coated plates prior to any treatments. For stimulation of protein synthesis cardiomyocytes were stimulated with phenylephrine (PE) (Sigma) for 4 hrs.
HeLa cells were grown in DMEM containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. 20% fetal bovine serum was used to stimulate the cells to measure protein synthesis.
Plasmids and Transfection
UAP56 wild-type (WT) plasmids were obtained from Origene. The single point mutations of WT UAP56 were created with the Quick-Change site directed mutagenesis kit (Stratagene) as previously described [14]. For transient plasmid expression, cells were transfected with Lipofectamine-2000 (Invitrogen) according to manufacturer instructions as described previously [15]. Cardiomyocyte transfection with UAP56 oligonucleotides siRNA (Dharmacon-Smart pool) to a final concentration of 50 nM was performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Briefly, 1 × 104 cells (in a 12-well plate) were transfected in 0.8 ml of serum-free and antibiotics-free DMEM containing 200 μl of Opti-MEM (Invitrogen), 3 μl of Lipofectamine RNAiMAX, and 50 nM of each siRNA. The medium was replaced 4 hours later with fresh DMEM containing 10% FBS. The cells were harvested 72 hours later for Western blotting.
Western blotting
After treatment, the cells were washed with PBS and harvested in modified RIPA buffer containing protease inhibitor cocktail (Sigma). 50 μg total protein lysates were separated on a 10% SDS-PAGE, transferred to a nitrocellulose membrane and immuno-blotted with a UAP56 (Santa Cruz), or a tubulin antibody (Sigma) followed by horseradish peroxidase-conjugated secondary antibody (Amersham Life Science).
[3H] Leucine Incorporation Assay
Cardiomyocytes were transfected with 50 nM of control or UAP56 siRNA for 72 hours and protein synthesis was measured after treating the cells with or without 50 μM PE for 4 hours. HeLa cells were treated with 20% FBS for 24 hours prior to measurement of protein synthesis. During the last one hour (HeLa cells) or two hours (cardiomyocytes) of incubation 1 μCi/ml of 3[H] leucine was added to the medium. The cells were washed with cold PBS three times and 10% trichloroacetic acid was added to each well and the precipitate was then collected on a glass micro-fiber filter under vacuum using a manifold. Filters were washed twice with cold 5% trichloroacetic acid followed by 95% ethanol, allowed to air dry and then suspended in 5-ml scintillation fluid. Acid precipitable counts per minute (cpm) were quantitated using a liquid scintillation counter.
Immunofluorescence
Cardiomyocytes were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton, and stained with UAP56 antibody (Santa Cruz) followed by anti-mouse Alexa Fluor 488 secondary antibody (Invitrogen). Nuclei were stained with DAPI (Sigma). Cells were visualized with an Olympus (BX-51) fluorescent microscope. Depicted images were adjusted evenly across the image to match contrast and brightness of the DAPI and UAP56 images. To assess cell size, cardiomyocytes were stained with a myocyte marker, sarcomeric α-actinin antibody (Sigma) followed by anti-rabbit Alexa 546 (Invitrogen) secondary antibody. Fluorescent images were taken at 40X magnification using an Olympus BX-51 fluorescent microscope. Cardiomyocyte total surface area was calculated using NIH Image J software, and expressed as the average of approximately 25 randomly selected cells per condition.
Statistics
Numeric data are expressed as means ± SEM or SD as indicated in the figure legends. Statistical analysis was performed with the StatView 5.0 package (ABACUS Concepts, Berkeley, Calif). Differences were analyzed with a 1-way or a 2-way repeated-measures ANOVA as appropriate, followed by Scheffe’s correction for multiple comparisons. A probability value of <0.05 was considered significant.
