Abstract
Objective
Differences exist among various populations with regards to hypertension prevalence, severity, progression and response to therapy. Such differences may be due to genetic or environmental factors. We characterized the genetic variation and haplotype diversity of four hypertension candidate genes (CLCNKA, CLCNKB, BSND, NEDD4L) in four different ethnic groups (Caucasian Americans, African-Americans, Han Chinese, and Mexican-Americans).
Methods
We genotyped 42 single nucleotide polymorphisms across the four genes in equal numbers of each ethnically defined population, then tested for linkage disequilibrium, computed allelic and haplotype frequencies, and compared data across the different ethnic groups.
Results
We identified significant genotype and allele frequency differences among ethnic groups. The strongest differences were observed between African-American and Mexican-Americans and between Caucasian and Mexican-Americans. In addition, haplotype blocks were defined for BSND, CLCNKA_B and NEDD4L in the four populations examined. Completely mismatched (‘yin yang’) haplotypes were also observed. We found that the number of inferred halpotypes varied gene to gene and in some instances between the populations for a given gene indicating substantial haplotype diversity. The haplotype diversity among the various ethnic populations observed in our study was greater than that reported in Perlegen database.
Conclusions
Haplotype diversity in hypertension candidate genes has important implications for designing and evaluating candidate gene or genome-wide blood pressure association studies that consider these genes.
Key Words: BSND, CLCNKA, CLCNKB, NEDD4L, Hypertension, Haplotype blocks, Population genetics
Introduction
The etiology of essential hypertension, which affects more than 60 million individuals in the United States, stems from a complex interaction of environmental and genetic factors [1]. However, not all populations have the same risk for developing hypertension [2,3,4]. For example, as compared with Caucasians, African-Americans have a higher prevalence of hypertension, suffer a disproportionate morbidity from the disease, and more often exhibit salt-sensitivity [4]. Identifying genetic factors that contribute to differences in hypertension risk will improve our understanding of this disease and may lead to improved clinical management.
Long-term control of blood pressure is mediated predominantly by maintenance of salt balance by the kidneys. This physiological control is enabled by renal tubular transport of Na and Cl. To date, most monogenic forms of abnormal blood pressure control involve genes that converge on this final common pathway, altering blood pressure by changing net renal salt reabsorption [1]. Essential hypertension is genetically more complex but may also represent subtle defects in regulating renal salt balance.
In the kidney approximately 30% of NaCl reabsorption takes place in the thick ascending limb of the loop of Henle (TAL) and this depends upon a chloride channel complex encoded by CLCNKB and BSND which mediate cell Cl efflux. The importance of these genes is illustrated by loss-of-function mutations causing Bartter syndrome which is characterized by severe salt-wasting, hypotension and metabolic alkalosis. A related chloride channel gene (CLCNKA) may also contribute to this inherited salt wasting condition [5,6,7]. Evidence also suggests that a ubiquitin ligase protein encoded by NEDD4L is an important regulator of renal tubular salt transport and further interacts with the TAL chloride channel complexes. Genetic variations in these genes could predispose certain populations, especially salt-sensitive subjects, to develop hypertension. Indeed, CLCNKA, CLCNKB, BSND and NEDD4L have all been proposed as candidate genes for hypertension due to their role in renal physiology and pathophysiology, but none have been extensively examined for polymorphisms or subjected to haplotype analysis.
In this study we examined allele, genotype and haplotype frequencies of these genes at 42 SNPs in four distinct ethnic populations. Understanding the haplotype and linkage disequilibrium (LD) structure of these genes in various populations will assist in evaluating candidate gene or genome-wide association studies that examine CLCNKA, CLCNKB, BSND, and NEDD4L in the setting of abnormal blood pressure.
Methods
Study Population, Bioinformatics and Genotyping
The study subjects were obtained from a large panel of anonymous, unrelated DNA samples from the Human Variation Collection of the NIGMS Repository held by the Coriell Institute. We specifically used sets of DNA samples obtained from four distinct ethnic groups residing in the United States including Caucasian Americans, African-Americans, Han Chinese of Los Angeles and Mexican-Americans. Using Ensembl (www.ensembl.org), we defined regions of each gene of interest within ENsMart (www.ensembl.org/EnsMart). We examined exons (coding regions, 5′UTR and 3′UTR), introns and 10 kb flanking of the genes. Since CLCNKA and CLCNKB are only 11 kb apart, they were considered as one gene (CLCNKA_B) in this study.
Once a region of interest was identified, we then searched the database for SNPs with minor allele frequency (MAF) >5%, particularly searching for non-synonymous, coding SNPs. We did not find many such variants due to either low frequency or, as in the case with BSND, no coding region polymorphisms were reported in the literature at the time we began our study. Using the Applied Biosystems (ABI) design pile line, our final SNP selection for the study involved SNPs with MAF >5% and SNPs selected from the literature or public databases. Given the LD pattern in these genes, our analyses should provide adequate coverage to gain an excellent understanding of the patterns of variation in each of these genes. Table 1 provides detailed information about each SNP we studied. We genotyped 42 SNPs across 4 genes in equal numbers (n = 85) of each population using SNPlex genotyping platform (Applied Biosystems). Genotyping products wereanalyzed using either an ABI 3730 DNA analyzer or an ABI Prism 7900 sequence detection system. Quality control measures to ensure genotyping accuracy included eight wells of ABI positive control, eight wells of no template control and sixteen wells of ABI allelic ladder.
