Abstract
MicroRNAs are endogenous repressors of gene expression. We examined microRNAs in the renal medulla of Dahl salt-sensitive rats and consomic SS-13BN rats. Salt-induced hypertension and renal injury in Dahl salt-sensitive rats, particularly medullary interstitial fibrosis, had been previously shown to be substantially attenuated in SS-13BN rats. Of 377 microRNAs examined, 5 were found differentially expressed between Dahl salt-sensitive rats and consomic SS-13BN rats receiving a high-salt diet. Real-time PCR analysis demonstrated that high-salt diets induced substantial up-regulation of miR-29b in the renal medulla of SS-13BN rats, but not in SS rats. miR-29b was predicted to regulate 20 collagen genes, matrix metalloproteinase-2 Mmp2, integrin β1 Itgb1, and other genes related to extracellular matrix. Expression of 9 collagen genes and Mmp2 was up-regulated by a high-salt diet in the renal medulla of SS rats, but not in SS-13BN rats, an expression pattern opposite to miR-29b. Knockdown of miR-29b in the kidneys of SS-13BN rats resulted in up-regulation of several collagen genes. miR-29b reduced expression levels of several collagen genes and Itgb1 in cultured rat renal medullary epithelial cells. Moreover, miR-29b suppressed the activity of luciferase when the reporter gene was linked to a 3’-untranslated segment of collagen genes Col1a1, Col3a1, Col4a1, Col5a1, Col5a2, Col5a3, Col7a1, Col8a1, Mmp2, or Itgb1, but not Col12a1. The result demonstrated broad effects of miR-29b on a large number of collagens and genes related to extracellular matrix, and suggested involvement of miR-29b in the protection from renal medullary injury in SS-13BN rats.
Keywords: gene expression, renal injury, hypertension, extracellular matrix, kidney
Introduction
A mammalian genome encodes several hundred microRNAs (miRNA). Each miRNA is computationally predicted to regulate dozens to hundreds of protein-coding genes.1,2 In most cases, miRNAs decrease the expression level of target genes by suppressing translation or reducing mRNA abundance.3,4,5 MiRNAs have been shown in recent years to be important regulators of several physiological processes and diseases in human and mammalian model organisms.
Evidence is beginning to emerge that suggests a role for miRNAs in the development of hypertension and renal injury.6 For example, the miRNA miR-155 has been shown to be involved in angiotensin II signaling.7,8 miR-192 and miR-377 may contribute to diabetic glomerular injury.9,10 MiRNAs in general appears to be critical for the development of podocytes.11,12,13
The role of miRNAs in salt-sensitive hypertension and renal injury remains unknown. The Dahl salt-sensitive (SS) rat is the most widely used genetic model of human salt-sensitive forms of hypertension and renal injury.14,15,16 SS-13BN rats are consomic rats in which chromosome 13 has been introgressed from Brown Norway rats into the SS genome.17 Salt-induced hypertension and renal injury are substantially attenuated in SS-13BN rats.17,18,19,20 Comparison of SS and SS-13BN rats has led to the discovery of several pathways, such as renal medullary oxidative stress,20 local glucocorticoid excess,21 and deficiencies in fumarate metabolism,22,23 that may contribute to salt-induced hypertension and renal injury. It appears that a tree-like regulatory network might be underlying the disease phenotypes in SS rats.18,21 An exciting possibility is that miRNAs could play an important role in this complex regulatory network.6,24
In the present study, we found that a miRNA, miR-29b, was elevated in the renal medulla of SS-13BN rats by a high-salt diet, but not in SS rats. We discovered evidence suggesting that miR-29 might be a master regulator of a large number of collagens and other genes related to extracellular matrix, which might be relevant to the development of prominent medullary interstitial fibrosis in SS rats.
Methods and Materials
Rats
Inbred SS and consomic SS-13BN rats, produced at the Medical College of Wisconsin,17 were maintained on the AIN-76A diet containing 0.4% NaCl (Dyets) as described previously.17,21,23,25,26 Male rats were used for experiments at approximately 6 weeks of age. The animal study was approved by the Institutional Animal Care and Use Committee at Medical College of Wisconsin.
MiRNA Isolation and Expression Profiling
Expression levels of 377 miRNAs were analyzed with a custom-made miRNA microarray as described previously.27 The renal medulla of SS and SS-13BN rats on the 0.4% NaCl diet or after 3 days on a 4% NaCl diet were examined with a total of 16 microarrays (n=4). Signal intensity data were processed and normalized as described previously.27 Criteria of differential expression were set according to permutation-based false positive rate as previously described.19,27,28
Measurement of Individual MiRNAs Using Modified Real-time PCR
Expression levels of individual miRNAs were quantified using modified real-time PCR with Taqman (Applied Biosystems) chemistry as described previously.27 Total RNA samples used for the real-time PCR analysis were independent of those used in the microarray experiment. 5S rRNA was used as an internal normalizer.
Bioinformatic Analysis
MiRNA target predictions were retrieved from three commonly used algorithms, TargetScan (http://www.targetscan.org/), PicTar (http://pictar.bio.nyu.edu/), and miRanda (http://microrna.sanger.ac.uk/targets/v4/). Sequence analysis of Col8a1 was performed using DNASTAR.
3’-Untranslated Region (UTR) Reporter Constructs
Reporter gene constructs containing the 3’-UTR region of a miRNA target gene were generated using molecular cloning techniques that we described previously.21,23,29,30 Briefly, a segment, typically a few hundred bases, of the 3’-UTR that included the binding sites for miR-29b was amplified from DNA samples extracted from rat tissues using PCR primers containing restriction endonuclease recognition sequences. Locations of the segments and primer sequences are shown in Supplemental Table S1 (please see http://hyper.ahajournals.org). The PCR product with flanking restriction nuclease sites was double-digested and inserted into the multiple cloning site of the pMIR-REPORT luciferase plasmid (Ambion) downstream from a firefly luciferase gene. The chimeric 3’-UTR-luciferase reporter construct was transformed into competent E. Coli (Invitrogen). Colonies that were confirmed by PCR to contain an insert of the expected size were cultured. The plasmids were extracted using a QIAprep Spin Miniprep kit (Qiagen) and sequenced to verify the insertion of the 3’-UTR region. Transfection-quality plasmids were prepared using a PureLink HiPure Plasmid Filter Purification Kit (Invitrogen).
