Abstract
Enduracidins (1, 2) and ramoplanin (3) are structurally and functionally closely related lipodepsipeptide antibiotics. They are active against multidrug resistant Gram-positive pathogens, including MRSA. Each peptide contains one chlorinated non-proteinogenic amino acid residue, Cl2-Hpg or Cl-Hpg. To investigate the timing of halogenation, the importance of chlorination on bioactivity and bioavailability of enduracidin, and to probe the substrate specificity and portability of the ramoplanin halogenase, we constructed the mutant strain SfΔ30 in which the enduracidin halogenase gene orf30 had been deleted and complemented it with the ramoplanin counterpart orf20. We also expressed orf20 in the enduracidin wild type producer. Metabolite analysis revealed SfΔ30 produced the novel analogues dideschloroenduracidins A (4) and B (5), while the recombinant strains SfΔ30R20 and SfR20 produced monodeschloroenduracidins A (6) and B (7), and a trichlorinated enduracidin (8), respectively. In addition, orf30 self-complementation yielded the strain SfΔ30E30 which is capable of producing six peptides including 6 and 7. MS/MS analysis positioned the single chlorine atom in 6 at Hpg13 and localized the third chlorine atom in 8 to Hpg11. Biological evaluation of these enduracidin analogues indicated that all retained activity against Staphylococcus aureus. Our findings lay the foundation for further utilization of enduracidin and ramoplanin halogenases in combinatorial biosynthesis.
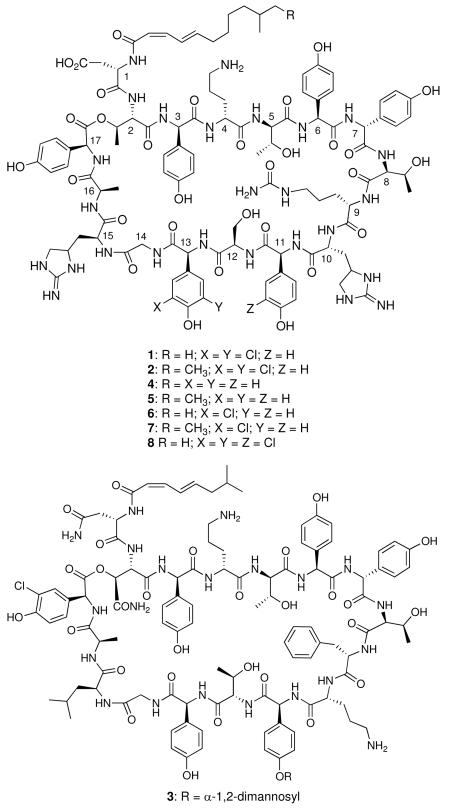
Enduracidins A (1) and B (2) are among 17 amino acid lipodepsipeptides discovered in the late 1960s from fermentations of the soil bacterium Streptomyces fungicidicus (ATCC21013).1, 2 The two peptides are analogues that differ by one carbon in the length of the attached lipid chain. Structurally, enduracidins are distinguished by a C12 or C13 2Z,4E branched fatty acid moiety and the presence of numerous nonproteinogenic amino acid residues, such as enduracididine (End), 4-hydroxyphenylglycine (Hpg), 3,5-dichloro-4-hydroxyphenylglycine (Cl2-Hpg), citrulline (Cit) and ornithine (Orn). Seven of the 17 amino acids have the D configuration and six of the residues are Hpg or a chlorinated derivative. Enduracidin (for simplicity, the peptides will be referred to singularly) exhibits potent in vitro and in vivo antibacterial activity against a wide spectrum of primarily Gram-positive organisms, including methicillin-resistant Staphylococcus aureus (MRSA).3-7 Minimal inhibitory concentrations (MICs) are as low as 0.05 μg/mL and the effect is bactericidal.4, 6 Typical MICs for vancomycin toward sensitive strains of S. aureus range from 0.5 to 2 μg/mL. Enduracidin has been shown to be effective in humans for treating urinary tract and skin infections caused by MRSA, but not chronic bone infections.8 No toxicity or side effects were reported.
The only peptide related to enduracidin that has been structurally characterized is ramoplanin (3, A-16686) reported in 1984 from the Actinoplanes strain ATCC33076.9, 10 The differences in the peptides are the shorter C8 unsaturated lipid tail, an α-1,2-dimannosyl moiety appended to Hpg11, and substitution of D-Orn and L-Leu for the D- and L-enduracididine residues at C-10 and C-15, respectively. Like enduracidin, ramoplanin exhibits potent bactericidal activity and was reported to be 4 to 8-fold more active than vancomycin versus 500 strains of Gram-positive species.11 Ramoplanin was shown to be active against Gram-positive pathogens including vancomycin-resistant enterococci (VRE) and MRSA.12, 13 Ramoplanin has recently been in development for the treatment of Clostridium difficile infections.14-16
Halogenation occurs widely in the biosynthesis of more than 4,000 natural products and it is a common structural feature in many peptide antibiotics such as enduracidin and ramoplanin.17 The steric and electronegative effects of chlorine can have significant consequences on bioactivity.17-20 Therefore, halogenase genes have been targeted for combinatorial biosynthesis to create structural analogues of natural product antibiotics via genetic engineering.18, 21-25 In our previous report on the characterization of the enduracidin gene cluster, we found a single putative halogenase gene (orf30) and predicted it to be responsible for chlorination of the Hpg residue at C-13 of enduracidin. In this report, we demonstrate production of enduracidin analogues by genetic manipulation of the enduracidin and ramoplanin halogenases genes in S. fungicidicus.