Table 1.
Gene and SNP information
| Gene | SNP | rs# | Position, bp | Alleles | Role |
|---|---|---|---|---|---|
| BSND (Chromosome 1) | 1 | rs5774206 | 55176280 | G/– | Promoter |
| 2 | rs1003767 | 55176341 | A/G | Promoter | |
| 3 | rs6682884 | 55186346 | A/C | Exon | |
| 4 | rs2864124 | 55186700 | C/T | 3' UTR | |
| 5 | rs943645 | 55187909 | C/T | 3' UTR | |
| 6 | rs17111474 | 55191988 | C/T | 3' UTR | |
| 7 | rs2479398 | 55192095 | A/G | 3' UTR | |
| 8 | rs2495506 | 55193790 | C/T | 3' UTR | |
| CLCNKA_B (Chromosome 1) | 1 | rs1048302 | 16086185 | A/C | Promoter |
| 2 | rs1739840 | 16087543 | A/G | Promoter | |
| 3 | rs1763597 | 16088603 | A/G | Promoter | |
| 4 | rs945425 | 16093718 | C/T | Promoter | |
| 5 | rs2003943 | 16111671 | C/G | Promoter | |
| 6 | rs6604910 | 16117309 | C/G | Intron (boundary) | |
| 7 | rs2050522 | 16124314 | C/T | Intron | |
| 8 | rs1414499 | 16136119 | C/T | 3' UTR | |
| NEDD4L (Chromosome 18) | 1 | rs1942563 | 53876954 | C/G | Intron |
| 2 | rs4461163 | 53877085 | G/T | Intron | |
| 3 | rs1977948 | 53879316 | A/G | Intron | |
| 4 | rs878396 | 53962056 | A/G | Intron | |
| 5 | rs6566942 | 53990160 | A/G | Intron | |
| 6 | rs4340401 | 53992303 | C/T | Intron | |
| 7 | rs1008899 | 54013845 | A/G | Intron | |
| 8 | rs4941366 | 54019884 | G/T | Intron | |
| 9 | rs513563 | 54025439 | C/T | Intron | |
| 10 | rs522155 | 54029322 | C/T | Intron | |
| 11 | rs192659 | 54046768 | A/G | Intron | |
| 12 | rs158866 | 54053673 | C/T | Intron | |
| 13 | rs158857 | 54060683 | A/G | Intron | |
| 14 | rs11152064 | 54105061 | C/T | Intron | |
| 15 | rs4941378 | 54107946 | C/G | Intron | |
| 16 | rs17064701 | 54109007 | C/G | Intron | |
| 17 | rs11664416 | 54121476 | C/T | Intron | |
| 18 | rs8089678 | 54151704 | A/G | Intron | |
| 19 | rs9675944 | 54170475 | A/G | Intron | |
| 20 | rs2075405 | 54188829 | A/G | Intron | |
| 21 | rs4940684 | 54223169 | A/G | 3' UTR |
Statistical Analysis
Statistical tests for differences in single locus allele and genotype frequencies between ethnic groups and deviations from Hardy Weinberg Equilibrium (HWE) were performed using Powermarker statistical software [8]. Statistical significance for these analyses was determined using Fisher's exact test. In the case of significant deviations from HWE, the direction of the deviations were assessed using Selander's index, with negative values indicating a deficiency of heterozygotes and positive values an excess [9]. Each SNP was assessed for allelic variation within populations as compared to between populations using Wright's fixation index (Fst) calculated using the algorithm of Weir and Cockerham [10]. Fst analyses were performed using software based on the protocols of Raymond and Rousett 1995 (Tools for Population Genetic Analysis, TFPGA, and version 1.3 available at (http://bioweb.usu.edu/mpmbio/index.htm).
Pairwise linkage disequilibrium (LD) was characterized and haplotype frequencies were calculated for all ethnic groups using Powermarker [11] and HaploView [12] statistical software. Standard summary statistics D′ and r2 were calculated using HaploView [13]. Haplotype blocks were assigned using the D′ confidence interval algorithm [14]. Both Powermarker and HaploView use an EM algorithm to determine haplotype frequency distributions when phase is unknown. The Powermarker haplotype trend regression (HTR) analysis was performed to test for haplotype frequency differences between ethnic groups. The test for association then uses an F test for a specialized additive model. This approach can be applied to both quantitative traits and dichotomous traits.