Transfection and Analysis of 3’-UTR Reporter Constructs
HeLa cells were cultured in 96-well CELLSTAR white µClear plates (Greiner Bio-One) with DMEM plus 5% FBS. Cells at 80–90% confluency were co-transfected with a 3’-UTR reporter construct (100 ng per well), a pMIR-REPORT β-galactosidase plasmid (50 ng per well), and control pre-miR oligonucleotides or the miR-29b mimic pre-miR-29b (10 pmol per well, from Ambion), using Lipofectamine 2000 (Invitrogen) following Invitrogen’s protocol.21,22,23,29,30 At 24 h after the transfection, luciferase and β-galactosidase activities in each well were measured using the Dual-Light system (ABI) following the manufacturer’s protocol for microplate assay. The light signals were acquired using the high-throughput Analyst HT 96.384 microplate reader (Molecular Devices). β-galactosidase activity was used to normalize luciferase activity to control for transfection efficiency and cell density.
Culture and transfection of rat medullary thick ascending limb cells
Rat medullary thick ascending limb (mTAL) cells were cultured as described previously.31 Transfection of the miR-29b mimic pre-miR-29b or the control pre-miR (Ambion) was performed using Lipofectamine 2000 (Invitrogen) as described previously.21,22,23,29,30,32
Real-time PCR
Real-time PCR using the SYBR Green chemistry was performed as described previously.28 18S rRNA was used as the internal normalizer. Sequences of the primers used were shown in Supplemental Table S2 (please see http://hyper.ahajournals.org.
Western blot
Western blot was performed as we described previously.21,22,23,27,29,30 Antibodies were from Santa Cruz Biotechnology. The density of a specific band was normalized by Coomassie blue staining of the entire lane.
In vivo administration of locked nucleic acid (LNA)-modified anti-miR in chronically instrumented rats
LNA-modified anti-miR has been recently reported to efficiently knock down specific microRNAs in mice and monkeys.33 We adopted the method for use in rats. SS-13BN rats were instrumented for chronic infusion through the femoral vein and blood pressure measurement through the femoral artery as we described previously.21,22,23,29 Following recovery from surgery and stable baseline recording of blood pressure, LNA anti-miR-29b or LNA scrambled anti-miR was injected intravenously (10 mg/kg body weight, dissolved in 2 ml of normal saline, and injected over 3 min). The rats were switched to the 4% NaCl diet after the injection, and tissue collected 14 days later for expression analysis.
Statistics
Data were analyzed using student t-test or multiple-group analysis of variance. P<0.05 was considered significant unless otherwise indicated. Data are shown as mean ± SEM.
Results
MiRNA expression profiles in the renal medulla of SS and SS-13BN rats
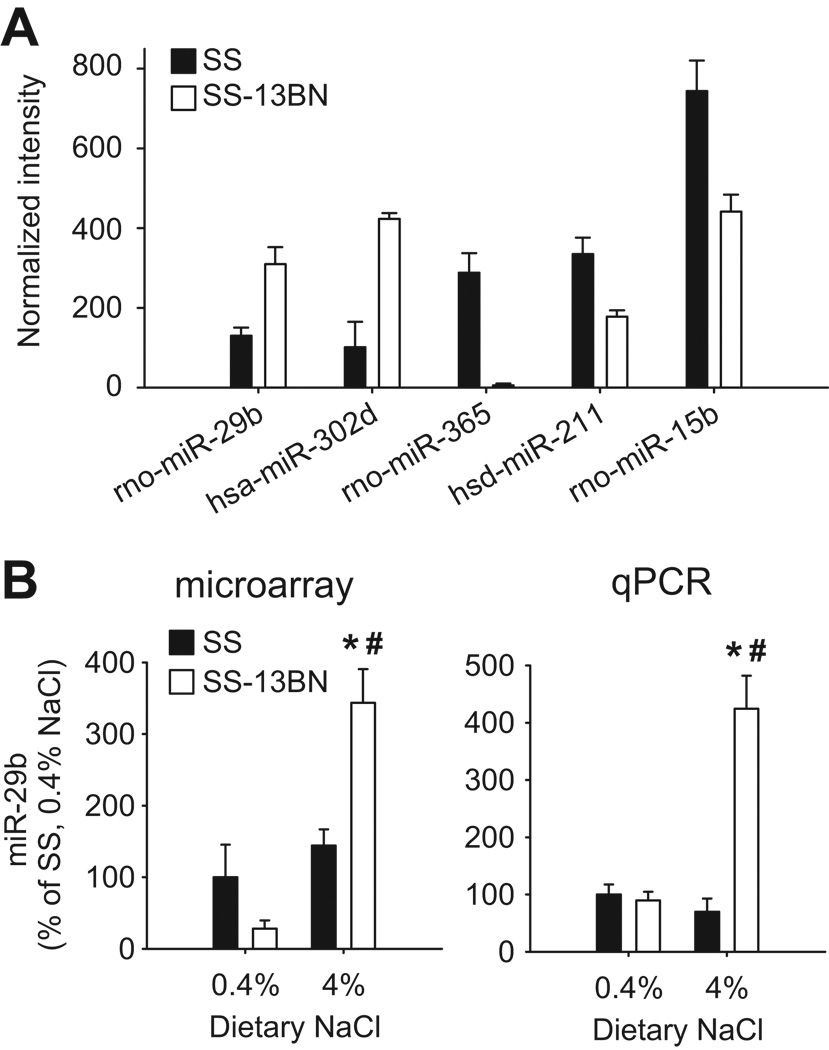
The abundance of 377 miRNAs in the renal medulla of 6 weeks old SS and SS-13BN rats was examined with miRNA microarrays. Five miRNAs were considered differentially expressed between the two rat strains after 3 days on the 4% NaCl diet (Figure 1A). Rno-miR-29b and hsa-miR-302d were expressed at lower levels in SS rats, while rno-miR-365, hsa-miR-211, and rno-miR-15b were higher in SS rats. The criteria of differential expression were P<0.05, signal difference > 150, and fold change > 50%. The overall, permutation-based false positive rate was 20%.
Figure 1. MiRNA expression in the renal medulla of SS and SS-13BN rats.
