Abstract
Parathyroid hormone-related protein (PTHrP) acts on the mammary mesenchyme and is required for proper embryonic mammary development. In order to understand PTHrP’s effects on mesenchymal cells, we profiled gene expression in WT and PTHrP−/− mammary buds, and in WT and K14-PTHrP ventral skin at E15.5. By cross-referencing the differences in gene expression between these groups, we identified 35 genes potentially regulated by PTHrP in the mammary mesenchyme, including 6 genes known to be involved in BMP signaling. One of these genes was MMP2. We demonstrated that PTHrP and BMP4 regulate MMP2 gene expression and MMP2 activity in mesenchymal cells. Using mammary bud cultures, we demonstrated that MMP2 acts downstream of PTHrP to stimulate ductal outgrowth. Future studies on the functional role of other genes on this list should expand our knowledge of how PTHrP signaling triggers the onset of ductal outgrowth from the embryonic mammary buds.
Keywords: parathyroid hormone-related protein, mammary development, ductal morphogenesis, oligonucleotide gene arrays, matrix metalloproteinase, epidermal appendage
Introduction
Mammary development in mice begins with the formation of bilateral mammary lines, which are multilayered ridges of epidermal cells stretching between the fore and hind limb buds (Hens and Wysolmerski, 2005; Robinson, 2007; Watson and Khaled, 2008). The mammary lines first appear on embryonic day 10 (E10), and between E10.5 and E12.5, cells from within the lines invaginate into the underlying mesenchyme to form mammary buds at 10 characteristic locations. Between E15 and E16, cells within the mammary buds proliferate and give rise to a cord of epithelial cells that grows through the condensed mammary mesenchyme and into the developing fat pad beneath the dermis. Once in the fat pad, the nascent mammary duct begins to bifurcate and by birth gives rise to approximately 10–15 primary branches. This rudimentary duct system persists until changing hormone levels at puberty stimulate a second round of ductal branching morphogenesis, which completes the formation of the typical virgin duct system (Hens and Wysolmerski, 2005; Robinson, 2007; Watson and Khaled, 2008).
The development of the embryonic mammary gland depends on sequential and reciprocal interactions between mammary epithelial cells and the surrounding mesenchymal cells (Hens and Wysolmerski, 2005; Robinson, 2007; Watson and Khaled, 2008). Our understanding of the interactions between these cells remains rudimentary, but recent work has shed light on some of the pathways involved in the specification of the mammary line and placodes, and the formation of the initial mammary buds. As in other organs, members of the FGF, hedgehog, WNT, TNFα and EGF signaling pathways are critical for the initiation of mammary placode and bud development (Mailleux et al., 2002; Chu et al., 2004; Eblaghie et al., 2004; Mustonen et al., 2004; Howard et al., 2005; Hatsell and Cowin, 2006; Veltmaat et al., 2006; Panchal et al., 2007). We know much less about the signals that initiate ductal development from the buds once they have formed. As discussed in the following paragraphs, two molecules contributing to this process are parathyroid hormone-related protein (PTHrP) and bone morphogenetic protein (BMP)-4.
PTHrP is a peptide growth factor named for its evolutionary and structural relationship to parathyroid hormone (PTH) (Wysolmerski, 2008). The high degree of homology between the amino-termini of PTHrP and PTH allows them both to bind and activate a common G-protein coupled receptor known as the Type 1 PTH/PTHrP receptor (PTHR1) (Wysolmerski, 2008). Both PTHrP and the PTHR1 are required for normal mammary gland development. Disruption of either gene in mice and loss of PTHR1 function in humans results in the absence of a mammary gland (Wysolmerski et al., 1998; Wysolmerski et al., 2001). Mammary epithelial cells express PTHrP as early as the placode stage and the PTHR1 is found on immature mesenchymal cells located beneath the embryonic epidermis. During bud formation, PTHrP from the epithelium interacts with the surrounding mesenchyme to contribute to the formation of the specialized dense mammary mesenchyme. In response to PTHrP, these mesenchymal cells maintain the mammary fate of the epithelial cells, trigger the overlying epidermis to form the nipple sheath and initiate ductal outgrowth and morphogenesis (Dunbar et al., 1999; Foley et al., 2001). If PTHrP signaling is disrupted, mammary epithelial cells differentiate into skin, no nipple is formed and morphogenesis is interrupted. Conversely, overexpression of PTHrP in basal keratinocytes using the K14 promoter (K14-PTHrP mice) leads to the conversion of the ventral dermis into condensed mammary mesenchyme, the suppression of hair follicle development and the acquisition of nipple-like characteristics by the ventral epidermis (Dunbar et al., 1999; Foley et al., 2001).
BMPs constitute a large family of secreted growth factors that signal through a heteromeric complex of type I and II receptor serine/threonine kinases (von Bubnoff and Cho, 2001; Derynck and Zhang, 2003). We recently showed that PTHrP and BMP signaling interact within the embryonic mammary mesenchyme and that this interaction appears to be important for the initiation of ductal morphogenesis (Hens et al., 2007). During the formation of the mammary buds, the subepidermal mesenchymal cells on the ventral surface of the embryo express BMP4. PTHrP from the bud epithelial cells acts to upregulate the expression of one of the receptors for BMP4, the BMPR1a, sensitizing the mesenchymal cells around the bud to the effects of BMP4. In response, the mesenchymal cells inhibit the formation of hair follicles around the primary mammary duct and also trigger the initiation of ductal branching morphogenesis. These actions are consistent with the reported effects of BMP4 on hair follicle formation (Botchkarev, 2003) and the regulation of branching morphogenesis in lung, salivary glands, kidneys, prostate and ureter (Bellusci et al., 1996; Miyazaki et al., 2000; Weaver et al., 2000; Lamm et al., 2001; Shi et al., 2001; Martinez et al., 2002; Dean et al., 2004; Eblaghie et al., 2006). Therefore, PTHrP regulates ductal outgrowth from the mammary bud, at least in part, by facilitating autocrine/paracrine BMP4 signaling within the mammary mesenchyme.
Previous studies have shown that PTHrP signaling is necessary for the mammary mesenchyme to initiate epithelial ductal development (Dunbar et al., 1999; Foley et al., 2001; Hens et al., 2007). Therefore, further characterization of PTHrP’s effects on these cells may identify additional pathways that participate in the epithelial-mesenchymal interactions regulating the initiation of ductal morphogenesis. In pursuit of this goal, we analyzed changes in gene expression in WT versus PTHrP−/− mammary buds and in K14-PTHrP versus WT ventral skin in the hopes of identifying genes upregulated or downregulated by PTHrP in the mammary mesenchyme. In this report, we detail the results of our gene expression experiments, which identified 35 genes that appear to be regulated by PTHrP in mammary mesenchyme cells. This list of genes lends further support to interactions between PTHrP and BMP signaling in mesenchymal cells. In addition, we demonstrate that one gene identified from this analysis, membrane metalloproteinase 2 (MMP2), is regulated by PTHrP and BMP4, and contributes to the outgrowth of embryonic mammary buds.
Results
Analysis of Gene Expression
Our goal was to define genes up-regulated and down-regulated by PTHrP within the mammary mesenchyme. WT buds contain these specially differentiated cells, while PTHrP−/− buds lack them. Likewise, K14-PTHrP ventral skin contains ectopic mammary mesenchyme, while normal ventral skin does not (Foley et al., 2001; Hens et al., 2007). Therefore, we reasoned that a comparison of the genes overexpressed in WT mammary buds vs. PTHrP−/− mammary buds with those overexpressed in K14-PTHrP ventral skin vs. WT ventral skin might reveal genes that were induced by PTHrP’s actions in the mammary mesenchyme. Similarly, genes both overexpressed in PTHrP−/− vs. WT mammary buds and overexpressed in WT vs. K14-PTHrP ventral skin should be those downregulated as a result of PTHrP signaling. We profiled mammary buds and ventral skin at E15.5, just before the normal buds would be expected to initiate ductal outgrowth (which fails to occur in PTHrP−/− buds) because we were particularly interested in defining pathways involved in initiating ductal morphogenesis.