Results
Hardy-Weinberg Equilibrium
Individual allele frequencies in four different ethnic populations are tabulated in table 2. The genotype differences mirrored the allele frequency differences. Statistically significant deviations from Hardy-Weinberg equilibrium (HWE) (table 2) were seen in BSND marker rs2864124 in Mexican Americans (p = 0.0002), CLCNKA_B markers rs2050522 in African Americans (p = 0.001) and Caucasians (p = 0.01), rs522155 in Caucasians (p = 0.05), rs158857 in Mexicans (p = 0.04), rs11152064 in African Americans (p = 0.02) and rs4940684 in Caucasians (p = 0.0007). Caucasians and Mexican Americans had very similar MAFs in CLCNKA_B and NEDD4L, with more variability in BSND. For rs2864124, rs2050522, rs4940684, and rs11152064 the deviations from HWE were due to a deficiency of heterozygotes with Selander's index values of −0.39, −0.44, −0.36 and −0.19, respectively. For both rs522155 and rs158857 the Selander's index values were 0.23, indicating an excess of heterozygotes.
Table 2.
Minor allele frequencies and Hardy-Weinberg equilibrium for tested SNPs
| Gene | rs# | Allele frequency |
HWE |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele | AA | CAU | HAN | MA | AA | CAU | HAN | MA | ||
|
BSND |
rs5774206 | G | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| rs1003767 | A | 0.37 | 0.71 | 0.35 | 0.53 | 1.00 | 0.19 | 1.00 | 0.01 | |
| rs6682884 | C | 0.42 | 0.41 | 0.31 | 0.16 | 0.24 | 0.61 | 0.17 | 0.68 | |
| rs2864124 | C | 0.43 | 0.42 | 0.31 | 0.18 | 0.22 | 0.26 | 0.23 | <0.001 | |
| rs943645 | T | 0.41 | 0.40 | 0.28 | 0.14 | 0.44 | 0.64 | 0.24 | 1.00 | |
| rs17111474 | T | 0.39 | 0.39 | 0.41 | 0.70 | 0.22 | 1.00 | 0.28 | 0.96 | |
| rs2479398 | G | 0.09 | 0.13 | 0.04 | 0.04 | 0.93 | 0.52 | 1.00 | 0.26 | |
| rs2495506 | T | 0.18 | 0.29 | 0.57 | 0.44 | 0.09 | 1.00 | 1.00 | 0.27 | |
|
CLCNKA_B |
rs1048302 | A | 0.20 | 0.29 | 0.04 | 0.40 | 1.00 | 0.18 | 0.17 | 1.00 |
| rs1739840 | A | 0.21 | 0.40 | 0.20 | 0.56 | 1.00 | 0.17 | 0.35 | 0.80 | |
| rs1763597 | G | 0.23 | 0.40 | 0.20 | 0.57 | 0.75 | 0.19 | 0.43 | 0.94 | |
| rs945425 | T | 0.20 | 0.30 | 0.05 | 0.40 | 1.00 | 0.32 | 0.27 | 0.43 | |
| rs2003943 | C | 0.15 | 0.33 | 0.60 | 0.26 | 0.38 | 0.31 | 1.00 | 1.00 | |
| rs6604910 | C | 0.31 | 0.67 | 0.73 | 0.77 | 0.77 | 0.45 | 0.76 | 0.73 | |
| rs2050522 | T | 0.33 | 0.95 | 1.00 | 0.93 | <0.001 | 0.01 | 1.00 | 1.00 | |
| rs1414499 | C | 0.39 | 0.51 | 0.02 | 0.46 | 0.80 | 0.52 | 1.00 | 0.91 | |
| NEDD4L | rs1942563 | G | 0.28 | 0.15 | 0.00 | 0.12 | 0.21 | 1.00 | 1.00 | 0.59 |
| rs4461163 | G | 0.18 | 0.15 | 0.00 | 0.11 | 0.44 | 0.68 | 1.00 | 0.59 | |
| rs1977948 | G | 0.27 | 0.16 | 0.00 | 0.13 | 0.77 | 1.00 | 1.00 | 0.62 | |
| rs878396 | A | 0.25 | 0.21 | 0.22 | 0.30 | 0.56 | 0.34 | 0.10 | 0.18 | |
| rs6566942 | G | 0.40 | 0.22 | 0.44 | 0.10 | 0.66 | 0.35 | 0.14 | 0.20 | |
| rs4340401 | T | 0.38 | 0.22 | 0.44 | 0.12 | 0.64 | 0.54 | 0.13 | 1.00 | |
| rs1008899 | A | 0.17 | 0.35 | 0.50 | 0.30 | 0.66 | 0.58 | 0.82 | 0.10 | |
| rs4941366 | G | 0.34 | 0.65 | 0.35 | 0.58 | 0.62 | 0.64 | 1.00 | 0.53 | |
| rs513563 | T | 0.34 | 0.57 | 0.35 | 0.52 | 0.14 | 0.08 | 0.81 | 0.82 | |