Results
UAP56 increases protein synthesis in HeLa cells
To assess the effect of UAP56 on protein synthesis, WT UAP56 was overexpressed in HeLa cells and [3H] leucine incorporation was measured. In cells stimulated with 20% FBS, WT UAP56 increased protein synthesis 2.0±0.27-fold compared with control vector (Figure 1A). We next examined the effect of UAP56 lysine 95 mutants on protein synthesis. Lysine 95 of UAP56 is part of motif I, a conserved DExD/H box protein motif that is important for the helicase and ATPase activity of UAP56 (Figure 1B) [16]. Binding of ATP to lysine 95 of UAP56 is required for mRNA export [11]. We found that overexpression of UAP56 K95M and K95Q mutants both inhibited serum induced protein synthesis compared with WT UAP56 (Figure 1C). These results demonstrated that lysine 95 of UAP56, critical for the ATPase and helicase activity of UAP56 [5], is important for protein synthesis.
Figure 1. Overexpression of UAP56 increases protein synthesis in HeLa cells.
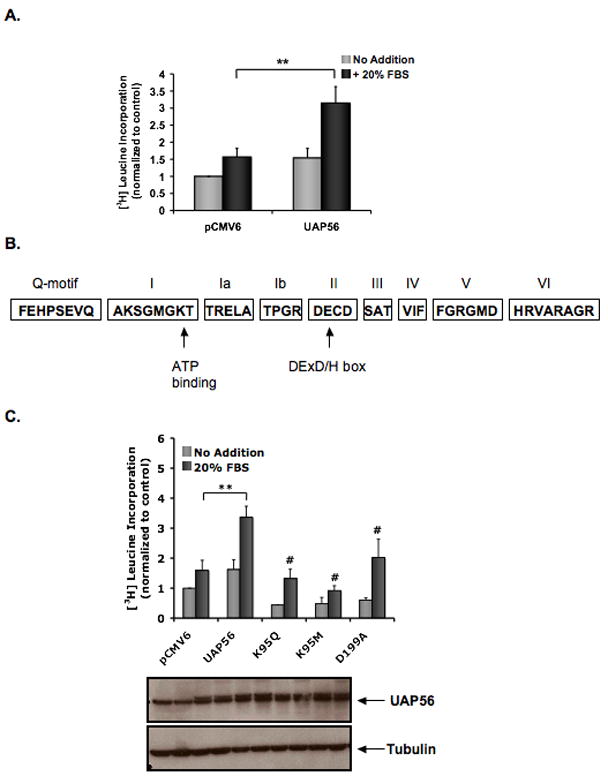
(A) Following transfection with UAP56 WT plasmid or control, cells were serum starved for 24 hours and then treated with or without 20% FBS for 24 hours. During the last 1 hour of incubation, cells were pulse labeled with [3H] leucine and then [3H] leucine incorporation assay was performed. (**p<0.01). (B) Schematic of the conserved DExD/H box motifs in UAP56. Not drawn to scale. (C) Following transfection with WT UAP56, control plasmid or UAP56 mutants, HeLa cells were serum starved for 24 hours and then treated with or without 20% FBS for 24 hours. [3H] leucine incorporation was measured as in A. Western blot demonstrating UAP56 protein levels after transfection and tubulin control is shown below. (**p<0.01; #p<0.01 compared with UAP56 plus 20% FBS). For figures, the data are the mean ± SD of two or more independent experiments performed in triplicate.
Helicase dead UAP56 mutant inhibits protein synthesis
Similar to motif I, motif II, the “DExD/H box” (Figure 1B), is also important for the helicase and ATPase activity of UAP56 [5]. We next examined whether mutation of motif II affects protein synthesis. Overexpression of the UAP56 mutant D199A (blocks the helicase activity of UAP56 but not its ATPase activity) [5] inhibited serum induced protein synthesis compared with WT UAP56 in HeLa cells (Figure 1C) suggesting that the RNA helicase activity of UAP56 is important for protein synthesis.
Localization of endogenous UAP56
We next examined the localization of endogenous UAP56 in cardiomyocytes. Immunofluorescence staining demonstrated that endogenous UAP56 is present in the nucleus of cardiomyocytes (Figure 2). UAP56 endogenous expression in cardiomyocytes was also confirmed by Western blot (Figure 3).