Using Affymetrix (Santa Clara, CA) oligonucleotide arrays, we defined 1,299 genes that were overexpressed in WT mammary buds as compared to PTHrP−/− buds and 499 genes that were overexpressed in PTHrP−/− buds as compared to WT mammary buds. We have deposited these gene lists with the NCBI Gene Expression Omnibus (GEO) (GSE17654). Functional annotation clustering of these large lists revealed significantly higher levels of expression of groups of genes involved in glycolysis and energy metabolism, mitochondrial function, protein biosynthesis and cytoskeletal and structural proteins in WT buds as compared to PTHrP−/− buds. PTHrP−/− mammary buds were enriched for the expression of genes associated with RNA processing/turnover and chromatin assembly/remodeling. While WT buds are on the verge of entering a rapid growth phase at E15.5, PTHrP−/− buds fail to grow out (Wysolmerski et al., 1998). Therefore, it is not surprising that PTHrP−/− buds downregulate genes associated with biosynthesis and energy production.
In PTHrP−/− buds, mammary epithelial cells appear to be shunted towards an epidermal fate and express some markers of superbasal keratinocytes (Foley et al., 2001). In order to examine this shift in cell fate further, we examined differences in gene expression between WT mammary buds and WT ventral skin and compared them to changes in gene expression in WT vs. PTHrP−/− mammary buds. There were 196 genes overexpressed in WT mammary buds as compared to WT skin. These genes presumably contribute towards defining the identity of the mammary gland as compared to the skin and hair follicles. Interestingly, 120 of the genes on this list were also overexpressed in WT buds as compared to PTHrP−/− buds. Thus, disruption of PTHrP signaling results in a reduction in the expression of genes involved in distinguishing buds from skin, supporting our previous conclusion that mammary buds in the PTHrP−/− embryos take on an epidermal identity. Functional annotation of these 196 genes identified three major groups. First, mammary buds appeared to express a particular subset of keratin genes (4, 6a, 8, 13, 18, 19) that were not expressed in the epidermis. Second, relative to skin, the mammary buds overexpressed a large number of transcription factor genes including many homeodomain containing factors and two genes, Tbx3 and GATA3, which have previously been shown to be expressed in embryonic mammary epithelial cells (Jerome-Majewska et al., 2005; Cho et al., 2006; Kouros-Mehr et al., 2006; Asselin-Labat et al., 2007). Finally, the mammary buds expressed higher levels of a series of genes contained within specific growth factor signaling pathways. The full gene lists can be accessed from the NCBI GEO site (GSE17654). Selected genes from the three functional classes that are both overexpressed in WT buds as compared to skin and in WT buds as compared to PTHrP−/− buds are listed in Table 1.
Table 1.
Selected genes from the overlap between those overexpressed in WT as compared to PTHrP−/− mammary buds and those overexpressed in WT mammary buds as compared to WT ventral skin.
| Keratins | Transcription Factors | Growth Factor Signaling |
|
|---|---|---|---|
| Keratin 4 | Single-minded 2 (Sim2) |
Myeloblastosis oncogene (Myb) |
Tachykinin 1 |
| Keratin 6a | Sry-box containing gene 4 (Sox4) |
Meis homeobox 2 (Meis2) |
pleiotrophin |
| Keratin 8 | Inhibitor of DNA binding 1 (Id1) |
Iroquois related homeobox 5 (Irx5) |
Sclerostin domain containing 1 (Sostdc1) |
| Keratin 13 | Inhibitor of DNA binding 4 (Id4) |
Iroquois related homeobox 6 (Irx6) |
Rspondin homolog (Rspo1) |
| Keratin 18 | High mobility group AT-hook 2 (Hmga2) |
T-box 3, (Tbx3) | Macrophage migration inhibitory factor (Mif) |
| Keratin 19 | OVO homolog-like 1 (Ovol1) |
Activating transcription factor 7 interacting protein (Atf7ip) |
|
We also identified genes that were more highly expressed in skin than in mammary buds and compared this list to the set of genes overexpressed in PTHrP−/− as compared to WT mammary buds. There were 289 genes more highly expressed in skin as compared to the mammary buds. Functional annotation of these genes identified a cluster of 10 different collagen genes as well as a series of extracellular matrix genes that were down-regulated in mammary buds. As before, there were groups of skin-associated growth factor signaling genes and transcription factor genes that appeared to be specifically down-regulated in mammary buds. 106 of these 289 genes were also more highly expressed in PTHrP−/− buds. The early formation of the mammary placodes occurs normally in the absence of PTHrP. Therefore, it is interesting to note that none of the collagen genes, and few of the extracellular matrix associated genes, upregulated in skin as compared to buds are also upregulated in PTHrP−/− buds. Most of the genes in common to these two comparisons are in the growth factor signaling and transcription factor clusters. Selected genes from this comparison are shown in Table 2 and the entire gene sets are available from the NCBI GEO (GSE17654). These data suggest that down-regulation of skin-associated basement membrane and extracellular matrix genes occurs early in the process of mammary placode formation, before the point at which PTHrP signaling contributes to mammary bud development.
Table 2.
Selected genes from the overlap between those overexpressed in PTHrP−/− as compared to WT mammary buds and those overexpressed in WT ventral skin as compared to WT mammary buds.
| Extracellular Matrix | Transcription Factors | Growth Factor Signaling |
|---|---|---|
| Collagen, Type IX, alpha 1 (Col9a1) |
Basic transcription factor 3 (Btf3) |
Neuropilin (Nrp) |
| EGF-like-domain, multiple 6 (Egfl6) |
Enhancer of yellow 2 homolog (Eny2) |
Wnt inhibitory factor 1 (Wif1) |
| Matrilin 2 (Matn2) | Down-regulator of transcription 1 (Dr1) |
WNT1 inducible signaling pathway protein 1 (Wisp1) |
| Reversion-inducing- cyteine-rich protein with kazal motifs (Reck) |
Activating transcription factor 4 (Atf4) |
Nephroblastoma overexpressed gene (Nov, CCN3) |
| Nidogen 1 (Nid1) | Transcription factor 4 (Tcf4) |
Kit oncogene (c-Kit) |
| BSpondin 1 (Spon1) | Nuclear DNA binding protein (C1d) |
Platelet derived growth factor receptor-beta (Pdgfrb) |
| Thrombospondin 1 (Thbs1) | Friend leukemia integration 1 (Fli1) |
Patched homolog 1 (Ptch1) |
| Epidermal growth factor- containing fibulin-like extracellular matrix protein 1 (Efemp1) |
Minichromosome maintenance deficient 6 (Mcm6) |
|
| Asporin (Aspn) |
In order to discover candidate PTHrP-responsive genes expressed within the mammary mesenchyme we identified those genes both overexpressed in WT buds vs. PTHrP−/− buds and overexpressed in K14-PTHrP ventral skin vs. WT ventral skin. We also examined those genes both overexpressed in PTHrP−/− buds vs. WT buds and overexpressed in WT ventral skin vs. K14-PTHrP ventral skin, in order to identify candidate genes repressed by PTHrP signaling within the mammary mesenchyme. This analysis revealed 30 genes potentially up-regulated by PTHrP in the mammary mesenchyme and 5 genes that were potentially down-regulated by PTHrP in the mesenchyme (see Table 3). The relative change in expression and estimated false discovery rate for each of these genes from the microarray data for each of the two comparisons are shown in Supplementary Table 1. Several of these genes (Rspo1, tachykinin 1, Hoxd9) have previously been shown to be expressed within the mesenchyme during embryonic mammary development (Weil et al., 1995; Chen and Capecchi, 1999; Nam et al., 2007). Interestingly, 6 of the 35 genes have been shown either to be targets of, or involved in, BMP signaling (Table 3) (Jaatinen et al., 2002; Laurikkala et al., 2003; Wang and Hirschberg, 2003; Gu et al., 2004; Zhou et al., 2004a; Li et al., 2008; Ohta et al., 2008; Shelton and Yutzey, 2008; Maeda et al., 2009). Finally, as noted in Table 3, 18 of these 35 genes have been implicated as having functions during normal mammary development or to have pathological importance in breast cancers (Shields et al., 2002; Zhou et al., 2004b; Becker et al., 2005; Farias et al., 2005; Mylonas et al., 2005; Patel et al., 2005; Byun et al., 2006; Shimo et al., 2006; Abad et al., 2007; Kwak et al., 2007; Makretsov et al., 2007; Moraes et al., 2007; Cho et al., 2008; Laffin et al., 2008; Martin et al., 2008; Swarbrick et al., 2008; Thuault et al., 2008; Vendrell et al., 2008).