| rs522155 | T | 0.36 | 0.61 | 0.45 | 0.60 | 0.34 | 0.05 | 0.83 | 0.23 | |
| rs192659 | G | 0.42 | 0.38 | 0.49 | 0.36 | 0.82 | 0.12 | 0.83 | 0.64 | |
| rs158866 | T | 0.24 | 0.52 | 0.52 | 0.46 | 0.35 | 1.00 | 0.28 | 0.50 | |
| rs158857 | G | 0.40 | 0.49 | 0.19 | 0.47 | 0.77 | 0.62 | 0.10 | 0.04 | |
| rs11152064 | C | 0.15 | 0.24 | 0.29 | 0.30 | 0.02 | 0.34 | 0.26 | 0.41 | |
| rs4941378 | C | 0.49 | 0.40 | 0.34 | 0.40 | 0.53 | 0.13 | 0.47 | 0.07 | |
| rs17064701 | C | 0.11 | 0.00 | 0.00 | 0.02 | 0.59 | 1.00 | 1.00 | 0.03 | |
| rs11664416 | C | 0.18 | 0.15 | 0.02 | 0.06 | 0.45 | 0.06 | 1.00 | 1.00 | |
| rs8089678 | G | 0.38 | 0.38 | 0.25 | 0.31 | 0.64 | 1.00 | 0.77 | 1.00 | |
| rs9675944 | G | 0.39 | 0.65 | 0.74 | 0.63 | 0.16 | 0.81 | 1.00 | 0.09 | |
| rs2075405 | G | 0.39 | 0.26 | 0.21 | 0.27 | 1.00 | 1.00 | 0.67 | 0.77 | |
| rs4940684 | A | 0.34 | 0.62 | 0.62 | 0.61 | 0.46 | <0.001 | 0.65 | 0.16 | |
AA = African Americans; CAU = Caucasians; HAN = Han Chinese; MA = Mexican Americans.
Differences in Allele Frequency
Allele frequency analysis revealed significant differences in frequency distributions among the four different ethnic groups (table 3). Notably, African Americans and Caucasians had few statistically significant differences (p < 0.05) in either the allele or genotype level in BSND with the exception of marker rs1003767. Other ethnic group comparisons, however, seem to have statistically significant differences at most markers in BSND. The greatest differences between ethnic groups were between Caucasian and Mexican Americans, and between African-Americans and Mexican Americans. Marker rs2864124 (BSND) also exhibited statistically significant allele and genotype frequency differences when the comparison included Mexicans which had a much smaller MAF than the other ethnic groups (minor allele frequency 0.18).
Table 3.
Allele frequency differences among ethnic groups
| GENE | rs# | p value for allele frequency differences |
|||||
|---|---|---|---|---|---|---|---|
| AA | AA | AA | CAU | CAU | HAN | ||
| CAU | HAN | MA | HAN | MA | MA | ||
| BSND | rs5774206 | Invariant | |||||
| rs1003767 | <0.001 | 0.71 | 0.01 | <0.001 | <0.001 | 0.002 | |
| rs6682884 | 0.76 | 0.04 | <0.001 | 0.06 | <0.001 | <0.001 | |
| rs2864124 | 0.91 | 0.02 | <0.001 | 0.02 | <0.001 | 0.01 | |
| rs943645 | 0.83 | 0.01 | <0.001 | 0.02 | <0.001 | <0.001 | |
| rs17111474 | 0.91 | 0.74 | <0.001 | 0.76 | <0.001 | 0.002 | |
| rs2479398 | 0.24 | 0.11 | 0.11 | 0.01 | 0.01 | 0.79 | |
| rs2495506 | 0.02 | <0.001 | <0.001 | <0.001 | <0.001 | 0.02 | |
| CLCNKA_B | rs1048302 | 0.10 | <0.001 | <0.001 | <0.001 | 0.04 | <0.001 |
| rs1739840 | <0.001 | 0.69 | <0.001 | <0.001 | 0.003 | <0.001 | |
| rs1763597 | <0.001 | 0.49 | <0.001 | <0.001 | 0.002 | <0.001 | |
| rs945425 | 0.05 | <0.001 | <0.001 | <0.001 | 0.05 | <0.001 | |
| rs2003943 | <0.001 | <0.001 | 0.02 | <0.001 | 0.13 | <0.001 | |
| rs6604910 | <0.001 | <0.001 | <0.001 | 0.33 | 0.04 | 0.33 | |
| rs2050522 | <0.001 | <0.001 | <0.001 | 0.004 | 0.47 | <0.001 | |
| rs1414499 | 0.03 | <0.001 | 0.2 | <0.001 | 0.39 | <0.001 | |
| NEDD4L | rs1942563 | 0.01 | <0.001 | 0.001 | <0.001 | 0.39 | <0.001 |
| rs4461163 | 0.44 | <0.001 | 0.05 | <0.001 | 0.32 | <0.001 | |