Results and Discussion
Generation of the In-frame Deletion Mutant Strain SfΔ30
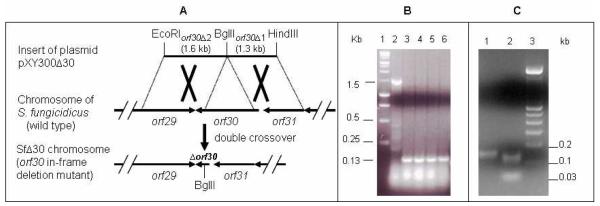
Sequence analysis of the enduracidin cluster revealed a single halogenase gene, orf30, which was predicted to introduce both chlorine atoms of the Cl2-Hpg13 residue in enduracidin.26 A halogenase deficient mutant was constructed by in-frame deletion via double crossover homologous recombination to decipher the timing of halogenation, the importance of this modification on antibiotic activity, and to explore the substrate specificity of these enzymes for use in combinatorial biosynthesis. The gene replacement delivery plasmid pXY300Δ30 designed to delete orf30 from the chromosome was constructed. After the in-frame deletion and error-free sequence of the insert were confirmed by restriction analysis and sequencing, pXY300Δ30 was conjugally introduced into the wild type S. fungicidicus. The rest of the procedures for double crossover homologous recombination were as described previously.26 Thiostrepton-resistant exconjugants were isolated and passed through high temperature (40 °C) selection. Independent markerless double crossover mutants showing thiostrepton-sensitive phenotype were obtained. The orf30 in-frame deletion mutants were further confirmed by PCR (Figure 1), sequencing and Southern analysis (data not shown).
Figure 1.
(A) Schematic presentation of the halogenase gene orf30 in-frame deletion mutation. (B) PCR analysis confirming chromosomal in-frame deletion of orf30 in the mutant strain SfΔ30 and BglII digestion of the gel-purified PCR products confirming the BglII site introduced into the chromosome of SfΔ30 (C). PCR products were separated on a 2% agarose gel. (B) Lane 1, molecular marker (Fermentas GeneRuler 1 kb DNA Ladder); Lane 2, PCR product from wild-type S. fungicidicus (1.5 kb); Lanes 3-6, PCR products from independent mutant colonies (0.13 kb). (C) Lane 1, gel-purified PCR product from SfΔ30; Lane 2, gel-purified PCR product digested with BglII; Lane 3, molecular marker (Fermentas GeneRuler 100 bp DNA Ladder).
Production of Dideschloroenduracidins A and B
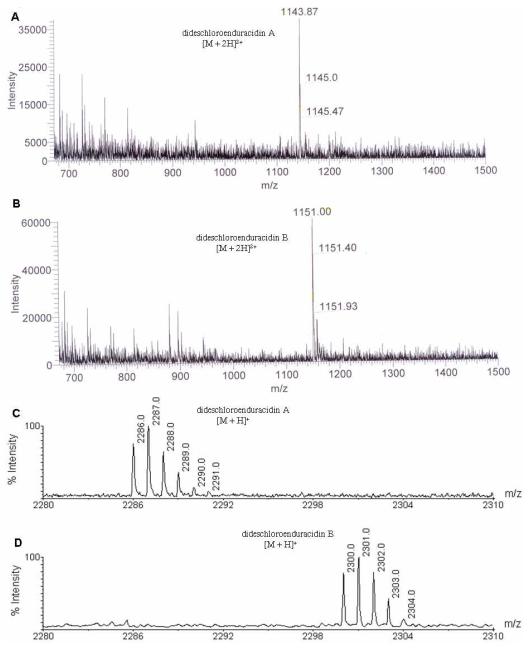
All media and growth conditions used for fermentation of the mutant strain SfΔ30, and methods for the metabolite preparation and analysis by HPLC were the same as reported previously.26 HPLC analysis of the metabolites produced by the mutant SfΔ30 revealed two new chromatographic peaks (Figure S1). These peaks were collected and analyzed by LC-MS ESI which confirmed they were the dideschloroenduracidins A (4, [M+2H]2+ = m/z 1143.87, Figure 2A) and B (5, [M+2H]2+ = m/z 1151.00, Figure 2B). The production of 4 and 5 by SfΔ30 was also confirmed by MALDI-TOF MS analysis with the purified compounds (Figure 2C and 2D). This serves to support the hypothesis that Orf30 introduces both chlorine atoms on Hpg13 of 1 and illustrates that prior chlorination of the free Hpg is not required for peptide assembly. Our results were similar to those observed when the disruption of the single halogenase gene in the balhimycin cluster, bhaA, led to complete loss of two chlorine atoms at residues 2 and 6 of this glycopeptide antibiotic.22 Further, the results are consistent with the previous prediction that a single halogenase, ComH, is involved in the formation of the Cl2-Hpg residues in complestatin.27 Regarding the timing of halogenation, in the balhimycin system, complementation studies with β-hydroxytyrosine (Hty) and chlorinated β-hydroxytyrosine suggested that halogenation occurs after completion of Hty biosynthesis and during peptide assembly on the NRPS.28 The formation of a chlorinated dipeptide when the balhimycin thioesterase was inserted behind module 2 to replace the C domain of module 3 in BpsA further supports that halogenation occurs on an NRPS-bound amino acid and not after completion of peptide formation.29 The timing of halogenation of enduracidin is presumed to be similar to that of balhimycin.