A. Renal medullary miRNAs differentially expressed between SS and SS-13BN rats on a 4% NaCl diet. Expression levels of 377 miRNAs were analyzed with miRNA microarrays (n=4). Differentially expressed miRNAs were shown. See the text for criteria of differential expression. B. MiR-29b in the renal medulla of SS-13BN rats, but not SS rats, was up-regulated by a 4% NaCl diet. Data from microarray or real-time PCR analyses were shown. Samples used for the PCR experiment were independent of those used in the microarray experiment. N=6; *, P<0.05 vs. SS-13BN 0.4%; #, P<0.05 vs. SS 4%.
In rats maintained on the 0.4% NaCl diet, no criteria for differential expression could be set that would yield an overall, array-wide false positive rate of less than 50%. Interestingly, a microRNA located on rat chromosome 13, miR-214, was readily detectable in SS rats, but barely so in SS-13BN rats.
Signal intensity data from all 16 microarrays, prior to any normalization or data processing, are shown in Supplemental Table S3 (please see http://hyper.ahajournals.org).
MiR-29 was predicted to regulate 20 collagens and several related genes
miR-29b, one of the miRNAs expressed at lower levels in the renal medulla of SS rats than in SS-13BN rats, is a member of the miR-29 family that also includes miR-29a and miR-29c. The mature sequences of the three isoforms differ at nucleotide positions 10, 18, 21, 22, or 23, but have identical seed regions in nucleotides 2 to 8. Consequently, the three isoforms have largely overlapping sets of predicted target genes.
As shown in Table 1, a highly prominent group of predicted targets of miR-29 was 20 collagens and several other genes related to extracellular matrix, such as Mmp2, Itgb1, Eln, Fbn1, Lamc1, and Sparc. The large number of collagens as predicted targets is unique to miR-29 as no other miRNA was predicted to target more than 11 of the 20 collagen genes.
Table 1.
miR-29 as a “master” regulator of several collagens and other genes related to extracellular matrix.
| Gene name |
protein name | TargetScan | PicTar | miRanda | up by salt in SS, not in SS-13BN |
down by miR-29b in mTAL |
confirmed by 3'-UTR assay |
previously confirmed* |
|---|---|---|---|---|---|---|---|---|
| Col1a1 | collagen type I alpha1 |
Yes | Yes | No | Yes | Yes | Yes | |
| Col1a2 | collagen type I alpha2 |
Yes | Yes | No | No | Yes | ||
| Col2a1 | collagen type II alpha1 |
Yes | Yes | Yes | ||||
| Col3a1 | collagen type III alpha1 |
Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Col4a1 | collagen type IV alpha1 |
Yes | Yes | No | Yes | Yes | Yes | Yes |
| Col4a2 | collagen type IV alpha2 |
Yes | Yes | No | No | Yes | Yes | |
| Col4a3 | collagen type IV alpha3 |
Yes | Yes | No | ||||
| Col4a4 | collagen type IV alpha4 |
Yes | Yes | Yes | ||||
| Col4a5 | collagen type IV alpha5 |
Yes | Yes | Yes | ||||
| Col5a1 | collagen type V alpha1 |
Yes | No | No | Yes | Yes | Yes | |
| Col5a2 | collagen type V alpha2 |
Yes | Yes | No | Yes | Yes | Yes | |
| Col5a3 | collagen type V alpha3 |
Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Col6a3 | collagen type VI alpha3 |
Yes | Yes | Yes | No | |||
| Col7a1 | collagen type VII alpha1 |
Yes | Yes | Yes | Yes | Yes | Yes | |
| Col9a1 | collagen type IX alpha1 |
Yes | Yes | Yes | ||||
| Col11a1 | collagen type XI alpha1 |
Yes | Yes | No | ||||
| Col15a1 | collagen type XV alpha1 |
Yes | Yes | No | No | Yes | ||
| Col16a1 | collagen type XVI alpha1 |
Yes | Yes | Yes | No | |||
| Col19a1 | collagen type XIX alpha1 |
Yes | Yes | No | ||||
| Col22a1 | collagen type XXII alpha1 |
Yes | Yes | No | ||||
| Col6a1 | collagen type VI alpha1 |
No | No | No | ||||
| Col6a2 | collagen type VI alpha2 |
No | No | Yes | No | |||
| Col8a1 | collagen type VIII alpha1 |
No† | No† | No† | Yes | Yes | ||
| Col9a2 | collagen type IX alpha2 |
No | No | No | ||||
| Col10a1 | collagen type X alpha1 |
No | No | Yes | ||||
| Col12a1 | collagen type XII alpha1 |
No | No | No | Yes | No | ||
| Col14a1 | collagen type XIV alpha1 |
No | No | No | ||||
| Col17a1 | collagen type XVII alpha1 |
No | No | No | ||||
| Col18a1 | collagen type XVIII alpha1 |
No | No | No | No | |||
| Col27a1 | collagen type XXVII alpha1 |
No | No | Yes | ||||
| Mmp2 | matrix metallopeptidase 2 |
Yes | Yes | No | Yes | Yes | ||
| Itgb1 | integrin beta 1 | Yes | Yes | No | No | Yes | Yes | |
| Eln | elastin | Yes | No | Yes | Yes | |||
| Fbn1 | fibrillin 1 | Yes | Yes | No | Yes | |||
| LAMC1 | laminin, gamma 1 | Yes | Yes | No | Yes | |||
| SPARC | osteonectin | Yes | Yes | No | Yes |
The predicted role of miR-29b in the regulation of collagens and other genes related to extracellular matrix is very interesting in the context of Dahl salt-sensitive rats. Extracellular matrix formation and renal medullary interstitial fibrosis are prominent features of SS rats, which are substantially attenuated in SS-13BN rats.17,19,20 The subsequent studies focused on assessing changes in collagens and related genes in Dahl salt-sensitive rats and the interaction between miR-29b and those genes.
Reciprocal expression of miR-29b and several collagens and related genes in the medulla
Real-time PCR analysis confirmed that the renal medullary abundance of miR-29b in SS-13BN rats was increased by more than 4 fold after 3 days on the 4% NaCl diet (Figure 1B). miR-29b levels in SS rats were not significantly altered by the high-salt intake. Similar changes were observed when rats were exposed to an 8% NaCl diet for 3 days (1.1 ± 0.2 fold of 0.4% NaCl for SS, n=4, not significant; 2.1 ± 0.3 fold for SS-13BN, n=4, P<0.05). Response of miR-29a or miR-29c in the renal medulla to 3 days of 8% NaCl diet was not different between SS and SS-13BN rats (n=4, not significant in any of the following comparisons). miR-29a levels after 3 days of 8% NaCl diet, compared to 0.4% NaCl diet, were 1.2 ± 0.2 fold in SS rats and 1.3 ± 0.2 fold in SS-13BN rats. miR-29c levels were 1.8 ± 0.3 fold in SS rats and 1.7 ± 0.3 fold in SS-13BN rats.