| rs1977948 | 0.03 | <0.001 | 0.003 | <0.001 | 0.43 | <0.001 | |
| rs878396 | 0.45 | 0.62 | 0.32 | 0.80 | 0.07 | 0.14 | |
| rs6566942 | <0.001 | 0.39 | <0.001 | <0.001 | 0.003 | <0.001 | |
| rs4340401 | 0.001 | 0.23 | <0.001 | <0.001 | 0.01 | <0.001 | |
| rs1008899 | 0.001 | <0.001 | 0.01 | 0.01 | 0.43 | <0.001 | |
| rs4941366 | <0.001 | 1.00 | <0.001 | <0.001 | 0.21 | <0.001 | |
| rs513563 | <0.001 | 1.00 | 0.002 | <0.001 | 0.28 | 0.002 | |
| rs522155 | <0.001 | 0.07 | <0.001 | 0.003 | 0.86 | 0.004 | |
| rs192659 | 0.50 | 0.19 | 0.34 | 0.05 | 0.83 | 0.02 | |
| rs158866 | <0.001 | <0.001 | <0.001 | 1.00 | 0.27 | 0.32 | |
| rs158857 | 0.24 | 0.002 | 0.32 | <0.001 | 0.73 | <0.001 | |
| rs11152064 | 0.08 | 0.004 | 0.01 | 0.31 | 0.25 | 0.80 | |
| rs4941378 | 0.09 | 0.003 | 0.08 | 0.31 | 0.82 | 0.27 | |
| rs17064701 | <0.001 | <0.001 | <0.001 | – | 0.06 | 0.13 | |
| rs11664416 | 0.37 | <0.001 | <0.001 | <0.001 | 0.01 | 0.18 | |
| rs8089678 | 0.82 | 0.01 | 0.20 | 0.01 | 0.18 | 0.23 | |
| rs9675944 | <0.001 | <0.001 | <0.001 | 0.06 | 0.83 | 0.04 | |
| rs2075405 | 0.06 | 0.004 | 0.05 | 0.42 | 0.78 | 0.29 | |
| rs4940684 | <0.001 | <0.001 | <0.001 | 1.00 | 0.83 | 0.83 | |
AA = African Americans; CAU = Caucasians; HAN = Han Chinese; MA = Mexican Americans.
Within CLCNKA_B,genotype analysis was similar for African-Americans and Caucasians with 3/8 SNPs showing significant differences. Similar to BSND genotype, other ethnic group comparisons had statistically significant differences at 6–7/8 markers within CLCNKA_B except for the Caucasians and Mexicans Americans comparison where there were no differences at any of the markers.
Caucasians and Mexicans Americans also did not have as many statistically significant genotype or allele frequency differences in the NEDD4L gene as the other comparisons, with only three of the markers demonstrating significant differences. A trend of note is that markers within two regions in NEDD4L were less frequently different between ethnic groups (region 1, chromosome position 53876954–53990160 bp: rs1942563, rs4461163, rs1977948, rs878396, rs6566942 and region 2, 54046768–54170475 bp: rs192659, rs158866, rs158857, rs11152064, rs4941378, rs17064701, rs11664416, rs8089678, rs9675944).
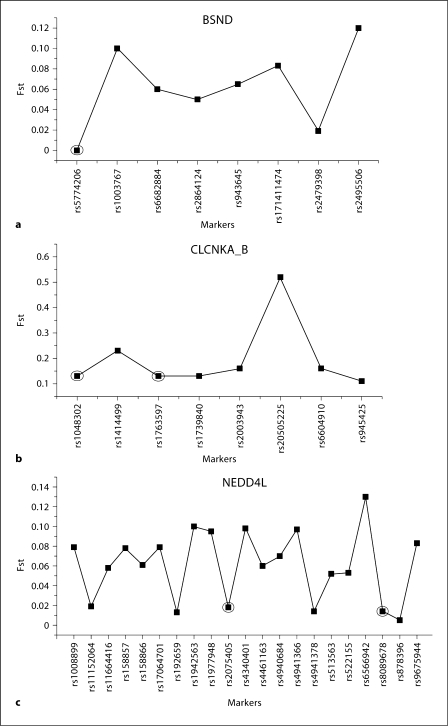
In order to assess the degree of similarity in genetic structure among the different populations, we calculated Fst for each SNP common across all populations (fig. 2). Fst values range from 0 to 1 and increases as the allele frequency difference between populations increases; a value >0.05 is typically considered a significant difference in allele frequencies between the populations. Five of 8 BSND SNPs were greater than 0.05, while 8/8 of the markers in CLCNKA_B were greater than 0.05 with rs2050522 having an extremely large peak at 0.5 because it was invariant in the Han Chinese. Fifteen of 21 SNPs had substantial differences between ethnic groups in NEDD4L with several of the markers having Fst values >0.07.
Fig. 2.
Fst values for BSND, CLCNKA_B and NEDD4L. Circles indicate > 50 % conserved markers. Only markers common to all populations were examined for Fst analyses.