Figure 2.
LC-ESI-MS analysis of partially purified dideschloroenduracidins A (A) and B (B). MALDI-TOF MS analysis of the peaks corresponding to dideschloroenduracidins A (C) and B (D).
Self-complementation of the orf30 In-frame Deletion Mutant
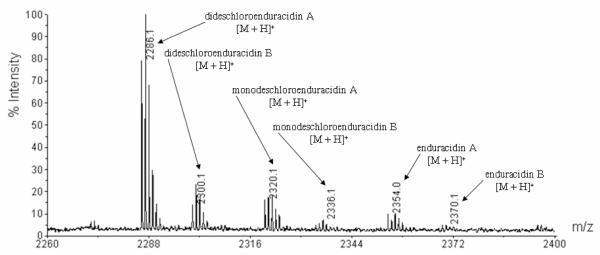
To prove orf30 is the only gene required for dichlorination of enduracidin, the construct pXY152aE30 was introduced into the in-frame deletion mutant SfΔ30. The resulting recombinant strain SfΔ30E30 carries an intact copy of orf30 integrated into the chromosome and its expression is controlled by the ermE* promoter.30 The strain was fermented in production medium and the metabolites were analyzed by MALDI-TOF MS. The results showed SfΔ30E30 produced six peptides: 1, 2, 4, 5, and monodeschloroenduracidins (6 and 7, Figure 3). The results unambiguously demonstrate that Orf30 alone is responsible for catalyzing the dichlorination observed in enduracidin. The poor efficiency of this self-complementation might be due to incomplete and/or less efficient processing of the substrate by the ectopic trans-complemented halogenase. The failure of complete restoration of chlorination of balhimycin with ectopic expression of the halogenase gene was suggested to be the outcome of the positional effect and/or different expression levels of bhaA in wild type and recombinant strains.22 It was previously noted that the wild type S. fungicidicus produced two minor metabolites enduracidins C and D as the monochlorinated species.31 These are presumably the same as 6 and 7 produced by the orf30 self-complementation strain SfΔ30E30 because they are made by the same halogenase. The success of the orf30 self-complementation motivated us to conduct the heterologous complementation of mutant SfΔ30 with the ramoplanin halogenase gene.
Figure 3.
MALDI-TOF MS analysis of the metabolites produced by the fermentation of the recombinant strain SfΔ30E30.
Constructing the Recombinant Strain SfΔ30R20
Sequence alignment of the enduracidin (Orf30) and ramoplanin (Orf20) halogenases showed an overall 62% amino acid identity (Figure S2). Depending on the factors governing substrate specificity, the ramoplanin halogenase may be able to complement the orf30 in-frame deletion mutant SfΔ30 and restore production of the parent peptide or generate other enduracidin derivatives. To evaluate this hypothesis, the integrative plasmid pXY152aR20 was constructed by cloning the ramoplanin orf20 into the integrative vector pSET152. pXY152aR20 was conjugally introduced into the mutant strain SfΔ30 to yield the recombinant strain SfΔ30R20. The expression of the ramoplanin orf20 in the orf30 deficient mutant was driven by the strong constitutive promoter ermE*. Ectopic integration of pXY152aR20 into the chromosome of SfΔ30 was confirmed by Southern analysis (Figure S3) and resistance to apramycin (AmR).
Production of Monodeschloroenduracidins A and B by Strain SfΔ30R20
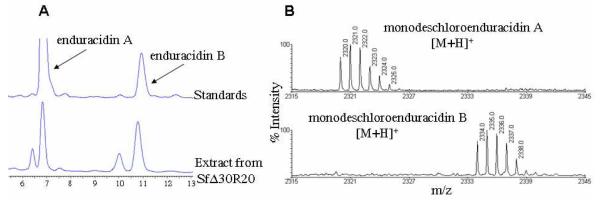
SfΔ30R20 was fermented in the enduracidin production medium.1 Metabolites were prepared from 8 day fermentation broth and mycelia, and analyzed by HPLC and mass spectrometry. HPLC detected two minor and two major peaks in the metabolite extracts (Figure 4A). The major peaks have similar retention times to enduracidin A and B standards while the two minor peaks clearly have shorter retention times than those of the standards. Analysis of the purified peaks by MALDI-TOF MS established that the two major peaks are monodeschloroenduracidins A and B, not enduracidins (Figure 4B) and the two minor peaks are dideschloroenduracidins (4 and 5). The production of monochlorinated species by SfΔ30R20 was also confirmed by LC-MS ESI analysis (Figure S4). In ramoplanin (3), a single chlorination occurs at Hpg residue 17. Production of monodeschloroenduracidins by the recombinant strain SfΔ30R20 suggested that the ramoplanin halogenase may only be capable of conducting a single chlorination at a specific amino acid residue. The observation that neither the restoration of the parent peptide enduracidins (1 and 2) nor a triply chlorinated species were detected in the fermentations of SfΔ30R20, appeared to be supportive of the above assumption.
Figure 4.
HPLC analysis of the crude extract from the fermentation of the recombinant strain SfΔ30R20 (A) and MALDI-TOF MS analysis of purified monodeschloroenduracidins A and B from SfΔ30R20 (B).