In the renal cortex, miR-29b was not significantly altered by 3 days of 8% NaCl diet in SS rats (1.2 ± 0.4 fold of 0.4% NaCl, n=6, not significant) or in SS-13BN rats (0.8 ± 0.2 fold of 0.4% NaCl, n=3, not significant).
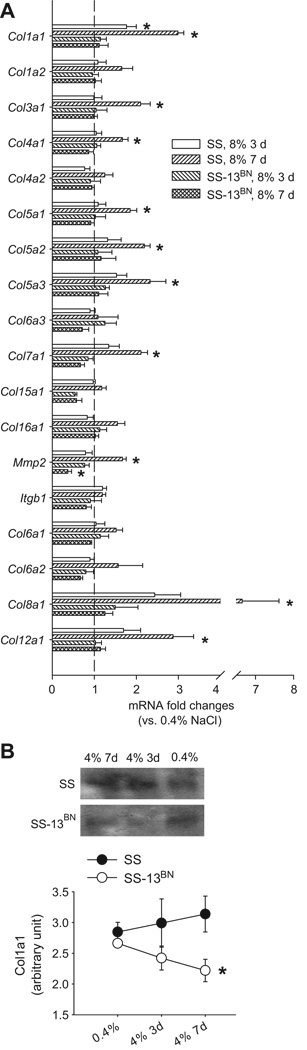
Taqman real-time PCR was performed to analyze the renal outer medullary mRNA abundance of 12 collagens as well as Mmp2 and Itgb1, which were predicted to be miR-29 targets, and 4 collagens not predicted to be miR-29 targets. Of the predicted targets, Col1a1, Col3a1, Col4a1, Col5a1, Col5a2, Col5a3, Col7a1, and Mmp2 were found to be up-regulated in SS rats, but not in SS-13BN rats, after 3 or 7 days on the 8% NaCl diet (Figure 2A). Col1a2, Col4a2, Col6a3, Col15a1, Col16a1, and Itgb1 were not significantly altered, although several of them tended to be up-regulated in SS rats. Of the genes not predicted to be targets of miR-29b, Col8a1 and Col12a1 were significantly up-regulated in SS rats after 7 days on the 8% NaCl diet, while Col6a1 and Col6a2 were not (Figure 2A). See the section “3’-UTR Reporter Analysis” for additional information about a target site in Col8a1.
Figure 2. Effect of a high-salt diet on expression levels of 16 collagen genes, matrix metallopeptidase 2, and integrin β1.
A. mRNA abundance was measured using real-time PCR in the renal outer medulla of male SS and SS-13BN rats. Rats were 6–7 weeks old and had been maintained on a 0.4% NaCl diet since birth or switched to an 8% NaCl diet for 3 or 7 days. Data were expressed as fold changes relative to the 0.4% NaCl group in each strain. N=6; *, P<0.05 vs. 0.4% NaCl of the respective strain. B. Protein abundance in the renal medulla estimated by Western blot. Rats were 6–7 weeks old and had been maintained on a 0.4% NaCl diet since birth or switched to a 4% NaCl diet for 3 or 7 days. N=5–6; *, P<0.05 vs. SS.
Exposure to the 4%, instead of 8%, NaCl diet significantly increased the renal medullary mRNA levels of Col1a2 (1.7 ± 0.2 fold, 7 days), Col4a1 (1.6 ± 0.2 fold, 3 days), and Col7a1 (2.3 ± 0.3 fold, 7 days) in SS rats (n=3–6, P<0.05 vs. 0.4% NaCl diet), but did not significantly affect these genes in SS-13BN rats. The 4% NaCl diet for 3 or 7 days increased Col1a1 mRNA in both SS and SS-13BN rats and did not cause significant changes in Col3a1, Col4a2, Col5a1, Col5a2, Col5a3, Col6a3, Col15a1, Col16a1, Mmp2, Igtb1, Col6a1, Col6a2, Col8a1, or Col12a1 in either rat strain.
Western blot analysis showed that the renal medullary protein abundance of Col1a1 was significantly higher in SS rats than in SS-13BN rats following 7 days of exposure to the 4% NaCl diet (Figure 2B).