Haplotype Diversity
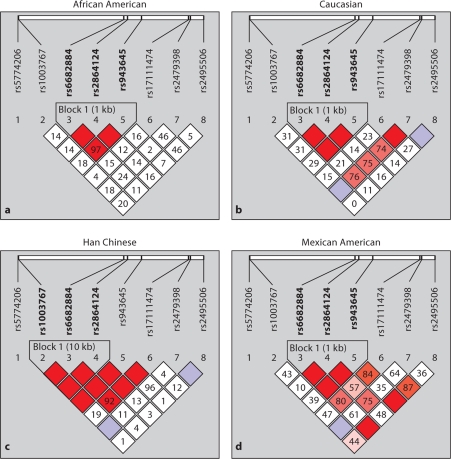
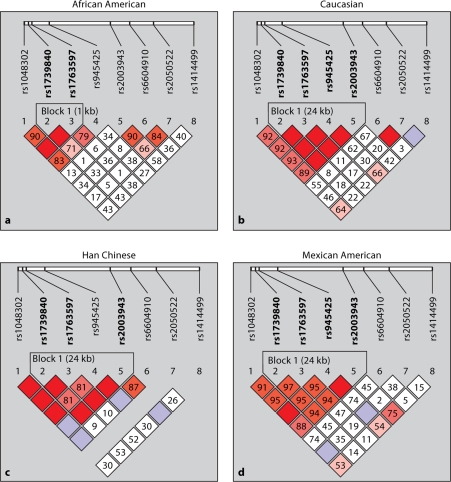
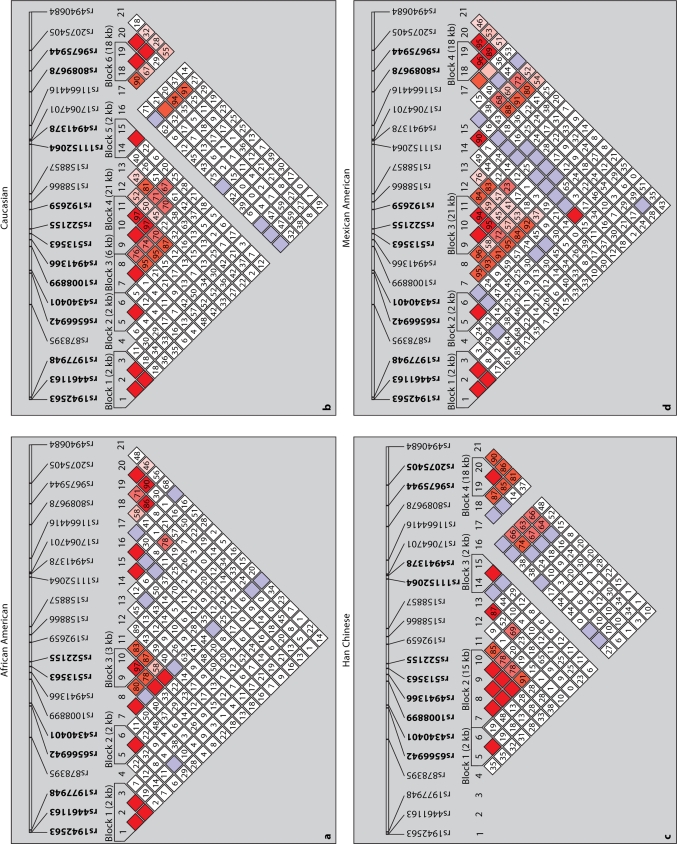
Haplotype blocks were defined in BSND, CLCNKA_B, and NEDD4L in the four populations (fig. 3, 4 and 5). BSND exhibited an LD block (1 kb), common to African Americans, Caucasians and Mexican Americans that included markers rs6682884, rs2864124 and rs943645 (fig. 3a, b and d). These markers were also in strong LD in the Han Chinese population, but did not define a block. Instead, another LD block (10 kb) was identified in the Han Chinese population that included rs103767, rs6682884 and rs2864124. Two of the markers (rs6682884 and rs2864124) had strong LD and were within haplotype blocks in all populations. CLCNKA_B (fig. 4) exhibited more variable LD, with Caucasian, Han Chinese and Mexicans populations showing strong LD. One block (24 kb) which included markers rs1739840, rs1763597, rs945425 and rs20039443 was common to these populations. Two of these markers defined a block (1 kb rs1739840 and rs1763597) within African Americans which exhibited weak linkage disequilibrium. NEDD4L (fig. 5) had several non-contiguous SNPs in LD with each other in several of the populations. One block was common to all of the populations examined (2 kb includes markers rs6566942 and rs4340401), another block common to African Americans and Caucasians (2 kb includes markers rs1942563, rs4461163 and rs1977948), and another block was common to Caucasians and Mexicans (21 kb includes markers rs513563, rs522155 and rs192659). Several of the markers were invariant in the Han Chinese population (rs1942563, rs4461163, rs1977948 and rs17064701).
Fig. 3.
Haploview generated LD map of the 8 SNPs within BSND for all four ethnic groups. Regions of high LD (D′ = 1) are shown in dark squares. Markers with low LD are shown in white. The number within each box indicates the D′ statistic value between the corresponding two SNPs.
Fig. 4.
Haploview generated LD map of the 8 SNPs within CLCNKA_B for all four ethnic groups. Regions of high LD (D′ = 1) are shown in dark squares. Markers with low LD are shown in white. The number within each box indicates the D′ statistic value between the corresponding two SNPs.
Fig. 5.