Constructing Recombinant Strain SfR20 and Metabolite Analysis
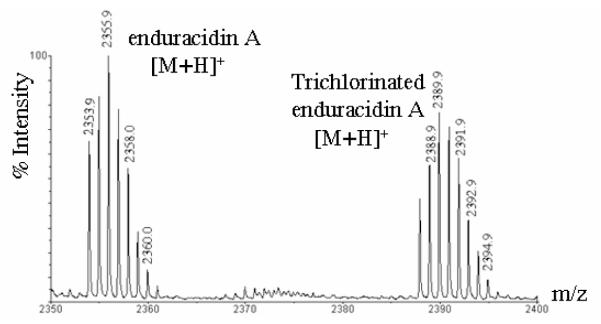
Because we had entertained the possibility that the ramoplanin Orf20 may catalyze the chlorination of Hpg17 of enduracidin, we also transformed wild type S. fungicidicus with the plasmid pXY152aR20. Ectopic integration of pXY152aR20 into the chromosome of S. fungicidicus was confirmed by Southern analysis (data not shown) and the AmR phenotype. The resulting strain, SfR20, was fermented in enduracidin production medium and the metabolite was analyzed by LC-MS ESI (Figure S5) and MALDI-TOF MS. Figure 5 shows that the predominant products were enduracidin A and a trichlorinated analogue (8).
Figure 5.
MALDI-TOF MS analysis of metabolites produced by recombinant strain SfR20.
Localization of the Chlorine Atom in Monodeschloroenduracidins Produced by SfΔ30R20
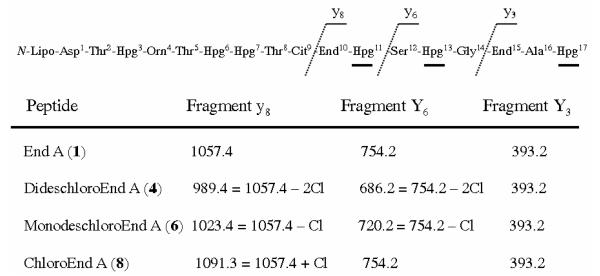
Because orf20 is the only halogenase gene in the ramoplanin cluster it is presumed responsible for the chlorination of Hpg17. Due to the close structural similarity of enduracidin and ramoplanin, we thought it possible that chlorination by the ramoplanin Orf20 may occur at Hpg17. However, the monodeschloroenduracidins detected in the self complemented recombinant strain SfΔ30E30 would be expected to be chlorinated at residue 13, as in the dichloro species found in the wild type organism. Therefore, it was necessary to independently identify the site of chlorination on the monochloro species produced by SfΔ30R20. To localize the position of the chlorine atom on the SfΔ30R20 peptide, the compound was subjected to tandem MS analysis. In the MS/MS spectrum of 1, prominent ions are observed for fragment peptides End15–Hpg17 (m/z 393.2), Ser12–Hpg17 (m/z 754.2) and End10–Hpg17 (m/z 1057.4) (Figure 6). Inspection of the MS/MS spectrum for the SfΔ30R20 monochloro peptide revealed that the ion at m/z 393.3 was still present, eliminating Hpg17 as chlorination site, but ions at m/z 1057.4 and 754.2 were absent. New fragment ions were seen at m/z 1023.4 and 720.2, corresponding to the addition of a single chlorine to Hpg13 in 6 and 7 (Figure 6).
Figure 6.
Localization of the chlorine atom on monodeschloroenduracidin A and chloroenduracidin A by MS/MS analysis.
The finding that Orf20 can halogenate Hpg13 raises the question of what factors govern substrate selection and what species is halogenated. The close structural relationship of enduracidin (1 and 2) and ramoplanin (3), including six Hpg residues, and the high sequence similarity of Orf20 and Orf30 along the entire length of the proteins, suggest that the primary determinant of halogenase substrate specificity may be dictated by local NRPS sequence. This would be consistent with the studies on balhimycin chlorination.29 If the halogenase substrate was an advanced NRPS-bound species or a nascent peptide, one might expect that Orf20 would preferentially chlorinate Hpg17 over Hpg13. Because the Orf20 is able to recognize and chlorinate Hpg13 of enduracidin it would be very interesting to create a ramoplanin orf20 deficient mutant and determine if it can be complemented by enduracidin orf30. These complementation experiments would allow one to explore the potential halogenating capacity of Orf30 and to evaluate if it has the ability to catalyze single and/or dichlorination of residues Hpg17 and Hpg13.
Localization of the Third Chlorine Atom on Trichlorinated Enduracidin
The presence of a trichlorinated peptide indicates that Orf20 must be capable of recognizing and chlorinating a residue other than Hpg13 as was observed with the SfΔ30R20 strain. To localize the third chlorine atom, we again relied on tandem MS analysis. This was especially important because the metabolite was produced in low yield and we were unable to completely separate the trichloro analogue from enduracidin A. MS/MS allowed us to work with partially purified peptides. In the MS/MS spectrum, prominent ions are observed for enduracidin A fragment peptides End15–Hpg17 (m/z 393.2), Ser12–Hpg17 (m/z 754.2) and End10–Hpg17 (m/z 1057.4). Inspection of the MS/MS spectrum for the SfR20 trichloro peptide revealed that the ion at m/z 1057.4 was absent and a new fragment appeared at m/z 1091.3, corresponding to the addition of chlorine to the End10–Hpg17 region. Because the ions at m/z 393.3 and 754.2 were still observed, the site of the third chlorination must be Hpg11 (Figure 6). Assuming no change in the basic halogenation mechanism of Orf20, chlorination is expected ortho to the phenolic hydroxy group in the analogues. Because there are no significant differences in antibacterial activities of the enduracidin analogues compared to the parent peptides, efforts to precisely localize the chlorine atoms were not pursued.