Table 3.
Genes predicted to be regulated by PTHrP in the embryonic mammary mesenchyme. 30 genes were predicted to be up-regulated and 5 were predicted to be down-regulated. Six genes are regulated by or involved in BMP signaling. 18 genes were previously reported to be involved in mammary physiology or breast cancer.
| Gene Symbol |
BMP Signaling |
Mammary/ Breast Cancer |
|---|---|---|
| Up-regulated by PTHrP | ||
| Myl9 | ||
| Sim2 | + | |
| CD99 | + | |
| Tac1 | + | |
| Pcbp4 | ||
| Nnat | + | |
| Hmga2 | + | |
| Abcf1 | ||
| Igfbp4 | + | + |
| Si | ||
| Palld | ||
| Mmp2 | + | + |
| Mrg1 | ||
| 6330403K07Rik | ||
| Serpine 2 | + | |
| Sgta | ||
| Tgfbi | + | |
| Sostdc1 | + | |
| Hoxd9 | + | |
| Rspo1 | ||
| Inhbb | + | + |
| Fxyd6 | ||
| Rbp1 | + | |
| Thy1 | + | |
| Numa1 | + | |
| Bcam | ||
| Timp2 | + | |
| Gna12 | ||
| Tagln | + | |
| Orai1 | ||
| Down-regulated by PTHrP | ||
| Ctgf | + | + |
| Egfl6 | ||
| Vcam1 | ||
| Ptch1 | + | |
| Mme | + | |
Given our previous data showing interactions between PTHrP and BMP4 signaling in the mammary mesenchyme, we first chose to validate the changes in gene expression of the 6 BMP-responsive genes in K14-PTHrP ventral skin versus WT ventral skin by quantitative real-time RT-PCR (qPCR). As shown in Fig. 1, overexpression of PTHrP altered the expression of these 6 genes in the same manner as predicted by the microarray.
Figure 1. Expression of candidate BMP- and PTHrP-regulated genes in WT and K14-PTHrP ventral skin.
Ventral skins were harvested from WT and K14-PTHrP embryos at E15.5. mRNA levels were measured by quantitative, real-time RT-PCR (qPCR). Each bar represents the average of 3 PCR reactions using pooled RNA from 3–5 WT and K14-PTHrP embryos.
PTHrP Regulates MMP2 Expression and Activity
Of the genes potentially regulated by PTHrP and listed in Table 3, one, MMP2, had previously been shown to be regulated by PTHrP in dermal fibroblasts (Maioli et al., 2002). Therefore, we chose to examine further the effects of PTHrP on MMP2 expression. As shown in Fig. 1, MMP2 mRNA levels were 3.4-fold higher in K14-PTHrP ventral skin as compared to WT ventral skin, validating the increase in expression suggested by the microarray data. In order to determine if the increase in MMP2 mRNA was a direct result of PTHrP signaling, we next examined the effects of PTHrP treatment on MMP2 expression in C3H10T1/2 cells, a pluripotent mesenchymal cell line that we had previously shown to be responsive to PTHrP (Hens et al., 2007). MMP2 gene expression has also been shown to be responsive to BMP signaling (Wang and Hirschberg, 2003). Given that PTHrP signaling interacts with BMP signaling in C3H10T1/2 cells and in the mammary mesenchyme in vivo, we examined the effect of BMP4 alone and together with PTHrP. As shown in Fig. 2A, both PTHrP and BMP treatment individually increased the expression of the MMP2 gene in these cells at 24-hours. Although it appeared that PTHrP treatment was somewhat more effective than BMP4 treatment, the effects of PTHrP were also more variable and there was no statistical difference between these treatments. The combination of both PTHrP and BMP4 did not appear to be much more effective at increasing MMP2 expression than treatment with PTHrP alone, but this combination did reach statistical significance as compared to controls. We also assessed the activity of secreted MMP2 in the conditioned media of C3H10T1/2 cells treated with PTHrP, BMP4 and the combination using gelatin zymography. As shown in Figure 2B, treatment with PTHrP and BMP4 both led to an increase in MMP2 activity. However, unlike the effects on mRNA levels, BMP4 appeared to be more effective than PTHrP at inducing MMP2 activity, and the combination was better than either treatment alone. In summary, both MMP2 gene expression and MMP2 activity are increased by PTHrP, BMP4, and the combination of PTHrP and BMP4.
Figure 2. MMP2 gene expression and activity is elevated in response to PTHrP and BMP treatment.
A) qPCR results for C3H10T1/2 cells treated with PTHrP, BMP4 or the combination of PTHrP and BMP4. Bars represent the average and SEM of three independent experiments. * denotes statistical significance as compared to no treatment (NT) by ANOVA (P < 0.05). # denotes that treatment with BMP4 was significantly different than NT when a direct comparison was made using a one-sample t test (P = 0.013). B) MMP activity in conditioned media from C3H10T1/2 cells treated with PTHrP, BMP4 or the combination of PTHrP and BMP4. MMP2 activity was measured by densitometric evaluation of gelatin zymography. Each bar represents the average and SEM of 8–10 individual experiments. Statistical significance was assessed by ANOVA employing the Newman-Keuls multiple comparison test. * denotes statistical significance as compared to NT (P < 0.001). # denotes statistical significance as compared to PTHrP (P < 0.001). § denotes statistical significance as compared to BMP4 (P < 0.05).