Knockdown of miR-29b up-regulated several ECM genes in the kidneys of SS-13BN rats
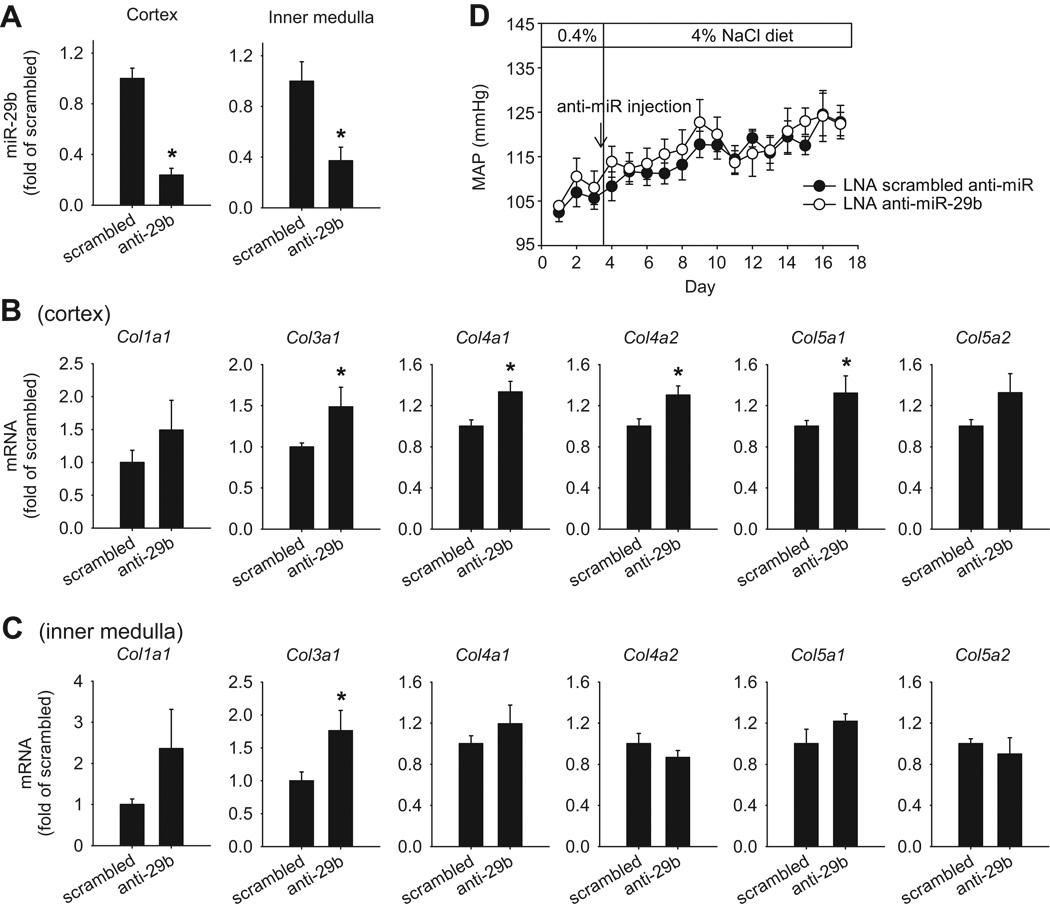
Intravenous administration of LNA anti-miR-29b (10 mg/kg body weight) in SS-13BN rats resulted in substantial and significant knockdown of miR-29b by 76% and 63% in the renal cortex and the renal inner medulla, respectively, compared to rats treated with a scrambled LNA anti-miR (Figure 3A). miR-29b levels in the outer medulla, however, were not significantly altered (1.1 ± 0.2 folds compared to rats treated with the scrambled, n=4 to 7, NS). In addition, LNA anti-miR-29b did not significantly suppress miR-29a (1.1 ± 0.4 folds in the cortex, 0.8 ± 0.1 folds in the inner medulla, compared to the scrambled) or miR-29c (0.9 ± 0.3 folds in the cortex, 0.8 ± 0.2 folds in the inner medulla) (n=4 to 7, NS).
Figure 3. Effect of LNA anti-miR-29b on SS-13BN rats in vivo.
LNA anti-miR-29b (n=4) or LNA scrambled anti-miR (n=7) was administered intravenously to chronically instrumented, conscious rats at the dose of 10 mg/kg body weight. Rats were then switched to a 4% NaCl diet and gene expression analyzed 14 days later. A. miR-29b expression in kidney regions. B. Collagen gene expression in the cortex. C. Collagen gene expression in the inner medulla. D. Mean arterial blood pressure. *, P<0.05 vs. scrambled.
mRNA levels of Col1a1, Col3a1, Col4a1, Col4a2, Col5a1, Col5a2, and Col5a3 were examined. Knockdown of miR-29b was associated with significant up-regulation of Col3a1, Col4a1, Col4a2, and Col5a1 in the renal cortex and Col3a1 in the inner medulla (Figure 3B). Col5a2 in the cortex and Col1a1 in the inner medulla tended to be up-regulated, but did not reach statistical significance (P=0.071 and 0.088, respectively). The up-regulation of ECM genes was not secondary to changes in arterial blood pressure since the treatment with LNA anti-miR-29b did not significantly affect mean arterial blood pressure (Figure 3C).
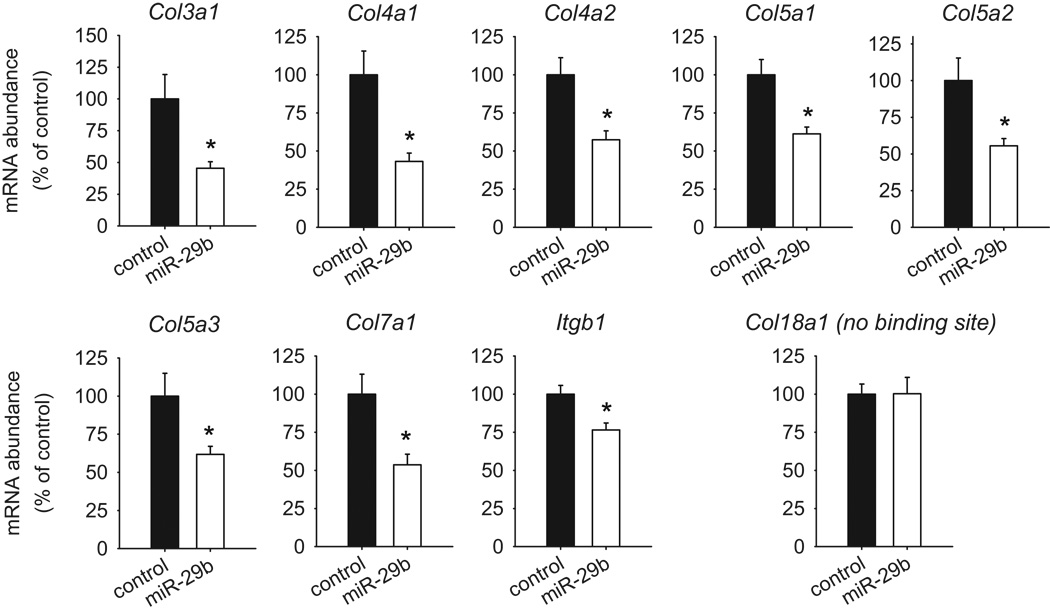
Suppression of collagens and related genes by miR-29b in cultured cells
Cultured rat mTAL cells were treated with control pre-miR oligonucleotides or the miR-29b mimic pre-miR-29b. mRNA levels of 18 collagen genes, Mmp2, and Itgb1 were measured with Taqman real-time PCR 48 hours later. The mTAL cells were used in this analysis because they were from the renal medulla of rats, although other cell types may also contribute to medullary interstitial fibrosis. The result showed that miR-29b, compared to control pre-miR, significantly reduced mRNA levels of Col3a1, Col4a1, Col4a2, Col5a1, Col5a2, Col5a3, Col7a1, and Itgb1 (Figure 4). miR-29b did not affect the mRNA level of Col18a1, which did not contain a miR-29b binding site and could be viewed as a negative control. miR-29b did not significantly affect the mRNA level of Col16a1 even though Col16a1 has a predicted miR-29b binding site. mRNA levels of Col1a1, Col1a2, Col6a1, Col6a2, Col6a3, Col12a1, Col14a1, Col15a1, Col17a1, and Mmp2 were not readily detectable in these cultured cells.