The introduction of a chlorine atom at Hpg11 of enduracidin by the ramoplanin halogenase was surprising. This unexpected outcome of the halogenation catalyzed by Orf20 further strengthened our assumption that local NRPS sequence plays an essential role in determining halogenase substrate specificity. This is in agreement with studies on balhimycin halogenation.29 Through a complete set of halogenase complementation experiments, we have demonstrated the substrate flexibility and partial equivalency of the ramoplanin and enduracidin halogenases. These findings expand our knowledge about the capacity, substrate specificity, and portability of halogenases, and the knowledge gained here may be extended to create novel peptide antibiotics from other chlorinated natural product secondary metabolite gene clusters via combinatorial biosynthesis.
Biological Evaluation of Enduracidin Analogue
It is well established that the loss or replacement of the chlorine atom(s) from various natural products can have a pronounced affect on biological activities.18-20, 32 Hence, the enduracidin A analogues having varying degrees of chlorination were evaluated for in vitro antibacterial activity in an agar diffusion assay using Staph. aureus as an indicator organism. Due to the low yield and difficulty in separating the trichlorinated 8 from 1, the extract containing compounds 1 and 8 was used in the bioassay. All the analogues tested retained activity against S. aureus. The monodeschloro compound 6 and the mixture of 1 and 8 showed no measurable difference in activity compared to 1. The dideschloro compound 4 produced a zone of inhibition that was slightly less than that observed for 1 but was within typical experimental variation (Figure S6).
Experimental Section
Bacterial Strains, Plasmids, Fosmids, Media and Culture Conditions
Streptomyces fungicidicus ATCC21013, Actinoplanes strain ATCC33076 and Escherichia coli S17-1 ATCC47055 were purchased from ATCC. E. coli XL1-Blue MR (Stratagene), EPI300™-T1R (Epicentre) were routinely used as hosts for E. coli plasmids, fosmids and E. coli-Streptomyces shuttle vectors. The pGEM-T easy cloning vector was from Promega. Plasmids pSET152 and pIJ773 were provided by Professor Keith Chater (JIC, Norwich, England). Plasmid pWHM860 harboring ermE*p was provided by Professor Bradley Moore (UCSD, San Diego). ISP2 (Difco ISP Medium 2), ISP4 and TSB (Bacto Tryptic Soy Broth) were purchased from VWR. Media and culture conditions for S. fungicidicus were previously described.1 All E. coli procedures were performed according to standard protocols.33
DNA Isolation and Manipulations
Isolation of chromosomal DNA from S. fungicidicus wild type and the mutant strains, and agarose gel electrophoresis were performed according to Kieser et al.34 QIAprep Spin Miniprep kits (Qiagen) were used to prepare plasmids and fosmids from E. coli strains. Restriction endonucleases, DNA ligase, DNA polymerase, Klenow enzyme and alkaline phosphatase were purchased from Roche, BioLabs and Invitrogen, and used according to the manufacturers’ recommendations. Primers used for PCR and DNA sequencing were synthesized from Fisher. PCR products and restriction fragments from agarose gels were purified using QIAquick Gel Extraction kits from Qiagen.
Construction of Plasmid pXY300Δ30 for In-frame Deletion of the orf30 Gene
pXY300Δ30 was constructed by cloning two fragments that flank the chromosomal sequence destined for deletion into the E. coli-Streptomyces shuttle conjugal temperature sensitive vector pXY300 containing the thiosptrepton resistance gene (tsr) for selection in Streptomyces.35 An “upstream” 1.3 kb and a “downstream” 1.6 kb flanking sequences, designated orf30Δ1 and orf30Δ2, respectively, were generated by PCR using fosmid pXYF148 as the template.26 Primers 30Δ1pf (5′-GTCAAGCTTGAGGAACTCGTGCTCG-3′, HindIII site is in bold) and 30Δ1pr (5′-CTGAGATCTACTCATTCGGCCTC-3′, BglII site is in bold) were used to amplify fragment orf30Δ1; primers 30Δ2pf (5′-GCGAGATCTGGAGAGTACGCCGGCGA-3′; BglII site is in bold) and 30Δ2pr (5′-CTGACGGACGCGAATTCCCTTGC-3′; EcoRI site is in bold) were used to amplify fragment orf30Δ2. The location of these fragments within the enduracidin cluster is shown in Figure 1A. These two PCR fragments were appropriately restricted and simultaneously ligated with vector pXY300, prepared by digestion with EcoRI and HindIII, to yield plasmid pXY300Δ30. The in-frame deletion and error-free insert of pXY300Δ30 were confirmed by sequencing.