Inhibition of MMP Activity Blocks the Outgrowth of Mammary Buds in Culture
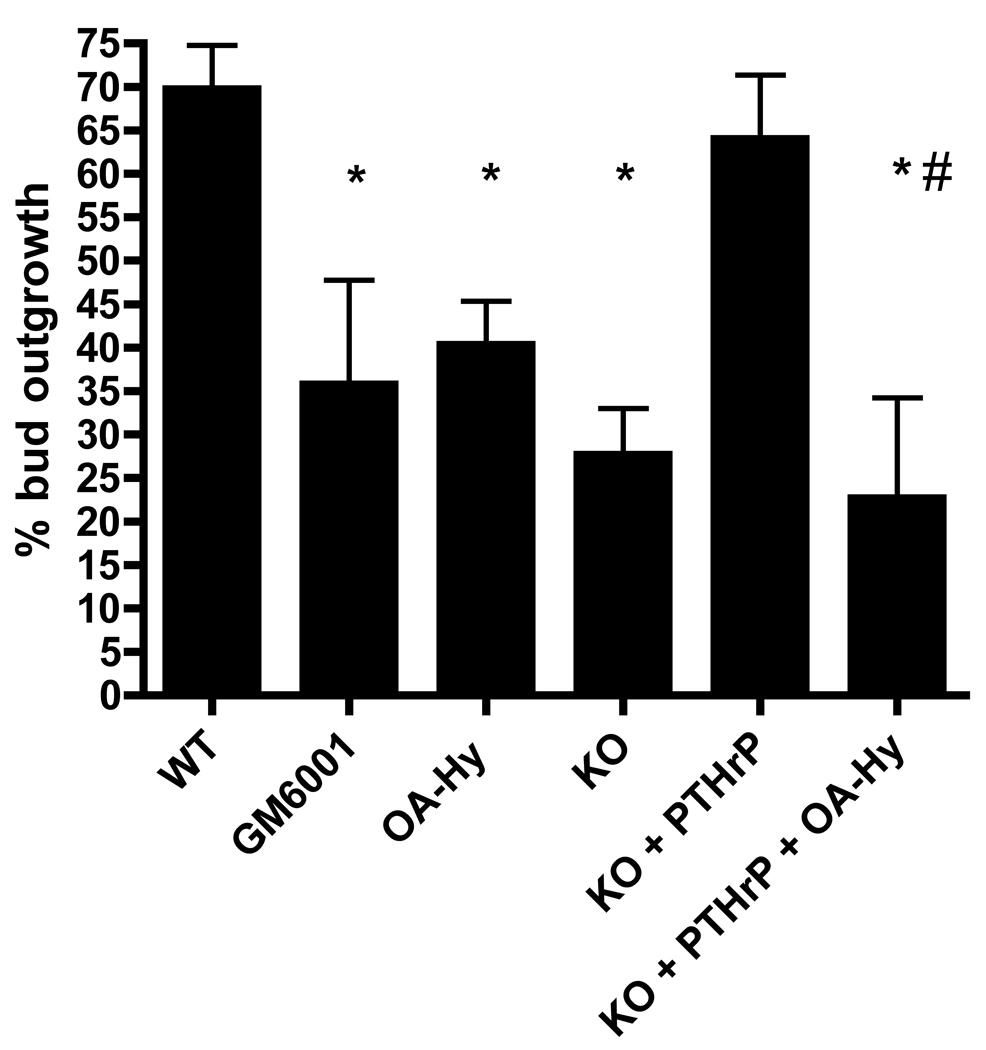
Freshly isolated mammary buds from E13 mouse embryos will initiate branching morphogenesis when cultured with ventral dermal mesenchyme ex vivo (Hens et al., 2007). Using this culture system, we have previously shown that PTHrP−/− mammary buds mimic the mammary phenotype of PTHrP−/− embryos in vivo. Moreover, addition of PTHrP to the media rescues the outgrowth of PTHrP−/− buds in culture. In order to examine the physiological relevance of MMP2 in mammary bud development, we used this same system to determine if MMP inhibitors could block the outgrowth of wild-type mammary buds in culture and/or the ability of PTHrP to rescue outgrowth of PTHrP−/− buds. Mammary buds dissected from WT or PTHrP−/− embryos were cultured in the presence or absence of MMP inhibitors and/or PTHrP for 5 days and outgrowth for each bud was scored as present or absent. As previously defined, buds were considered to have initiated ductal growth if the length of the initial sprout was more than the width of the bud itself and if at least one branch point had formed (Hens et al., 2007). We used two different MMP inhibitors for these experiments. As shown in Fig. 3, treatment of the bud cultures with GM6001, a broad-spectrum MMP inhibitor (Simian et al., 2001), caused a reduction in the percentage of WT mammary buds initiating outgrowth. We performed similar experiments using OA-Hy, a more specific MMP2 antagonist (Berton et al., 2001). Treatment with OA-Hy inhibited outgrowth of WT buds as well (Fig. 3). Once again, we demonstrated that PTHrP−/− buds display impaired ductal outgrowth ex vivo that could be corrected by treatment with exogenous PTHrP (Fig. 3). Importantly, when we treated PTHrP−/− buds with both PTHrP and OA-Hy, the ability of exogenous PTHrP to rescue the outgrowth of PTHrP−/− buds was impaired. The fact that MMP2 antagonists inhibited ductal outgrowth from WT mammary buds in culture suggests that MMP2 activity contributes to the initiation of ductal outgrowth. Furthermore, the ability of MMP2 inhibitors to antagonize the PTHrP-mediated rescue of PTHrP−/− bud outgrowth suggests that MMP2 acts downstream of PTHrP during the process of ductal outgrowth.
Figure 3. Inhibition of MMP2 activity blocks WT bud outgrowth and the ability of PTHrP to rescue outgrowth of PTHrP−/− mammary buds.
Mammary buds were cultured and treated with the general MMP inhibitor, GM6001, or a specific MMP2 inhibitor, OA-Hy, for 5 days. Bars represent the percentage of buds (average ± SEM) demonstrating ductal outgrowth under the culture conditions noted on the graph. Each experiment was performed at least 3 times; the total numbers of buds were as follows: WT = 56 buds, GM6001 = 26 buds, OA-Hy = 22 buds, KO = 15 buds, KO + PTHrP = 15 buds, KO + PTHrP + OA-Hy = 14 buds. Statistical significance was determined by ANOVA employing the Newman-Keuls multiple comparison test. * denotes a significant difference compared to WT. # denotes a significant difference compared to KO + PTHrP. The differences between KO + PTHrP and KO were not statistically significantly by ANOVA, but in a two-way comparison, they reached statistical significance (T-test p value = 0.016).
Discussion
Loss of PTHrP led to a significant downregulation in the expression of a large number (1299) of genes. The most significantly affected genes clustered into functional groups involved in the regulation of glycolysis and energy production, mitochondrial function, protein biosynthesis and structural proteins. An additional 499 genes were expressed at significantly higher levels in PTHrP−/− buds as compared to WT buds. Functional groups within this list of genes included those involved in the regulation of RNA processing and turnover, and chromatin assembly/remodeling. Loss of PTHrP signaling leads to the failure of mammary bud outgrowth (Wysolmerski et al., 1998; Foley et al., 2001; Hens et al., 2007). Thus, these differences in gene expression may reflect the different growth potentials of the buds taken from the two genotypes. Given that WT buds are on the verge of initiating a rapid phase of cell proliferation and morphogenesis, it is likely that they upregulate biosynthetic pathways and the infrastructure involved in energy production. However, PTHrP−/− buds, which will not grow out, fail to upregulate these pathways and cells may instead turn over their RNA pool and condense their chromatin as they enter a quiescent/senescent state. While it is likely that these changes are secondary to the changes in cell fate brought about by the loss in PTHrP signaling, it has also been reported that PTHrP can exert intracrine functions by entering the nucleus and binding to RNA (Wysolmerski, 2008). Moreover, a recent study suggested that the loss of nuclear trafficking of PTHrP in vivo led to widespread cellular senescence (Miao et al., 2008). Therefore, it is possible that some of the changes in the expression of genes associated with protein biosynthesis, RNA processing and/or chromatin remodeling may be direct effects of nuclear PTHrP.
Mammary buds are epidermal appendages and embryonic mammary epithelial cells are recruited from the developing epidermis (Hens and Wysolmerski, 2005; Robinson, 2007; Watson and Khaled, 2008). However, relatively little is known about the mechanisms though which mammary epithelial cells and skin cells diverge from their common epidermal ancestors. Therefore, we also defined genes that were differentially expressed between WT mammary buds and the surrounding ventral skin in the hopes of highlighting pathways involved in mammary epithelial fate determination. It should be noted that since hair follicles are beginning to form in the ventral skin at E15, this comparison between mammary bud and skin fate includes both potential differences between mammary buds and hair follicles as well as between mammary buds and interfollicular epidermis. We identified 196 genes that were overexpressed in mammary buds as compared to skin and 289 genes that were more highly expressed in skin as compared to buds. These data suggest that the suppression of skin-like characteristics may be as important to establishing a mammary identity as the activation of mammary-specific pathways. Functional clustering suggested that changes in transcription factors and growth factor pathways were associated with the differences in cell fate. Interestingly, a specific cohort of keratin genes was upregulated in buds as compared to skin and a large group of collagen and other extracellular matrix genes were upregulated in skin as compared to buds. Further studies will be needed to understand the importance of individual genes within the different functional groups but several have already been implicated in the regulation of mammary bud formation. For example, keratins 8 and 18 had previously been noted to distinguish mammary epithelial from skin cells (Stingl et al., 1998). We found both Tbx3 and GATA3 to be upregulated in mammary buds and both have been shown to be necessary for mammary development (Eblaghie et al., 2004; Jerome-Majewska et al., 2005; Cho et al., 2006; Kouros-Mehr et al., 2006; Asselin-Labat et al., 2007). Lastly, we found Patched 1 to be upregulated in skin as compared to mammary buds. Patched 1 participates in the hedgehog signaling pathway and is, itself, a target of hedgehog signaling (Jiang and Hui, 2008). Two recent studies suggested that suppression of hedgehog signaling is important for mammary bud formation and the establishment or maintenance of mammary epithelial fate in embryos (Hatsell and Cowin, 2006; Gritli-Linde et al., 2007). The downregulation of patched 1 gene expression in buds as compared to ventral skin is consistent with these data.