Figure 2. UAP56 is present in cardiomyocytes.
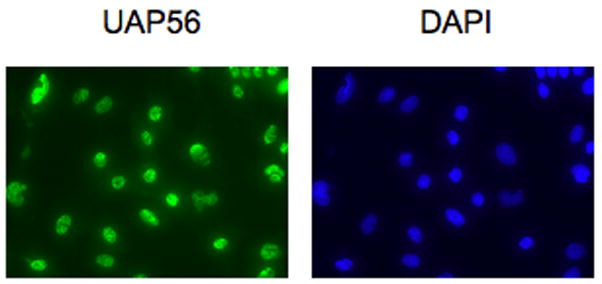
Cardiomyocytes were fixed, permeabilized, and stained with UAP56 antibody followed by Alexa Fluor 488 secondary antibody (green). Nuclei were stained with DAPI (blue).
Figure 3. Knockdown of UAP56 expression inhibits PE induced protein synthesis in cardiomyocytes.
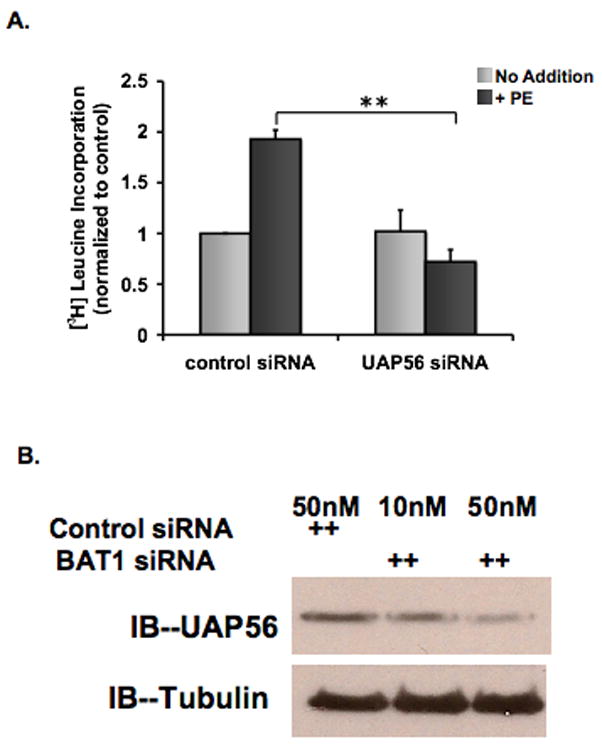
(A) Cardiomyocytes were transfected with control or UAP56 siRNA for 72 hours and treated with or without 50 μM PE for 4 hours. During the last two hours of incubation, cells were pulse labeled with [3H] leucine and then [3H] leucine incorporation was measured. (B) Rat cardiomyocytes were treated with control siRNA (50 nM) or UAP56 siRNA (10 or 50 nM) for 72 hours, and whole cell lysates were harvested for Western blotting demonstrating that UAP56 siRNA reduces UAP56 protein expression. (**p<0.01).
Knockdown of UAP56 inhibits protein synthesis in cardiomyocytes
We then examined the effect of UAP56 on protein synthesis in cardiomyocytes. Cardiomyocytes were transfected with 50 nM of UAP56 or control siRNA for 72 hours and treated with or without phenylephrine (50 μM) for four hours. Cells were then harvested and [3H] leucine incorporation was measured. UAP56 siRNA inhibited PE induced protein synthesis by 62.7± 5.6% compared with control siRNA (p<0.01) (Figure 3).
Knock down of UAP56 inhibits cardiomyocyte growth
We next examined whether or not the effect of UAP56 on protein synthesis in cardiomyocytes was associated with an effect on cardiomyocyte growth. Cardiomyocytes were treated with UAP56 siRNA or control siRNA and then treated with or without PE 50 μM for four hours. PE increased cardiomyocyte size by 3.9 ± 0.5 fold compared to control and this increase was inhibited by UAP56 siRNA (Figure 4). These data suggest that UAP56 is an important mediator of PE induced protein synthesis and cardiac hypertrophy.