Figure 4. miR-29b suppressed the expression of several collagens and integrin β1 in medullary epithelial cells.
Cultured rat medullary thick ascending limb cells were treated with pre-miR-29b or control scrambled pre-miR for 48 hours before mRNA abundance was measured using real-time PCR. N=7; *, P<0.05 vs. cells treated with control pre-miR.
3’-UTR reporter analysis
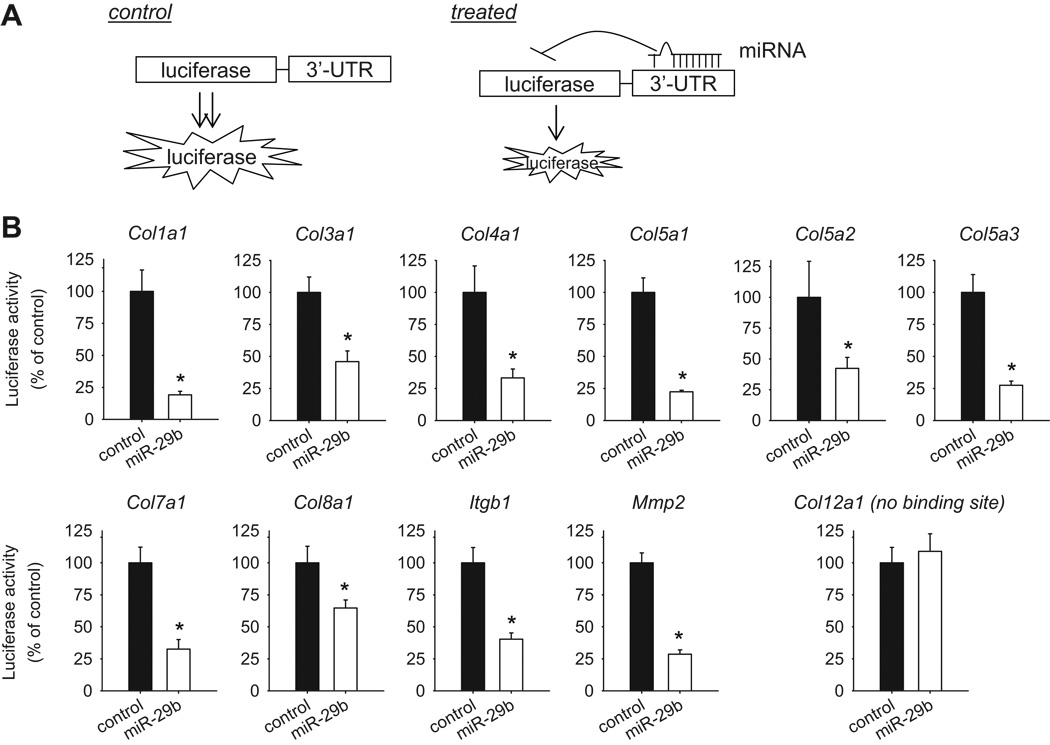
3’-UTR reporter analysis (Figure 5A) was performed to directly examine any 3’-UTR-mediated interactions between miR-29b and 9 collagen genes as well as Mmp2 and Itgb1. The miR-29b mimic, compared to a control miRNA mimic, significantly reduced luciferase activity when the reporter gene was linked to a 3’-UTR segment of Col1a1, Col3a1, Col4a1, Col5a1, Col5a2, Col5a3, Col7a1, Col8a1, Mmp2, or Itgb1 (Figure 5B).
Figure 5. miR-29b interacted with the 3’-UTR of several collagen genes, matrix metallopeptidase 2, and integrin β1.
A. 3’-UTR reporter assay. A 3’-UTR segment was cloned into a gene expression construct downstream from a luciferase reporter gene. Interactions between a miRNA and the 3’-UTR segment would be expected to decrease luciferase activity. B. miR-29b decreased luciferase activity when the reporter gene was linked to a 3’-UTR segment of several collagens, matrix metallopeptidase 2, and integrin β1. Hela cells were transfected with a 3’-UTR reporter construct, a β-galactosidase construct (for normalization), and pre-miR-29b or control scrambled pre-miR. Luciferase and β-galactosidase activities were analyzed 24 hours later. N=4–8; *, P<0.05 vs. cells treated with control pre-miR.
Col8a1 is not a predicted target for miR-29b according to TargetScan, PicTar, or miRanda (Table 1). A 3’-UTR segment of 183 bases was used for rat Col8a1 in those prediction algorithms. A segment of 2,655 bases, however, was used for mouse Col8a1. Bioinformatic analysis of rat genomic DNA indicated the presence of a putative 3’-UTR segment largely homologous to the mouse segment. Importantly, the 8-base sequence in the mouse 3’-UTR that is complementary to the seed region of miR-29b is also present in the putative rat 3’-UTR. Primers (Supplemental Table S1) were designed to clone a 673-base segment of the predicted 3’-UTR from rat Col8a1 mRNA, which was used in the 3’-UTR reporter analysis. The result indicated that Col8a1 was a target of miR-29b (Figure 5B).
Col12a1 does not have a predicted binding site for miR-29b. The 3’-UTR analysis confirmed that Col12a1 was not a target of miR-29b (Figure 5B). The result of Col12a1 could be viewed as a negative control for the 3’-UTR analysis.
MiR-29 as a “master” regulator of collagens and related genes
Table 1 summarized the data supporting a “master” regulatory role of miR-29 in the expression of collagens and other genes related to extracellular matrix in the renal medulla of SS and SS-13BN rats. It included computationally predicted interactions, up-regulation of collagens and Mmp2 in the renal medulla in SS rats (as opposed to the up-regulation of miR-29b in SS-13BN rats) in response to the 8% NaCl diet, suppression of collagens and Itgb1 in cultured renal medullary cells by a miR-29b mimic, and miR-29b-target interactions demonstrated by 3’-UTR reporter analysis.