We are most interested in those genes both regulated by PTHrP and important to mammary bud formation. Table 1 and Table 2 list selected genes from the overlap between those both overexpressed in WT vs. PTHrP−/− buds and mammary buds vs. skin (Table 1), as well as the overlap between the lists of genes overexpressed in WT skin and PTHrP−/− buds (Table 2). The majority of genes (61%) that were overexpressed in mammary buds as compared to skin were also overexpressed in WT buds as compared to PTHrP−/− buds. Since loss of PTHrP signaling involved the downregulation of many genes involved in defining mammary epithelial cell fate, these findings are consistent with previous data suggesting that loss of PTHrP signaling caused mammary epithelial cells to acquire an epidermal cell fate (Foley et al., 2001). In contrast, only 37% of the genes overexpressed in skin vs. buds overlapped with the genes overexpressed in PTHrP−/− buds vs. WT buds, suggesting that much of the program involved in the suppression of epidermal identity plays out normally in the absence of PTHrP signaling, perhaps during the initial formation of the mammary placode. It is especially interesting that a large number of transcription factors were up- or down-regulated in mammary buds as compared to skin and were also affected by alterations in PTHrP signaling. Many of these genes (Sim2, Sox4, Id1, Id4, Hmga2, Tbx3, Btf3, Tcf4 and Atf4) are expressed in normal mammary and/or breast cancer cells (Graham et al., 1999; Beger et al., 2001; Bagheri-Yarmand et al., 2003; Davies et al., 2006; Laffin et al., 2008; Ohta et al., 2008; Swarbrick et al., 2008; Thuault et al., 2008). Interestingly, Atf4 has been reported to impair mammary epithelial differentiation in vivo, so its down-regulation in mammary buds may be functionally important (Bagheri-Yarmand et al., 2003).
Our main interest in profiling PTHrP−/− buds was to identify genes regulated by PTHrP in the mammary mesenchyme. Table 3 lists 35 such candidates. Several aspects of this list are noteworthy. First, 3 of these genes, Rspondin 1, tachykinin 1, and Hoxd9, had previously been shown to be expressed specifically within the mammary mesenchyme, suggesting that this approach was able to identify mesenchymal genes (Weil et al., 1995; Chen and Capecchi, 1999; Nam et al., 2007). Second, 6 of the 35 genes (Table 3) have been shown to be modulators or targets of BMP signaling (Jaatinen et al., 2002; Laurikkala et al., 2003; Wang and Hirschberg, 2003; Gu et al., 2004; Zhou et al., 2004a; Mylonas et al., 2005; Shimo et al., 2006; Li et al., 2008; Ohta et al., 2008; Shelton and Yutzey, 2008; Maeda et al., 2009). We had previously reported that PTHrP signaling sensitizes the mammary mesenchyme to the autocrine/paracrine actions of BMP4 and the microarray data support the notion that PTHrP modulates mesenchymal BMP signaling. It is interesting that the data suggest upregulation of Sostdc1, both a target of BMP signaling and a BMP antagonist. Sostdc1 has been shown to regulate epidermal appendage patterning by mediating lateral inhibition of hair follicle and tooth development around already developing structures (Pummila et al., 2007; Narhi et al., 2008; Munne et al., 2009). Thus, it is possible that induction of Sostdc1 helps to shape the lateral reach of the mammary mesenchyme. Finally, a number of genes on this list are also potential modulators and/or targets of the Wnt signaling pathway. Although we previously did not detect active canonical Wnt signaling in mammary mesenchyme in vivo (Chu et al., 2004), a more recent report suggested that Wnt signaling may indeed be active in the mesenchyme around developing buds (Boras-Granic et al., 2006). Therefore, we are currently reassessing whether PTHrP signaling regulates mesenchymal Wnt signaling in addition to BMP signaling.
Our analysis suggested that PTHrP signaling might regulate MMP2. We confirmed that MMP2 mRNA levels were higher in K14-PTHrP ventral skin as compared to WT ventral skin by qRT-PCR. We had previously shown that PTHrP and BMP4 cooperate to regulate Msx2 gene expression in the pluripotent mesenchymal cell line, C3H10T1/2 cells (Hens et al., 2007), and, therefore, we examined the interaction between PTHrP and BMP4 in regulating MMP2 gene expression and activity in these cells. As shown in Fig. 2, MMP2 expression and activity are regulated by BMP4 in these cells, and PTHrP and BMP4 cooperate to regulate MMP2 expression and activity. In order to determine if MMP2 might mediate some of the effects of PTHrP on ductal development, we examined the effects of two different MMP inhibitors on ductal outgrowth from embryonic mammary buds cultured ex vivo. Both GM6001, a broad-spectrum MMP inhibitor, and OA-Hy, a more specific MMP2 antagonist (Berton et al., 2001; Simian et al., 2001), inhibited ductal outgrowth. Inhibition of MMP2 activity with OA-Hy was also able to prevent exogenous PTHrP from rescuing ductal outgrowth in cultures of PTHrP−/− mammary buds, suggesting that MMP2 participates in the induction of ductal outgrowth in response to PTHrP signaling. MMP activity has previously been shown to regulate branching morphogenesis at several sites and the addition of MMP inhibitors to cultures of embryonic submandibular glands, lung and ureteric buds ex vivo has been reported to inhibit branching morphogenesis (Lelongt et al., 1997; Kheradmand et al., 2002; Steinberg et al., 2005). Inhibition of MMP activity with GM6001 has also been shown to inhibit the formation of branching in three-dimensional cultures of mammary epithelial cells (Simian et al., 2001). MMP2 has specifically been shown to promote ductal extension but suppress ductal side-branching in the mammary gland during puberty in intact mice (Wiseman et al., 2003). Stromally derived MMP2 has also been thought to contribute to breast cancer cell invasion and the establishment of metastases (Jezierska and Motyl, 2009). Therefore, it would appear reasonable that MMP2 contributes to the initiation of ductal morphogenesis from the embryonic mammary bud. However, disruption of the MMP2 gene in mice does not prevent the outgrowth of the initial mammary duct system in vivo (Wiseman et al., 2003). The explanation for this discrepancy is not evident from our experiments, but it may be that other MMPs can compensate for the developmental loss of MMP2 in vivo. Therefore, while our experiments suggest that MMP2 participates in bud outgrowth, it does not appear to be absolutely required for this response.
In summary, we identified a large number of genes that are either downregulated or upregulated in mammary buds in response to the loss of PTHrP. Many of these changes are likely related to alterations in cellular metabolism brought about by the impending growth failure of PTHrP−/− mammary buds. By comparing these large lists against differences in gene expression in WT mammary buds as compared to WT ventral skin, and against differences between K14-PTHrP and WT skin, we have also identified much smaller lists of genes enriched for growth factor pathways and transcription factors. These changes in gene expression, which are likely more directly related to PTHrP signaling support prior work demonstrating that PTHrP modulates mesenchymal BMP signaling and suggest that PTHrP may contribute to ductal outgrowth, at least in part, by modulating MMP2. Further studies into the functions of these pathways should enhance our understanding of embryonic mammary gland development.
Experimental Procedures
Animals
Animal experiments were approved by the Yale University IACUC. CD-1 wild-type mice were acquired from Charles River. K14-PTHrP, PTHrP−/− and PTH1R−/− mice were generated from established breeding colonies and identified as described previously (Dunbar et al., 1999; Foley et al., 2001).