Figure 4. Knockdown of UAP56 expression inhibits PE induced cardiomyocyte growth.
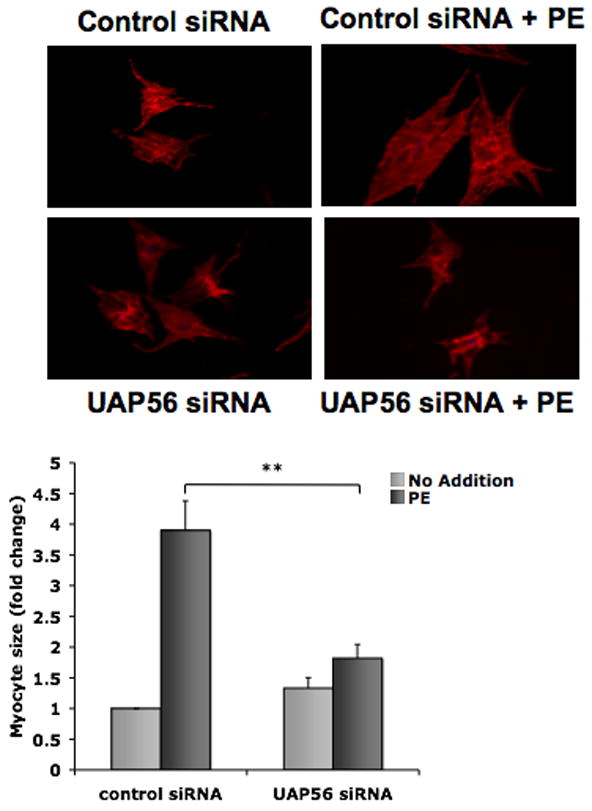
Cardiomyocytes were transfected with control or UAP56 siRNA for 72 hours and then treated with or without 50 μM PE for 4 hours. To assess cell size, cardiomyocytes were stained with a myocyte marker, sarcomeric α-actinin antibody, followed by Alexa 546 secondary antibody and fluorescence imaging was performed. Data are mean ± SEM. Cell area was measured for approximately 25 randomly chosen cells per condition. (**p<0.01).
Discussion
This is the first report to show that UAP56 is an important regulator of protein synthesis and cellular hypertrophy. The results presented here demonstrate that overexpression of UAP56 increases protein synthesis while the knockdown of UAP56 inhibits it. The results with UAP56 mutants also suggest that ATP binding and the ATPase and helicase activity of UAP56 are important in the regulation of protein synthesis. These findings are consistent with other reports showing that UAP56 plays an important role in RNA splicing and mRNA export from the nucleus to the cytoplasm.
Our findings further define the role of DExD/H box proteins and RNA helicases in regulating protein synthesis. While there is not complete understanding of the function of DExD/H box proteins, a clearer picture is beginning to emerge. DExD/H box proteins share several conserved motifs of which the DExD/H box (named after the amino acid sequence) comprises motif II [16, 17]. Most DExD/H box proteins hydrolyze nucleoside triphosphates (NTPs, such as ATP) into nucleoside diphosphates (NDPs, such as ADP) and some DExD/H box proteins possess RNA or DNA helicase activity [17]. DExD/H box proteins are associated with most processes involving RNA from transcription to RNA decay [16, 18]. The eukaryotic initiation factor eIF4A is the prototypical DExD/H box helicase [19]. Eukaryotic initiation factors are important in several steps of translation initiation including recruitment of mRNA to the small (40S) ribosome subunit and recruitment of the initiator methyionyl-tRNA (Met-tRNA1) that recognizes the start codon at the beginning of the coding region [20]. The eukaryotic initiation factor eIF4E binds to the 5′-cap structure of mRNA [20, 21] and also binds the scaffold protein eIF4G and eIF4A. The complex of eIF4A/eIF4G/eIF4E is known as eIF4F [20, 21]. eIF4A unwinds RNA secondary structures in the 5′untranslated region (5′ UTR) of mRNA and promotes the scanning process in which the preinitiation complex, including the 40S subunit and Met-tRNA1, scan the 5′ UTR of the mRNA for the start codon [20-22]. ATP hydrolysis by eIF4A is required for translation initiation [23]. Similarly, the DExD/H box protein Ded1p, an RNA helicase is also required for translation initiation [24]. We have now demonstrated that UAP56 is another RNA helicase that is important for protein synthesis. Similar to our findings with UAP56 mutants, mutations of motif I and motif II of eIF4A in yeast have been shown to block translation as well as cell growth [25].