Haploview generated LD map of the 21 SNPs within NEDD4L for all four ethnic groups. Regions of high LD (D′ = 1) are shown in dark squares. Markers with low LD are shown in white. The number within each box indicates the D′ statistic value between the corresponding two SNPs.
The haplotype frequencies in each group are reported for each gene (tables 4 and 5). BSND and CLCNKA_B haplotype analyses were run with all common markers, while NEDD4L haplotype analyses were run with markers with Fst values >0.07. The reason that only NEDD4Lmarkers with Fst values >0.07 were examined is because Fst analyses suggest that these markers best differentiate these populations and this approach reduces the number of possible haplotypes for this large gene. Haplotype blocks were identified in BSND (rs6682884, rs2864124 and rs28674124) with only ATC and CCT being present; in CLCNKA_B the haplotype for rs1739840 and rs1763597 revealed GA as having the larger frequency in African Americans, Caucasians and Han Chinese and AG having the larger frequency in Mexicans. NEDD4L also had a block (rs6566942 and rs4340401) with a larger frequency of AC across populations. Analysis of haplotype frequencies between pairs of populations revealed statistically significant haplotype frequencies for all comparisons (p <0.001) except for NEDD4L Caucasians vs. Mexicans and a marginally significant difference for CLCNKA_B Caucasians v Mexicans.
Table 4.
Frequencies of common haplotypes
| A | BSND |
Frequency |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | AA | CAU | HAN | MA | |
| H1 | G | A | A | T | C | C | A | C | 0.10 | 0.17 | 0.09 | 0.14 |
| H2 | G | A | A | T | C | C | A | T | 0.02 | 0.07 | 0.05 | 0.00 |
| H3 | G | A | A | T | C | T | A | C | 0.03 | 0.10 | 0.02 | 0.18 |
| H4 | G | A | A | T | C | T | A | T | 0.02 | 0.11 | 0.17 | 0.11 |
| H5 | G | A | C | C | T | C | A | C | 0.07 | 0.04 | 0.00 | 0.04 |
| H6 | G | A | C | C | T | C | G | C | 0.02 | 0.12 | 0.00 | 0.01 |
| H7 | G | A | C | C | T | T | A | C | 0.05 | 0.06 | 0.00 | 0.01 |
| H8 | G | A | C | C | T | T | A | T | 0.05 | 0.02 | 0.00 | 0.00 |
| H9 | G | G | A | T | C | C | A | C | 0.16 | 0.05 | 0.04 | 0.01 |
| H10 | G | G | A | T | C | C | A | T | 0.00 | 0.00 | 0.09 | 0.00 |
| H11 | G | G | A | T | C | C | G | C | 0.06 | 0.02 | 0.02 | 0.00 |
| H12 | G | G | A | T | C | T | A | C | 0.15 | 0.05 | 0.09 | 0.06 |
| H13 | G | G | A | T | C | T | A | T | 0.03 | 0.00 | 0.10 | 0.28 |
| H14 | G | G | C | C | T | C | A | C | 0.14 | 0.09 | 0.14 | 0.08 |
| H15 | G | G | C | C | T | C | A | T | 0.03 | 0.05 | 0.14 | 0.00 |
| SUM1 | 0.93 | 0.95 | 0.95 | 0.92 | ||||||||
| BSND Block | Frequency | |||||
|---|---|---|---|---|---|---|
| 3 | 4 | 5 | AA | CAU | HAN | MA |
| A | T | C | 0.57 | 0.57 | 0.67 | 0.78 |
| C | C | T | 0.36 | 0.38 | 0.28 | 0.14 |
| B | CLCNKA_B |
Frequency |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | AA | CAU | HAN | MA | |
| H1 | A | G | A | C | C | C | T | C | 0.00 | 0.00 | 0.00 | 0.01 |
| H2 | C | G | A | C | G | G | C | T | 0.27 | 0.00 | 0.00 | 0.01 |
| H3 | C | G | A | C | G | C | T | T | 0.09 | 0.05 | 0.01 | 0.01 |
| H4 | A | A | G | T | G | G | C | C | 0.06 | 0.03 | 0.00 | 0.02 |
| H5 | C | A | G | C | G | G | T | T | 0.01 | 0.07 | 0.08 | 0.03 |
| H6 | C | G | A | C | G | G | T | T | 0.00 | 0.07 | 0.18 | 0.04 |
| H7 | A | A | G | T | G | C | T | T | 0.00 | 0.04 | 0.03 | 0.07 |
| H8 | C | G | A | C | G | G | T | C | 0.02 | 0.10 | 0.01 | 0.08 |
| H9 | C | A | G | C | G | C | T | T | 0.01 | 0.02 | 0.08 | 0.12 |
| H10 | C | G | A | C | C | C | T | T | 0.11 | 0.22 | 0.58 | 0.18 |
| H11 | A | A | G | T | G | C | T | C | 0.03 | 0.20 | 0.01 | 0.27 |
| H12 | C |
G |
A |
C |
G |
G |
C |
C |
0.23 |
0.01 |
0.00 |
0.00 |
| SUM1 | 0.84 | 0.81 | 0.97 | 0.85 | ||||||||