Microarray Analysis
Embryonic mammary buds and ventral skin were dissected from WT, PTHrP−/− and K14-PTHrP transgenic embryos harvested on E15. Dissections were performed on an icepack and tissue samples stored in RNA Later (Ambion, Austin, TX) at −20°C. After genotyping the embryos, like samples were pooled and RNA was isolated using the RNeasy Mini Protocol with a Qiashredder column (Qiagen, Valencia, CA). Mechanical disruption of tissue was performed with a Kontes Pellet Pestle and a cordless motor (Thermo Fisher Scientific, Waltham, MA) prior to homogenizing with the Qiashredder. MessageClean (GenHunter Corporation, Nashville, TN) was used to remove contaminating DNA. On average, 120 mammary buds were required to produce approximately 20 ug total RNA. Biotinylated cRNA was generated from total RNA and hybridized to Affymetrix MOE430A oligonucleotide arrays (Affymetrix, Santa Clara, CA). Each experimental condition was hybridized in triplicate. Sample preparation and raw data collection were performed as described (Master et al., 2002). Data were normalized using GC-RMA (Zhijin et al., 2004) in R. Genes were filtered by retaining only probe sets with at least one MAS5 present call and more than 1.2 fold dynamic range across all samples. Differential expression of genes was assessed by Cyber-T (Baldi and Long, 2001). No specific cutoff threshold was used, but instead gene lists were generated at false discovery rate (FDR) of 10% for comparisons among the ventral skin sample groups, and at FDR of 5% for all other comparisons. The false discovery rates were estimated as part of the Cyber-T algorithm, by modeling t-test p-values as a mixture of beta (differentially expressed genes) and uniform (other genes) distributions. A p-value cutoff was chosen for each individual comparison such that the estimated proportion of non-changing genes fell below the desired percentage. Functional annotation clustering was performed using the DAVID bioinformatics database and resource of the National Institute of Allergy and Infectious Diseases (NIAID, NIH, Bethesda, MD).
Real-time PCR
Ventral skin was harvested from E15.5 embryos and stored in RNA later at –20°C until embryos were genotyped. Samples from like embryos were then pooled and total RNA was isolated using the Qiagen RNeasy kit with QiaShredder column (Qiagen Corp., Valencia, CA). RNA was further purified by Dnase 1 digestion using the MessageClean kit (GenHunter Corp., Nashville, TN). Total RNA was prepared from C3H10T1/2 cells using Trizol reagent (Invitrogen, Carlsbad, CA) followed by DNAse digestion as above. Quantitative RT-PCR (qRT-PCR) was performed with the Opticon II DNA engine (JM Research, Waltham, MA) using standard methods (Hens et al., 2007). The following primer sets were used: MMP2, forward 5’-CAAAAACAAGAAGACATACATCTT-3’ and reverse 5’-GCTTCCAAACTTCACGCTC-3’; INHBB, forward 5'-TCAGCTTTGCAGAGACATGG-3' and reverse 5'-ACCTTGACCCGTACCTTCCT-3'; IGFBP4, forward 5'-CGTCCTGTGCCCCAGGGTTCCT-3' and reverse 5'-GAAGCTTCACCCCTGTCTTCCG-3'; SOSTDC1, forward 5'-GAGGCAGGCATTTCAGTAGC-3' and reverse 5'-ATAGCCTCCTCCGATCCAGT-3'; TGFBI, forward 5'-GACTGCTGACCCTCGCTCT-3' and reverse 5'-GTTGGTGCCAATGACCTTCT-3'; and CTGF, forward 5'-CAAAGCAGCTGCAAATACCA-3' and reverse 5'-GGCCAAATGTGTCTTCCAGT-3'. SYBR-Green-based qRT-PCR was performed with Brilliant SYBR-Green qRT-PCR master mix (Stratagene, Agilent Technololgies, Santa Clara, CA). Samples were normalized for relative quantification of expression by the 2−ΔΔCT method using GAPDH as an internal control (relative quantification of gene expression: ABI Prism 7700 sequence detection system, user bulletin 2, revision B). Samples were run in triplicate. cDNA was prepared using the ABI PRISM as per the manufacturer’s instructions.
Cell Culture and Zymography
C3H10T1/2 cells (a gift from Dr. Mark Horowitz, New Haven, CT) were cultured on 100 mm Falcon dishes (Thermo Fisher Scientific, Waltham, MA) in DMEM supplemented with 10% fetal bovine serum (FBS), and 0.2 mM L-glutamine. Culture media was changed to 0.1% FBS 24-hours before treatment with either 50 ng/ml recombinant BMP4 (R&D Systems, Minneapolis, MN), 10−7 M PTHrP(1–34) (Sigma-Aldrich, St. Louis, MO), or both. Cells were harvested for RNA expression analysis after 24 hrs whereas MMP activity was assessed in conditioned media harvested after 4 days of treatment. Gelatin zymography was performed using 10% Biorad-Ready gelatin gels following the manufacturer’s protocol (Biorad Laboratories, Hercules, CA). Gels were counterstained in Coomassie Brilliant blue R-250 staining solution for 1 hr, and de-stained in 10% acetic acid/50% methanol/40% de-ionized water until the clear bands of protease activity were apparent. Gels were then photographed and densitometry was performed to quantify protease activity using a Kodak Digital Science Image Station 440 CF (Kodak Corp., Rochester, NY).
Mammary bud culture
All dissections were performed in DMEM at 4°C. Mammary buds and ventral mesenchyme were microdissected from E13 wild-type and PTHrP−/− embryos. Individual mammary buds were placed on tufts of ventral mesenchyme on Whatman 13 mm nuclepore Track-etched membranes (8 µm pore size; Thomas Scientific, Swedesboro, NJ). The filters were cultured on EC587-40 mesh screen grills (Thomas Scientific, Swedesboro, NJ) in six-well plates containing 10% FBS in DMEM/F12 media with antibiotics. Cultures were treated with 10−7 M PTHrP (1–34) (Sigma-Aldrich, St. Louis, MO), 27 nM GM6001 or 2.55 µM cis-9-Octadecenoyl-N-hydroxylamide (OaHy) (Calbiochem, San Diego, CA). Media was changed every other day and after 5 days of culture, bud outgrowths were fixed in acid alcohol and stained in carmine alum. Stained tissue was then dehydrated and mounted in Permount (Thermo Fisher Scientific, Waltham, MA) for viewing.
Statistical Analysis
GraphPad Prism, Version 4.0 for MacIntosh was used for statistical analysis (GraphPad Software, San Diego, CA). Comparisons of three or more groups used one-way analysis of variance with the Newman-Keuls multiple comparison test. Comparisons of 2 groups used a one-sample Student’s t test when one of the groups was arbitrarily set equal to one (fold-stimulation calculations) and a paired Student’s t test when both groups varied freely.