Discussion
The present study discovered the first miRNA possibly involved in the renal injury in Dahl salt-sensitive rats, and demonstrated a broad role of miR-29b in the regulation of a large number of collagens and other genes related to extracellular matrix.
Renal medullary interstitial fibrosis is a prominent pathological feature of SS rats, especially when the rats are on high-salt diets. Medullary interstitial fibrosis, along with salt-sensitive hypertension, is substantially attenuated in consomic SS-13BN rats.17,18,19,20 Up-regulation of miR-29b may be an important part of the molecular mechanism underlying the attenuation of medullary fibrosis in SS-13BN rats. The notion is supported by reciprocal expression of miR-29b and a large number of collagens and Mmp2 in vivo, as well as up-regulation of several collagens in vivo following miR-29b knockdown and suppression of several collagens by miR-29b in vitro. Several collagens targeted by miR-29b are components of, or associated with, basement membrane or intercellular matrix.34 Matrix metallopeptidase 2 might be pro-fibrotic in the kidney.35 Moreover, an anti-fibrotic role of miR-29b in the renal medulla would be consistent with the recently reported anti-fibrotic role of miR-29b in a mouse model of cardiac fibrosis.36
A previous study of miRNA expression in SS rats did not find differential expression of any miRNAs between SS and Lewis rats.37 A subset of miRNAs was examined in that study using whole kidney extracts and mostly pooled samples. Although the present study also indicated that the differential expression of miRNAs associated with SS rats was limited, we were able to identify miRNAs that appeared relevant to the disease phenotypes in SS rats. This may be attributed to the use of consomic rats as control, a focus specifically on the renal medulla, a more complete coverage of miRNAs, or different techniques of expression analysis.
An important question that remains to be addressed is why miR-29b is up-regulated in SS-13BN rats, but not in SS rats. One copy of the miR-29b gene, miR-29b-2, is located on chromosome 13 of the rat genome, the only chromosome in SS-13BN that is derived from the Brown Norway rat. This could suggest that genetic differences might exist between SS and Brown Norway genomes in the regulatory sequence for the miR-29b-2 gene. However, the miR-29b-2 gene is separated on the chromosome from miR-29c only by 448 base pairs. The other copy of the miR-29b gene, miR-29b-1, is located on chromosome 4, separated from miR-29a by 292 base pairs. But miR-29c or miR-29a did not seem to follow the same expression pattern as miR-29b. Together, this suggests that the differential regulation of miR-29b in SS and SS-13BN rats could involve post-transcriptional processing in addition to any transcriptional changes.
The regulation of extracellular matrix genes by miR-29b shown by the present study and other recent studies36,38,39 is one of the first experimentally proven cases in which one miRNA acts as a master regulator of a large group of related genes. Each miRNA has been predicted in silico to regulate a large number of protein-coding genes.1,2 It has led to the exciting theory that miRNAs may serve as hubs in complex regulatory networks by fine-tuning multiple components of a network.6,24 But experimental evidence supporting the theory has been sparse. Recent studies of cardiac tissue, bone cells, and tumors have led to the findings that miR-29b targets Col1a1, Col1a2, Col3a1, Col4a2, Col5a3, elastin, and fibrilin 1,36,39 and that miR-29c targets COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL15A1, laminin γ1, and osteonectin.38 The present study, which was focused on the renal medulla of Dahl salt-sensitive rats, has shown that miR-29b targets Col5a1, Col5a2, Col7a1, Col8a1, Mmp2, and Itgb1, in addition to confirming the targeting of Col1a1, Col3a1, Col4a1, and Col5a3. Together, these findings (see Table 1) support a master role of miR-29 in the regulation of extracellular matrix gene expression, which might be important in a number of diseases including hypertensive renal injury.
MiRNAs may suppress target expression through translational repression or induction of mRNA degradation. miR-29 decreased the mRNA abundance of collagens and other extracellular matrix genes as shown in the present study as well as previous publications.36,38 It appears that induction of mRNA degradation is a major mode of action for miR-29b.
Despite the broad effect of miR-29 on collagens and related genes, it is clear from Table 1 that miR-29 is not the only regulator of these genes in the medulla of SS and SS-13BN rats. Collagen expression and medullary fibrosis in the SS model are likely determined by a number of antagonistic forces. miR-29b appears to be one of the forces opposing fibrosis, while other mechanisms may drive fibrosis. This may explain why up-regulation of miR-29b or knockdown of miR-29b in vivo can be associated with a mosaic of target ECM gene expression, depending on the state of the rest of the regulatory network. For example, Col1a2, Col4a2, and Col15a1 are targets of miR-29 in vitro based on the present study and/or previous publications.36,38,39 However, these genes were not significantly up-regulated in SS rats within 7 days of 8% NaCl exposure. Col12a1 was up-regulated in SS rats, but it is not a target of miR-29. Knockdown of miR-29b in the renal cortex of SS-13BN rats was associated with up-regulation of several collagen genes, even though miR-29b was not differentially regulated in the renal cortex as it was in the medulla, suggesting that baseline levels of miR-29b may already be important in controlling the expression of ECM genes. LNA anti-miR-29b remarkably suppressed miR-29b in the cortex and the inner medulla, but not in the outer medulla, preventing us from directly assessing the in vivo role of outer medullary miR-29b. The reason for the regional differences in the efficiency of knockdown is not clear. In most cases, the in vivo and in vitro data were consistent as Col1a1, Col3a1, Col4a1, Col5a1, Col5a2, Col5a3, Col7a1, Col8a1, and Mmp2 are targets of miR-29 in vitro and also up-regulated in SS rats in vivo. However, even in these cases, the lack of miR-29b up-regulation in SS rats likely played a permissive role in the up-regulation of those genes, instead of being a primary stimulus.
Perspectives
The findings of the present study suggest that the function and regulation of miRNA may contribute to the molecular mechanisms underlying the development of tissue injury associated with hypertension. In addition, the “master” role of miR-29 as a regulator of collagens and genes related to extracellular matrix has significant implications for understanding the general biology of miRNAs.