Our findings further define the role that UAP56 plays in RNA synthesis and function. The different stages of RNA metabolism including transcription, splicing, RNA export and translation, are linked and in some instances one protein may affect more that one stage of RNA synthesis. For example, the splicing factor ASF/SF2 is an important mediator of translation initiation and mRNA export [26, 27]. Similarly, UAP56 affects several stages of RNA synthesis and function. Along with the THO complex and the export factor ALY, UAP56 is part of the TREX (transcription/export) complex [28]. TREX is recruited to activated genes during transcription and travels the length of the gene with RNA polymerase II during transcription elongation [28]. UAP56 is required for RNA splicing and spliceosome assembly [12]. UAP56 also recruits the export factor REF/ALY to mRNA [29] and is critical for mRNA export from the nucleus with knockdown of UAP56 resulting in the accumulation of polyadenylated mRNAs in the nucleus [10, 30]. We have now shown that UAP56 also plays a role in protein synthesis. While the splicing factor ASF/SF2 regulates translation and is present in the nucleus and cytoplasm, our findings and prior findings demonstrate the presence of UAP56 in the nucleus [11]. This does not however preclude the possible presence of UAP56 in the cytoplasm. In Drosophila, Meignin et al. have reported that while HEL/UAP56 is predominantly in the nucleus, it is also present in the cytoplasm where it directs mRNA cytoplasmic localization and post translational modification [31]. They did not detect UAP56 co-localized with mRNA in the cytoplasm and hypothesized that UAP56 may only transiently associate with RNA in the cytoplasm [31].
In conclusion, we have shown that UAP56 is an important regulator of protein synthesis and growth in cardiomyocytes. Our findings suggest that the RNA helicase activity of UAP56 is important in this regulation. Our findings further define the role of DExD/H box proteins in regulating RNA synthesis and function and suggest that UAP56 may be a key target for regulating cardiac hypertrophy.
Acknowledgments
Sources of Funding
This study was supported by NIH grant HL80938 to J.D.A.