Supplementary Material
Acknowledgments
Grant Information:
Grant Sponsor: NIH (NIDDK); Grant Number: DK055501
References
- Abad PC, Lewis J, Mian IS, Knowles DW, Sturgis J, Badve S, Xie J, Lelievre SA. NuMA influences higher order chromatin organization in human mammary epithelium. Mol Biol Cell. 2007;18:348–361. doi: 10.1091/mbc.E06-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bagheri-Yarmand R, Vadlamudi RK, Kumar R. Activating transcription factor 4 overexpression inhibits proliferation and differentiation of mammary epithelium resulting in impaired lactation and accelerated involution. J Biol Chem. 2003;278:17421–17429. doi: 10.1074/jbc.M300761200. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Becker M, Sommer A, Kratzschmar JR, Seidel H, Pohlenz HD, Fichtner I. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther. 2005;4:151–168. [PubMed] [Google Scholar]
- Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, Wong-Staal F. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci U S A. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- Berton A, Rigot V, Huet E, Decarme M, Eeckhout Y, Patthy L, Godeau G, Hornebeck W, Bellon G, Emonard H. Involvement of fibronectin type II repeats in the efficient inhibition of gelatinases A and B by long-chain unsaturated fatty acids. J Biol Chem. 2001;276:20458–20465. doi: 10.1074/jbc.M011664200. [DOI] [PubMed] [Google Scholar]
- Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA. Bone morphogenetic proteins and their antagonists in skin and hair follicle biology. J Invest Dermatol. 2003;120:36–47. doi: 10.1046/j.1523-1747.2003.12002.x. [DOI] [PubMed] [Google Scholar]
- Byun HJ, Hong IK, Kim E, Jin YJ, Jeoung DI, Hahn JH, Kim YM, Park SH, Lee H. A splice variant of CD99 increases motility and MMP-9 expression of human breast cancer cells through the AKT-, ERK-, and JNK-dependent AP-1 activation signaling pathways. J Biol Chem. 2006;281:34833–34847. doi: 10.1074/jbc.M605483200. [DOI] [PubMed] [Google Scholar]
- Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci U S A. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KW, Kim JY, Song SJ, Farrell E, Eblaghie MC, Kim HJ, Tickle C, Jung HS. Molecular interactions between Tbx3 and Bmp4 and a model for dorsoventral positioning of mammary gland development. Proc Natl Acad Sci U S A. 2006;103:16788–16793. doi: 10.1073/pnas.0604645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, Gurney A, Lewicki J, Clarke MF. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- Chu EY, Hens J, Andl T, Kairo A, Yamaguchi TP, Brisken C, Glick A, Wysolmerski JJ, Millar SE. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- Davies CR, Morris JS, Griffiths MR, Page MJ, Pitt A, Stein T, Gusterson BA. Proteomic analysis of the mouse mammary gland is a powerful tool to identify novel proteins that are differentially expressed during mammary development. Proteomics. 2006;6:5694–5704. doi: 10.1002/pmic.200600202. [DOI] [PubMed] [Google Scholar]
- Dean C, Ito M, Makarenkova HP, Faber SC, Lang RA. Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development. 2004;131:4155–4165. doi: 10.1242/dev.01285. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Dunbar ME, Dann P, Robinson GW, Hennighausen L, Zhang JP, Wysolmerski JJ. Parathyroid hormone-related protein signaling is necessary for sexual dimorphism during embryonic mammary development. Development. 1999;126:3485–3493. doi: 10.1242/dev.126.16.3485. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol. 2006;291:67–82. doi: 10.1016/j.ydbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Song SJ, Kim JY, Akita K, Tickle C, Jung HS. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J Anat. 2004;205:1–13. doi: 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias EF, Ong DE, Ghyselinck NB, Nakajo S, Kuppumbatti YS, Mira y, Lopez R. Cellular retinol-binding protein I, a regulator of breast epithelial retinoic acid receptor activity, cell differentiation, and tumorigenicity. J Natl Cancer Inst. 2005;97:21–29. doi: 10.1093/jnci/dji004. [DOI] [PubMed] [Google Scholar]
- Foley J, Dann P, Hong J, Cosgrove J, Dreyer BE, Rimm D, Dunbar ME, Philbrick WM, Wysolmerski JJ. Parathyroid hormone-related protein maintains mammary epithial fate and triggers nipple skin differentiation during embryonic bresat development. Development. 2001;128:513–525. doi: 10.1242/dev.128.4.513. [DOI] [PubMed] [Google Scholar]
- Graham JD, Hunt SM, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol. 1999;22:295–304. doi: 10.1677/jme.0.0220295. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, Hallberg K, Harfe BD, Reyahi A, Kannius-Janson M, Nilsson J, Cobourne MT, Sharpe PT, McMahon AP, Linde A. Abnormal hair development and apparent follicular transformation to mammary gland in the absence of hedgehog signaling. Dev Cell. 2007;12:99–112. doi: 10.1016/j.devcel.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K, Zhang L, Jin T, Rutherford RB. Identification of potential modifiers of Runx2/Cbfa1 activity in C2C12 cells in response to bone morphogenetic protein-7. Cells Tissues Organs. 2004;176:28–40. doi: 10.1159/000075025. [DOI] [PubMed] [Google Scholar]
- Hatsell SJ, Cowin P. Gli3-mediated repression of Hedgehog targets is required for normal mammary development. Development. 2006;133:3661–3670. doi: 10.1242/dev.02542. [DOI] [PubMed] [Google Scholar]
- Hens JR, Dann P, Zhang JP, Harris S, Robinson GW, Wysolmerski J. BMP4 and PTHrP interact to stimulate ductal outgrowth during embryonic mammary development and to inhibit hair follicle induction. Development. 2007;134:1221–1230. doi: 10.1242/dev.000182. [DOI] [PubMed] [Google Scholar]
- Hens JR, Wysolmerski JJ. Key stages of mammary gland development: molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7:220–224. doi: 10.1186/bcr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B, Panchal H, McCarthy A, Ashworth A. Identification of the scaramanga gene implicates Neuregulin3 in mammary gland specification. Genes Dev. 2005;19:2078–2090. doi: 10.1101/gad.338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaatinen R, Bondestam J, Raivio T, Hilden K, Dunkel L, Groome N, Ritvos O. Activation of the bone morphogenetic protein signaling pathway induces inhibin beta(B)-subunit mRNA and secreted inhibin B levels in cultured human granulosa-luteal cells. J Clin Endocrinol Metab. 2002;87:1254–1261. doi: 10.1210/jcem.87.3.8314. [DOI] [PubMed] [Google Scholar]
- Jerome-Majewska LA, Jenkins GP, Ernstoff E, Zindy F, Sherr CJ, Papaioannou VE. Tbx3, the ulnar-mammary syndrome gene, and Tbx2 interact in mammary gland development through a p19Arf/p53-independent pathway. Dev Dyn. 2005;234:922–933. doi: 10.1002/dvdy.20575. [DOI] [PubMed] [Google Scholar]
- Jezierska A, Motyl T. Matrix Metalloproteinase-2 involvement in breast cancer progression: a mini-review. Med Sci Monit. 2009;15:RA32–RA40. [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand F, Rishi K, Werb Z. Signaling through the EGF receptor controls lung morphogenesis in part by regulating MT1-MMP-mediated activation of gelatinase A/MMP2. J Cell Sci. 2002;115:839–848. doi: 10.1242/jcs.115.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;28:259–266. doi: 10.1093/carcin/bgl122. [DOI] [PubMed] [Google Scholar]
- Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, Schedin P, Porter WW. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm ML, Podlasek CA, Barnett DH, Lee J, Clemens JQ, Hebner CM, Bushman W. Mesenchymal factor bone morphogenetic protein 4 restricts ductal budding and branching morphogenesis in the developing prostate. Dev Biol. 2001;232:301–314. doi: 10.1006/dbio.2001.0187. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Kassai Y, Pakkasjarvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Lelongt B, Trugnan G, Murphy G, Ronco PM. Matrix metalloproteinases MMP2 and MMP9 are produced in early stages of kidney morphogenesis but only MMP9 is required for renal organogenesis in vitro. J Cell Biol. 1997;136:1363–1373. doi: 10.1083/jcb.136.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TS, Komota T, Ohshima M, Qin SL, Kubo M, Ueda K, Hamano K. TGF-beta induces the differentiation of bone marrow stem cells into immature cardiomyocytes. Biochem Biophys Res Commun. 2008;366:1074–1080. doi: 10.1016/j.bbrc.2007.12.095. [DOI] [PubMed] [Google Scholar]