Supplementary Material
Acknowledgments
Sources of Fundings
The study was supported by NIH grants HL077263, HL085267, DK084405 (ML), HL-029587, and HL-082798 (AWC).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
None
References
- 1.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 Suppl:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Makeyev EV, Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang M, Liu Y, Mladinov D, Cowley AW, Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z. MicroRNA: A New Frontier in Kidney and Blood Pressure Research (invited review) Am J Physiol Renal Physiol. 2009;297:F553–F558. doi: 10.1152/ajprenal.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 8.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3' untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. Faseb J. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rapp JP. Dahl salt-susceptible and salt-resistant rats. Hypertension. 1982;4:753–763. doi: 10.1161/01.hyp.4.6.753. [DOI] [PubMed] [Google Scholar]
- 15.Tobian L. Salt and hypertension. Lessons from animal models that relate to human hypertension. Hypertension. 1991;17:I52–I58. doi: 10.1161/01.hyp.17.1_suppl.i52. [DOI] [PubMed] [Google Scholar]
- 16.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 17.Cowley AW, Jr, Roman RJ, Kaldunski ML, Dumas P, Dickhout JG, Greene AS, Jacob HJ. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension. 2001;37:456–461. doi: 10.1161/01.hyp.37.2.456. [DOI] [PubMed] [Google Scholar]
- 18.Liang M, Lee NH, Wang H, Greene AS, Kwitek AE, Kaldunski ML, Luu TV, Frank BC, Bugenhagen S, Jacob HJ, Cowley AW., Jr Molecular networks in Dahl salt-sensitive hypertension based on transcriptome analysis of a panel of consomic rats. Physiol Genomics. 2008;34:54–64. doi: 10.1152/physiolgenomics.00031.2008. [DOI] [PubMed] [Google Scholar]
- 19.Mori T, Polichnowski A, Glocka P, Kaldunski M, Ohsaki Y, Liang M, Cowley AW., Jr High Perfusion Pressure Accelerates Renal Injury in Salt-Sensitive Hypertension. J Am Soc Nephrol. 2008;19:1472–1482. doi: 10.1681/ASN.2007121271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor NE, Glocka P, Liang M, Cowley AW., Jr NADPH oxidase in the renal medulla causes oxidative stress and contributes to salt-sensitive hypertension in Dahl S rats. Hypertension. 2006;47:692–698. doi: 10.1161/01.HYP.0000203161.02046.8d. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Singh RJ, Usa K, Netzel BC, Liang M. Renal Medullary 11β-Hydroxysteroid Dehydrogenase Type 1 in Dahl Salt-Sensitive Hypertension. Physiol Genomics. 2008;36:52–58. doi: 10.1152/physiolgenomics.90283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Z, Greene AS, Usa K, Matus IR, Bauwens J, Pietrusz JL, Cowley AW, Jr, Liang M. Renal regional proteomes in young Dahl salt-sensitive rats. Hypertension. 2008;51:899–904. doi: 10.1161/HYPERTENSIONAHA.107.109173. [DOI] [PubMed] [Google Scholar]
- 23.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW, Jr, Liang M. A Novel Role of Fumarate Metabolism in Dahl Salt-Sensitive Hypertension. Hypertension. 2009;54:255–260. doi: 10.1161/HYPERTENSIONAHA.109.129528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang M. MicroRNA: A new entrance to the broad paradigm of systems molecular medicine. Physiol Genomics. 2009;38:113–115. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang M, Yuan B, Rute E, Greene AS, Zou AP, Soares P, MCQuestion GD, Slocum GR, Jacob HJ, Cowley AW., Jr Renal medullary genes in salt-sensitive hypertension: a chromosomal substitution and cDNA microarray study. Physiol Genomics. 2002;8:139–149. doi: 10.1152/physiolgenomics.00083.2001. [DOI] [PubMed] [Google Scholar]
- 26.Liang M, Yuan B, Rute E, Greene AS, Olivier M, Cowley AW., Jr Insights into Dahl salt-sensitive hypertension revealed by temporal patterns of renal medullary gene expression. Physiol Genomics. 2003;12:229–237. doi: 10.1152/physiolgenomics.00089.2002. [DOI] [PubMed] [Google Scholar]
- 27.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. microRNA-target pairs in rat kidneys identified through microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison J, Knoll K, Hessner MJ, Liang M. Effect of high glucose on gene expression in mesangial cells: upregulation of the thiol pathway is an adaptational response. Physiol Genomics. 2004;17:271–282. doi: 10.1152/physiolgenomics.00031.2004. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Mladinov D, Pietrusz JL, Usa K, Liang M. Glucocorticoid Response Elements and 11β-Hydroxysteroid Dehydrogenases in the Regulation of Endothelial Nitric Oxide Synthase Expression. Cardiovasc Res. 2009;81:140–147. doi: 10.1093/cvr/cvn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Park F, Pietrusz JL, Jia G, Singh RJ, Netzel BC, Liang M. Suppression of 11β-hydroxysteroid dehydrogenase type 1 with RNA interference substantially attenuates 3T3-L1 adipogenesis. Physiol Genomics. 2008;32:343–351. doi: 10.1152/physiolgenomics.00067.2007. [DOI] [PubMed] [Google Scholar]
- 31.Eng B, Mukhopadhyay S, Vio CP, Pedraza PL, Hao S, Battula S, Sehgal PB, McGiff JC, Ferreri NR. Characterization of a long-term rat mTAL cell line. Am J Physiol Renal Physiol. 2007;293:F1413–F1422. doi: 10.1152/ajprenal.00426.2006. [DOI] [PubMed] [Google Scholar]
- 32.Liang M, Pietrusz JL. Thiol-related genes in diabetic complications: a novel protective role for endogenous thioredoxin 2. Arterioscler Thromb Vasc Biol. 2007;27:77–83. doi: 10.1161/01.ATV.0000251006.54632.bb. [DOI] [PubMed] [Google Scholar]
- 33.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 34.Prockop DJ, Kivirikko KI. Collagens: molecular biology, diseases, and potentials for therapy. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 35.Cheng S, Pollock AS, Mahimkar R, Olson JL, Lovett DH. Matrix metalloproteinase 2 and basement membrane integrity: a unifying mechanism for progressive renal injury. FASEB J. 2006;20:1898–1900. doi: 10.1096/fj.06-5898fje. [DOI] [PubMed] [Google Scholar]
- 36.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naraba H, Iwai N. Assessment of the microRNA system in salt-sensitive hypertension. Hypertens Res. 2005;28:819–826. doi: 10.1291/hypres.28.819. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.