Construction of the Integrative Plasmid pXY152aR20 for Heterologous Complementation
To express the ramoplanin halogenase gene orf20 in enduracidin halogenase deficient mutant, we cloned orf20 into an integrative plasmid pXY152a.36 orf20 was PCR-amplified by using a pair of primers (R20pf: 5′-GAGGACATATGGCTGCTCAACCGGAAGAG-3′, NdeI site is in bold; R20pr: 5′-ATCTGAATTCAGATCTCAGTCGGCGGCGACCCACGT-3′, EcoRI and BglII sites are in bold) and the genomic DNA from ramoplanin producer Actinoplanes strain ATCC33076. The PCR product was then cloned into vector pGEM-T easy to afford plasmid pGEMTE-R20. Error-free orf20 was confirmed by sequencing, then excised by digestion with NdeI and EcoRI and ligated into the similarly restricted vector pXY152a to yield the integrative plasmid pXY152aR20.
Construction of the Integrative Plasmid pXY152aE30 for orf30 Self-complementation
To self-complement the halogenase deficient mutant, we cloned orf30 into plasmid pXY152aR20 by substitution of orf20.26, 37 orf30 was PCR-amplified using a pair of primers (E30pf: 5′-TCAGCATATGCCGGGAGGCCGAATGAGTAC-3′, NdeI site is in bold; E30pr: 5′-TCGCAGATCTCAGGACGGATGCGGCTCCTC-3′, BglII site is in bold) and the genomic DNA from enduracidin producer S. fungicidicus ATCC21013. The PCR product was gel-purified and cloned into vector pGEM-T easy to generate plasmid pGEMTE-E30. Error-free orf30 was confirmed by sequencing, excised by digestion with NdeI and BglII, then ligated into the similarly restricted pXY152aR20 to yield the integrative plasmid pXY152aE30.
PCR Screening Protocol
Spores from independent mutant candidate colonies were inoculated in TSB liquid culture. After growth over night, mycelia were harvested by centrifugation and washed twice with TE buffer (10 mM Tris, 1 mM EDTA), pH 8.0. Mycelia were re-suspended in sterile H2O and used as PCR template. PCR reaction mixture in the final volume of 100 μL contained 60 μL mycelia, 150 pmol of each primer, 20 μL 5X AccuPrime GC-Rich buffer A (Invitrogen) and 1 μL polymix (added at 80 °C) from Expand Long Template PCR System (Roche). PCR was performed as follows: 1 cycle at 95 °C for 3 min, 30 cycles at 95 °C for 1 min, at 55 °C for 1 min and at 72 °C for 2 min. The reaction was terminated with one extension cycle at 72 °C for 10 min. PCR products were gel-purified and sequenced.
DNA Sequencing and Analysis
DNA sequencing was performed at Oregon State University Center for Genome Research and Biocomputing (CGRB) using the Amplitaq dye-terminator sequencing system (Perkin Elmer) and Applied Biosystems automated DNA sequencers (models 373 and 377). Nucleotide sequences were determined for both strands. Sequence analysis was carried out using the VectorNTI (Invitrogen) software packages. Nucleotide and amino acid sequence similarity comparisons were carried out in public databases using the BLAST program.38
Southern Hybridization
Genomic DNA from S. fungicidicus, mutant SfΔ30 and recombinant strains SfR20, SfΔ30E30 and SfΔ30R20 was digested with appropriate restriction endonucleases, electrophoresed in 0.8% to 1% agarose gels and transferred onto nylon membranes (Roche). Digoxigenin-labeled probes were used in Southern analyses. DNA probes were prepared using Digoxigenin-labeled dUTP and hybridization was revealed using a Digoxigenin-DNA detection kit (Roche).
Production and Isolation of Metabolites
Fermentation conditions for the production of enduracidins from the wild type strain, the mutant SfΔ30, and the recombinant strain SfΔ30R20 and SfR20 were as previously described.1 Briefly, 60 g of fresh mycelia were washed with 120 mL deionized H2O, resuspended in 120 mL MeOH and sonicated with microprobe tip for 1 min at 15 watts (Microson Ultrasonic Homogenizer, Model XL2000, Misonix Inc). The suspension was shaken at 230 rpm at 18 °C for 3 h and then centrifuged at 2000 g for 20 min. The supernatant was collected and evaporated to near dryness under reduced pressure, and resuspended in 10 mL of 90% MeOH. The solution was adjusted to pH 4.3 with 1 M HCl, and centrifuged at 2000 g for 20 min. The supernatant serves as the crude extracts for downstream analysis. Under the above culture and preparation conditions, wild type S. fungicidicus produced an average of 8 mg/L of enduracidins. The production of dideschloroenduracidins and monodeschloroenduracidins by the corresponding mutants dropped to 4 mg/L. Production of trichlorinated enduracidins by the recombinant SfR20 decreased by approximately 10-fold, to an average yield of 0.8 mg per liter.
The crude extracts were further purified by passage through a Varian HF Mega Bond Elut-C18 column that was sequentially eluted with H20, 25%, 50%, 100% CH3CN and 50% CH3CN in 50 mM phosphate buffer, pH 4.3. Fractions were concentrated and analyzed by HPLC and bioassay. The fractions confirmed to contain the desired compounds were combined and further purified by semi-preparative HPLC with C18 column (Gemini 10 μM, 250 × 10 mm) from Phenomenex. Solvent A was H20 with 0.1% TFA, and solvent B was CH3CN with 0.1%TFA. The flow rate was 5 mL/min starting with 25% B and 75% A, increasing to 65% B over 30 min, and then held for a further 10 min. Separate 1 min fractions were collected over the program. Fractions were analyzed by HPLC, those fractions containing the pure compounds were pooled for MS analysis and bioassay.