- Maeda A, Nishida T, Aoyama E, Kubota S, Lyons KM, Kuboki T, Takigawa M. CCN Family 2/Connective Tissue Growth Factor Modulates BMP Signalling as a Signal Conductor, Which Action Regulates the Proliferation and Differentiation of Chondrocytes. J Biochem. 2009;145:207–216. doi: 10.1093/jb/mvn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, Kato S, Dickson C, Thiery JP, Bellusci S. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129:53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- Maioli E, Fortino V, Torricelli C, Arezzini B, Gardi C. Effect of parathyroid hormone-related protein on fibroblast proliferation and collagen metabolism in human skin. Exp Dermatol. 2002;11:302–310. doi: 10.1034/j.1600-0625.2002.110403.x. [DOI] [PubMed] [Google Scholar]
- Makretsov NA, Hayes M, Carter BA, Dabiri S, Gilks CB, Huntsman DG. Stromal CD10 expression in invasive breast carcinoma correlates with poor prognosis, estrogen receptor negativity, and high grade. Mod Pathol. 2007;20:84–89. doi: 10.1038/modpathol.3800713. [DOI] [PubMed] [Google Scholar]
- Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS ONE. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Mishina Y, Bertram JF. BMPs and BMP receptors in mouse metanephric development: in vivo and in vitro studies. Int J Dev Biol. 2002;46:525–533. [PubMed] [Google Scholar]
- Master SR, Hartman JL, D'Cruz CM, Moody SE, Keiper EA, Ha SI, Cox JD, Belka GK, Chodosh LA. Functional microarray analysis of mammary organogenesis reveals a developmental role in adaptive thermogenesis. Mol Endocrinol. 2002;16:1185–1203. doi: 10.1210/mend.16.6.0865. [DOI] [PubMed] [Google Scholar]
- Miao D, Su H, He B, Gao J, Xia Q, Zhu M, Gu Z, Goltzman D, Karaplis AC. Severe growth retardation and early lethality in mice lacking the nuclear localization sequence and C-terminus of PTH-related protein. Proc Natl Acad Sci U S A. 2008;105:20309–20314. doi: 10.1073/pnas.0805690105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y, Oshima K, Fogo A, Hogan BL, Ichikawa I. Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J Clin Invest. 2000;105:863–873. doi: 10.1172/JCI8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes RC, Zhang X, Harrington N, Fung JY, Wu MF, Hilsenbeck SG, Allred DC, Lewis MT. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- Munne PM, Tummers M, Jarvinen E, Thesleff I, Jernvall J. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development. 2009;136:393–402. doi: 10.1242/dev.025064. [DOI] [PubMed] [Google Scholar]
- Mustonen T, Ilmonen M, Pummila M, Kangas AT, Laurikkala J, Jaatinen R, Pispa J, Gaide O, Schneider P, Thesleff I, Mikkola ML. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- Mylonas I, Jeschke U, Shabani N, Kuhn C, Friese K, Gerber B. Inhibin/activin subunits (inhibin-alpha, -betaA and -betaB) are differentially expressed in human breast cancer and their metastasis. Oncol Rep. 2005;13:81–88. [PubMed] [Google Scholar]
- Nam JS, Turcotte TJ, Yoon JK. Dynamic expression of R-spondin family genes in mouse development. Gene Expr Patterns. 2007;7:306–312. doi: 10.1016/j.modgep.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Narhi K, Jarvinen E, Birchmeier W, Taketo MM, Mikkola ML, Thesleff I. Sustained epithelial beta-catenin activity induces precocious hair development but disrupts hair follicle down-growth and hair shaft formation. Development. 2008;135:1019–1028. doi: 10.1242/dev.016550. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Nakagawa K, Imai Y, Katagiri T, Koike T, Takaoka K. Cyclic AMP enhances Smad-mediated BMP signaling through PKA-CREB pathway. J Bone Miner Metab. 2008;26:478–484. doi: 10.1007/s00774-008-0850-8. [DOI] [PubMed] [Google Scholar]
- Panchal H, Wansbury O, Parry S, Ashworth A, Howard B. Neuregulin3 alters cell fate in the epidermis and mammary gland. BMC Dev Biol. 2007;7:105. doi: 10.1186/1471-213X-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel HJ, Ramkissoon SH, Patel PS, Rameshwar P. Transformation of breast cells by truncated neurokinin-1 receptor is secondary to activation by preprotachykinin-A peptides. Proc Natl Acad Sci U S A. 2005;102:17436–17441. doi: 10.1073/pnas.0506351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pummila M, Fliniaux I, Jaatinen R, James MJ, Laurikkala J, Schneider P, Thesleff I, Mikkola ML. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 2007;134:117–125. doi: 10.1242/dev.02708. [DOI] [PubMed] [Google Scholar]
- Robinson GW. Cooperation of signalling pathways in embryonic mammary gland development. Nat Rev Genet. 2007;8:963–972. doi: 10.1038/nrg2227. [DOI] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Dev Biol. 2008;317:282–295. doi: 10.1016/j.ydbio.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zhao J, Anderson KD, Warburton D. Gremlin negatively modulates BMP-4 induction of embryonic mouse lung branching morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1030–L1039. doi: 10.1152/ajplung.2001.280.5.L1030. [DOI] [PubMed] [Google Scholar]
- Shields JM, Rogers-Graham K, Der CJ. Loss of transgelin in breast and colon tumors and in RIE-1 cells by Ras deregulation of gene expression through Raf-independent pathways. J Biol Chem. 2002;277:9790–9799. doi: 10.1074/jbc.M110086200. [DOI] [PubMed] [Google Scholar]
- Shimo T, Kubota S, Yoshioka N, Ibaragi S, Isowa S, Eguchi T, Sasaki A, Takigawa M. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J Bone Miner Res. 2006;21:1045–1059. doi: 10.1359/jbmr.060416. [DOI] [PubMed] [Google Scholar]
- Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, Hoffman MP. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 2005;132:1223–1234. doi: 10.1242/dev.01690. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- Swarbrick A, Roy E, Allen T, Bishop JM. Id1 cooperates with oncogenic Ras to induce metastatic mammary carcinoma by subversion of the cellular senescence response. Proc Natl Acad Sci U S A. 2008;105:5402–5407. doi: 10.1073/pnas.0801505105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, Rice R, Spencer-Dene B, Mailleux AA, Rice DP, Thiery JP, Bellusci S. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. 2006;133:2325–2335. doi: 10.1242/dev.02394. [DOI] [PubMed] [Google Scholar]
- Vendrell JA, Robertson KE, Ravel P, Bray SE, Bajard A, Purdie CA, Nguyen C, Hadad SM, Bieche I, Chabaud S, Bachelot T, Thompson AM, Cohen PA. A candidate molecular signature associated with tamoxifen failure in primary breast cancer. Breast Cancer Res. 2008;10:R88. doi: 10.1186/bcr2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bubnoff A, Cho KW. Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev Biol. 2001;239:1–14. doi: 10.1006/dbio.2001.0388. [DOI] [PubMed] [Google Scholar]
- Wang S, Hirschberg R. BMP7 antagonizes TGF-beta -dependent fibrogenesis in mesangial cells. Am J Physiol Renal Physiol. 2003;284:F1006–F1013. doi: 10.1152/ajprenal.00382.2002. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- Weil M, Itin A, Keshet E. A role for mesenchyme-derived tachykinins in tooth and mammary gland morphogenesis. Development. 1995;121:2419–2428. doi: 10.1242/dev.121.8.2419. [DOI] [PubMed] [Google Scholar]
- Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ, Soloway P, Itohara S, Werb Z. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003;162:1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysolmerski JJ. Parathyroid hormone-related protein. In: CJ Rosen., editor. Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism. Washington, D.C: American Society for Bone and Mineral Research; 2008. pp. 127–133. [Google Scholar]
- Wysolmerski JJ, Cormier S, Philbrick WM, Dann P, Zhang JP, Roume J, Delezoide AL, Silve C. Absence of functional type 1 parathyroid hormone (PTH)/PTH-related protein recpetors in humans is associated with abnormal breast development and tooth impaction. Journal of Clinical Endocrinology and Metabolism. 2001;86:1788–1794. doi: 10.1210/jcem.86.4.7404. [DOI] [PubMed] [Google Scholar]
- Wysolmerski JJ, Philbrick WM, Dunbar ME, Lanske B, Kronenberg HM, Karaplis AC, Broadus AE. rescue of the parathyroid hormone-related protein knockout mouse demonstrates that parathyroid hormone-related protein is essential for mammary gland development. Development. 1998;125:1285–1294. doi: 10.1242/dev.125.7.1285. [DOI] [PubMed] [Google Scholar]
- Zhijin W, Rafael A, Robert G, Francisco M-M, Forrest S. A model-based background background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- Zhou S, Glowacki J, Yates KE. Comparison of TGF-beta/BMP pathways signaled by demineralized bone powder and BMP-2 in human dermal fibroblasts. J Bone Miner Res. 2004a;19:1732–1741. doi: 10.1359/JBMR.040702. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yu C, Miao X, Tan W, Liang G, Xiong P, Sun T, Lin D. Substantial reduction in risk of breast cancer associated with genetic polymorphisms in the promoters of the matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 genes. Carcinogenesis. 2004b;25:399–404. doi: 10.1093/carcin/bgh020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.