Footnotes
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunter JJ, Chien KR. Signaling pathways for cardiac hypertrophy and failure. N Engl J Med. 1999 Oct 21;341(17):1276–83. doi: 10.1056/NEJM199910213411706. [DOI] [PubMed] [Google Scholar]
- 2.Mathew J, Sleight P, Lonn E, Johnstone D, Pogue J, Yi Q, et al. Reduction of cardiovascular risk by regression of electrocardiographic markers of left ventricular hypertrophy by the angiotensin-converting enzyme inhibitor ramipril. Circulation. 2001 Oct 2;104(14):1615–21. doi: 10.1161/hc3901.096700. [DOI] [PubMed] [Google Scholar]
- 3.Hannan RD, Jenkins A, Jenkins AK, Brandenburger Y. Cardiac hypertrophy: a matter of translation. Clin Exp Pharmacol Physiol. 2003 Aug;30(8):517–27. doi: 10.1046/j.1440-1681.2003.03873.x. [DOI] [PubMed] [Google Scholar]
- 4.Peelman LJ, Chardon P, Nunes M, Renard C, Geffrotin C, Vaiman M, et al. The BAT1 gene in the MHC encodes an evolutionarily conserved putative nuclear RNA helicase of the DEAD family. Genomics. 1995 Mar 20;26(2):210–8. doi: 10.1016/0888-7543(95)80203-x. [DOI] [PubMed] [Google Scholar]
- 5.Shen J, Zhang L, Zhao R. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem. 2007 Aug 3;282(31):22544–50. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- 6.Allcock RJ, Williams JH, Price P. The central MHC gene, BAT1, may encode a protein that down-regulates cytokine production. Genes Cells. 2001 May;6(5):487–94. doi: 10.1046/j.1365-2443.2001.00435.x. [DOI] [PubMed] [Google Scholar]
- 7.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992 Jul;11(7):2643–54. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spies T, Blanck G, Bresnahan M, Sands J, Strominger JL. A new cluster of genes within the human major histocompatibility complex. Science. 1989 Jan 13;243(4888):214–7. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- 9.Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997 Jul 15;11(14):1864–72. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 10.Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, et al. The DExD/H box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001 Oct 30;11(21):1716–21. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 11.Kota KP, Wagner SR, Huerta E, Underwood JM, Nickerson JA. Binding of ATP to UAP56 is necessary for mRNA export. J Cell Sci. 2008 May 1;121(Pt 9):1526–37. doi: 10.1242/jcs.021055. [DOI] [PubMed] [Google Scholar]
- 12.Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008 Jul 1;22(13):1796–803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan C, Ding B, Shishido T, Woo CH, Itoh S, Jeon KI, et al. Activation of extracellular signal-regulated kinase 5 reduces cardiac apoptosis and dysfunction via inhibition of a phosphodiesterase 3A/inducible cAMP early repressor feedback loop. Circ Res. 2007 Mar 2;100(4):510–9. doi: 10.1161/01.RES.0000259045.49371.9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akaike M, Che W, Marmarosh NL, Ohta S, Osawa M, Ding B, et al. The hinge-helix 1 region of peroxisome proliferator-activated receptor gamma1 (PPARgamma1) mediates interaction with extracellular signal-regulated kinase 5 and PPARgamma1 transcriptional activation: involvement in flow-induced PPARgamma activation in endothelial cells. Mol Cell Biol. 2004 Oct;24(19):8691–704. doi: 10.1128/MCB.24.19.8691-8704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao DF, Monia B, Dean N, Berk BC. Protein kinase C-zeta mediates angiotensin II activation of ERK1/2 in vascular smooth muscle cells. J Biol Chem. 1997 Mar 7;272(10):6146–50. doi: 10.1074/jbc.272.10.6146. [DOI] [PubMed] [Google Scholar]
- 16.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006 Feb 15;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Schwer B. A new twist on RNA helicases: DExD/H box proteins as RNPases. Nat Struct Biol. 2001 Feb;8(2):113–6. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 18.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004 Mar;5(3):232–41. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 19.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, et al. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–2. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006 Oct;21:362–9. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 21.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 22.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009 Feb 6;136(3):447–60. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blum S, Schmid SR, Pause A, Buser P, Linder P, Sonenberg N, et al. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7664–8. doi: 10.1073/pnas.89.16.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997 Mar 7;275(5305):1468–71. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 25.Schmid SR, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991 Jul;11(7):3463–71. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci U S A. 2004 Jun 29;101(26):9666–70. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008 Apr 25;30(2):179–89. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002 May 16;417(6886):304–8. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 29.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, et al. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001 Oct 11;413(6856):644–7. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 30.Kapadia F, Pryor A, Chang TH, Johnson LF. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 2006 Dec 15;384:37–44. doi: 10.1016/j.gene.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Meignin C, Davis I. UAP56 RNA helicase is required for axis specification and cytoplasmic mRNA localization in Drosophila. Dev Biol. 2008 Mar 1;315(1):89–98. doi: 10.1016/j.ydbio.2007.12.004. [DOI] [PubMed] [Google Scholar]