HPLC, LC-MS and MS Analyses of Enduracidins
Enduracidin A and B standards were purchased from MP Biomedicals, LLC (Aurora, Ohio, USA). Prior to HPLC analysis, samples were filtered through a 0.45 μm syringe filter. HPLC was performed using a Phenomenex Gemini C18 column, 5 μm, 4.6 × 150 mm with isocratic elution in 30% CH3CN and 70% 50 mM NaH2PO4, pH 4.5 at a flow rate of 1.0 mL/min. The UV region from 200-300 nm was scanned with a photodiode array detector or 230 nm was monitored with a variable wavelength detector. Two LC-MS systems were used to obtain the data reported here. The first is a ThermoFinnigan LCQ Advantage LC-MS system equipped with autosampler and photodiode array detector, and controlled by the software Xcalibur 1.3 on PC computer. The LC-MS analyses with this instrument were carried out as the same conditions mentioned above for HPLC except that the mobile phase for isocratic elution was replaced with 30% CH3CN and 70% H20 containing 0.1% TFA and the positive mode electrospray ionization was used for MS detection. The second system is a ThermoFinnigan LTQ-FT Ultra mass spectrometer, equipped with an electrospray ionization source run in positive mode combined with a Waters CapLC system. LC-ESI-MS analyses with this instrument were performed with a reversed phase C18 50 × 0.2 mm 5 μm Michrom Magic column, using the mobile phase CH3CN/H2O with 0.1% formic acid and a linear gradient from 5% to 60% in 20 min, then to 90% in 10 min, at a flow rate of 4 μL/min. MALDI-TOF MS analyses were performed using an Applied Biosystems ABI4700 TOF/TOF mass spectrometer in reflector mode with an accelerating voltage of 20 kV. Samples were mixed in a 1:4 ratio with α-cyano-4-hydroxycinnamic acid (HCCA) in 50% CH3CN and 0.1% TFA. An aliquot of 0.5 μL of the sample solution was applied to the sample plate and air dried.
Evaluation of Antibacterial Activity
Staphylococcus aureus ATCC29213 was used as an indicating microorganism in bioassay and inoculated in LB broth. After growth at 37 °C overnight, 100 μL of the culture mixed with 5 mL of the top agar (mixture of equal volume of nutrient agar and nutrient broth) was then overlaid onto a nutrient agar plate in which appropriately spaced wells were made by cutting out the agar plugs. Enduracidin standards and analogues were dissolved in 50% MeOH at the concentration of 20 μg/mL and 100 μL of each solution was loaded into the wells. After incubating the plates at 37 °C for 16 hours, the zones of inhibition were observed, compared and the plates photographed.
Supplementary Material
Acknowledgment
This work was supported by NIH Grants R01GM69320 (MZ) and R01AI073784 (XY), the OSU College of Pharmacy and General Research Fund (XY), and the Medical Research Foundation of Oregon (XY). Prof. P. Proteau is thanked for helpful discussions and critical reading of this manuscript. We thank Prof. K. Chater (JIC, Norwich, England) for providing plasmids pSET152 and pIJ773, and Prof. B. Moore (UCSD, San Diego) for providing plasmid pWHM860. Y. Wang is supported by the State Scholarship Fund through the China Scholarship Council of the Ministry of Education of the P.R. China. The Mass Spectrometry Facility at OSU is supported by NIEHS grant P30 ES00210.
Footnotes
Supporting Information Available. HPLC analysis comparing wild-type S. fungicidicus metabolites with the extract from mutant SfΔ30, alignment of enduracidin and ramoplanin halogenases, Southern hybridization confirming the recombinant strain SfΔ30R20, LC-MS data for monodeschloroenduracidins and trichlorinated enduracidins, as well as biological activity data, are available. This information is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Higashide E, Hatano K, Shibata M, Nakazawa K. J. Antibiot. 1968;21:126–137. [PubMed] [Google Scholar]
- (2).Asai M, Muroi M, Sugita N, Kawashima H, Mizuno K. J. Antibiot. 1968;21:138–146. doi: 10.7164/antibiotics.21.138. [DOI] [PubMed] [Google Scholar]
- (3).Goto S, Kuwahara S, Okubo N, Zenyoji H. J. Antibiot. 1968;21:119–125. doi: 10.7164/antibiotics.21.119. [DOI] [PubMed] [Google Scholar]
- (4).Tsuchiya K, Kondo M, Oishi T, Yamazaki I. J. Antibiot. 1968;21:147–153. [PubMed] [Google Scholar]
- (5).Kawakami M, Nagai Y, Fujii T, Mitsuhashi S. J. Antibiot. 1971;24:583–586. doi: 10.7164/antibiotics.24.583. [DOI] [PubMed] [Google Scholar]
- (6).Yourassowsky E, Monsieur R. Chemotherapy. 1972;17:182–187. doi: 10.1159/000220852. [DOI] [PubMed] [Google Scholar]
- (7).Komatsuzawa H, Suzuki J, Sugai M, Miyake Y, Suginaka H. J. Antimicrob. Chemother. 1994;33:1155–1163. doi: 10.1093/jac/33.6.1155. [DOI] [PubMed] [Google Scholar]
- (8).Peromet M, Schoutens E, Yourassowsky E. Chemotherapy. 1973;19:53–61. doi: 10.1159/000221439. [DOI] [PubMed] [Google Scholar]
- (9).Cavalleri B, Pagani H, Volpe G, Selva E, Parenti F. J. Antibiot. 1984;37:309–317. doi: 10.7164/antibiotics.37.309. [DOI] [PubMed] [Google Scholar]
- (10).Ciabatti R, Kettenring JK, Winters G, Tuan G, Zerilli L, Cavalleri B. J. Antibiot. 1989;42:254–267. doi: 10.7164/antibiotics.42.254. [DOI] [PubMed] [Google Scholar]
- (11).Jones RN, Barry AL. Diagn. Microbiol. Infect. Dis. 1989;12:279–282. doi: 10.1016/0732-8893(89)90029-1. [DOI] [PubMed] [Google Scholar]
- (12).Walker S, Chen L, Hu Y, Rew Y, Shin D, Boger DL. Chem. Rev. 2005;105:449–476. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
- (13).Fulco P, Wenzel RP. Expert. Rev. Anti. Infect. Ther. 2006;4:939–945. doi: 10.1586/14787210.4.6.939. [DOI] [PubMed] [Google Scholar]
- (14).Monaghan T, Boswell T, Mahida YR. Postgrad. Med. J. 2009;85:152–162. doi: 10.1136/gut.2007.128157. [DOI] [PubMed] [Google Scholar]
- (15).Bartlett JG. Curr. Infect. Dis. Rep. 2009;11:21–28. doi: 10.1007/s11908-009-0004-8. [DOI] [PubMed] [Google Scholar]
- (16).Hamburger JB, Hoertz AJ, Lee A, Senturia RJ, McCafferty DG, Loll PJ. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13759–13764. doi: 10.1073/pnas.0904686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gribble GW. J. Chem. Ed. 2004:1441–1449. [Google Scholar]
- (18).Bister B, Bischoff D, Nicholson GJ, Stockert S, Wink J, Brunati C, Donadio S, Pelzer S, Wohlleben W, Sussmuth RD. Chembiochem. 2003;4:658–662. doi: 10.1002/cbic.200300619. [DOI] [PubMed] [Google Scholar]
- (19).Eustaquio AS, Pojer F, Noel JP, Moore BS. Nat. Chem. Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gerebtzoff G, Li-Blatter X, Fischer H, Frentzel A, Seelig A. Chembiochem. 2004;5:676–684. doi: 10.1002/cbic.200400017. [DOI] [PubMed] [Google Scholar]
- (21).Donadio S, Sosio M. Curr. Top. Med. Chem. 2008;8:654–666. doi: 10.2174/156802608784221541. [DOI] [PubMed] [Google Scholar]
- (22).Puk O, Huber P, Bischoff D, Recktenwald J, Jung G, Sussmuth RD, van Pee KH, Wohlleben W, Pelzer S. Chem. Biol. 2002;9:225–235. doi: 10.1016/s1074-5521(02)00101-1. [DOI] [PubMed] [Google Scholar]
- (23).Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Chem. Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- (24).Eustaquio AS, Gust B, Li SM, Pelzer S, Wohlleben W, Chater KF, Heide L. Chem. Biol. 2004;11:1561–1572. doi: 10.1016/j.chembiol.2004.09.009. [DOI] [PubMed] [Google Scholar]
- (25).Eustaquio AS, Gust B, Luft T, Li SM, Chater KF, Heide L. Chem. Biol. 2003;10:279–288. doi: 10.1016/s1074-5521(03)00051-6. [DOI] [PubMed] [Google Scholar]
- (26).Yin X, Zabriskie TM. Microbiology. 2006;152:2969–2983. doi: 10.1099/mic.0.29043-0. [DOI] [PubMed] [Google Scholar]
- (27).Chiu HT, Hubbard BK, Shah AN, Eide J, Fredenburg RA, Walsh CT, Khosla C. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8548–8553. doi: 10.1073/pnas.151246498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Puk O, Bischoff D, Kittel C, Pelzer S, Weist S, Stegmann E, Sussmuth RD, Wohlleben W. J. Bacteriol. 2004;186:6093–6100. doi: 10.1128/JB.186.18.6093-6100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wohlleben W, Stegmann E, Sussmuth RD. Methods Enzymol. 2009;458:459–486. doi: 10.1016/S0076-6879(09)04818-6. [DOI] [PubMed] [Google Scholar]
- (30).Bibb MJ, Janssen GR, Ward JM. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- (31).Sugita N, Naito K, Asai M, Suzuki T, Higashide E, Mizuno K, K. T. 1972. pp. 313–330.
- (32).Gerhard U, Mackay JP, Maplestone RA, Williams DH. J. Am. Chem. Soc. 1993;115:232–237. [Google Scholar]
- (33).Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- (34).Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich, England: 2000. [Google Scholar]
- (35).Yin X, O’Hare T, Gould SJ, Zabriskie TM. Gene. 2003;312:215–224. doi: 10.1016/s0378-1119(03)00617-6. [DOI] [PubMed] [Google Scholar]
- (36).Fei X, Yin X, Zhang L, Zabriskie TM. J. Nat. Prod. 2007;70:618–622. doi: 10.1021/np060605u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).McCafferty DG, Cudic P, Frankel BA, Barkallah S, Kruger RG, Li W. Biopolymers. 2002;66:261–284. doi: 10.1002/bip.10296. [DOI] [PubMed] [Google Scholar]
- (38).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.