| CLCNKA_B Block | Frequency | ||||
|---|---|---|---|---|---|
| 2 |
3 |
AA |
CAU |
HAN |
MA |
| G | A | 0.72 | 0.45 | 0.78 | 0.33 |
| A | G | 0.11 | 0.36 | 0.2 | 0.51 |
| C | NEDD4L2 |
Frequency |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 6 | 7 | 8 | 13 | 16 | 18 | 21 | AA | CAU | HAN | MA | |
| H1 | C | A | A | C | A | T | A | G | A | A | 0.03 | 0.09 | 0.10 | 0.12 |
| H2 | C | A | A | C | A | T | A | G | A | G | 0.00 | 0.04 | 0.09 | 0.00 |
| H3 | C | A | A | C | A | T | A | G | G | G | 0.00 | 0.05 | 0.00 | 0.10 |
| H4 | C | A | G | T | A | T | A | G | A | A | 0.00 | 0.00 | 0.14 | 0.00 |
| H5 | C | A | G | T | A | T | A | G | G | G | 0.00 | 0.04 | 0.12 | 0.02 |
| H6 | C | A | A | C | G | G | A | G | A | A | 0.09 | 0.08 | 0.12 | 0.05 |
| H7 | C | A | A | C | G | G | A | G | A | G | 0.00 | 0.05 | 0.00 | 0.05 |
| H8 | C | A | A | C | G | G | A | G | G | G | 0.09 | 0.03 | 0.05 | 0.00 |
| H9 | C | A | G | T | G | G | A | G | A | A | 0.00 | 0.03 | 0.06 | 0.00 |
| H10 | C | A | G | T | G | T | A | G | A | G | 0.12 | 0.00 | 0.00 | 0.00 |
| H11 | C | A | A | C | G | G | G | G | A | A | 0.00 | 0.11 | 0.04 | 0.23 |
| H12 | C | A | A | C | G | G | G | G | G | A | 0.04 | 0.09 | 0.02 | 0.02 |
| H13 | C | A | A | C | G | G | G | G | G | G | 0.00 | 0.05 | 0.00 | 0.04 |
| H14 | C | A | G | T | G | G | G | G | G | A | 0.00 | 0.05 | 0.00 | 0.00 |
| H15 | C | A | G | T | G | T | G | G | G | G | 0.05 | 0.00 | 0.00 | 0.00 |
| H16 | G | G | G | T | G | G | G | G | A | A | 0.00 | 0.06 | 0.00 | 0.00 |
| SUM1 | 0.41 | 0.78 | 0.73 | 0.63 | ||||||||||
| NEDD4L Block |
Frequency |
||||
|---|---|---|---|---|---|
| 5 | 6 | AA | CAU | HAN | MA |
| A | C | 0.25 | 0.59 | 0.42 | 0.61 |
| G | T | 0.17 | 0.18 | 0.32 | 0.02 |
Marker numbers refer to table 1.
Bold print and italics indicate markers that predict each other.
The sum of the percentage of haplotypes accounting for common haplotypes in each population.
NEDD4L haplotype analysis contains only markers common to all populations and with Fst > 0.07.
Table 5.
Haplotype trend regression test for haplotype frequency differences
| Gene | p value for haplotype frequency differences |
|||||
|---|---|---|---|---|---|---|
| AA | AA | AA | CAU | CAU | HAN | |
| CAU | HAN | MA | HAN | MA | MA | |
| BSND | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| CLCNKA_B | <0.001 | <0.001 | <0.001 | <0.001 | 0.01 | <0.001 |
| NEDD4L | <0.001 | <0.001 | <0.001 | <0.001 | 0.45 | <0.001 |
AA = African Americans; CAU = Caucasians; HAN = Han Chinese; MA = Mexican Americans.
The ‘yin yang’ haplotypes are defined as two high-frequency haplotypes composed of completely mismatching SNP alleles, i.e., nucleotides differ at every SNP in the haplotype pair [15]. We detected such haplotype phenomenon in all the genes studied (table 4). In the BSND gene, one pair of ‘yin yang’ haplotypes, ATC and CCT, was observed with overall frequencies of 57–78% and 14–38%, respectively in the various groups. A pair of ‘yin yang’ haplotypes, GA and AG, was also observed in the CLCNKA_Bgene with frequencies of 33–78% and 11–51% respectively in the various groups. Lastly, we observed ‘yin yang’ haplotype in NEDD4L, AC and GT, with frequencies of 25–61% and 2–32% respectively among the four populations.
Discussion
In this study we examined allele and genotype frequencies, LD and haplotypes among African, Caucasian Chinese, and Mexican Americans in CLCNKA, CLCNKB, BSND and NEDD4L (fig. 1) using experimental and computational analyses. These populations vary with regard to incidence, prevalence and risk factors in the progression of hypertension. For example, African-Americans have a higher prevalence and suffer a disproportionate morbidity from the disease and more often exhibit salt sensitivity. Understanding genetic factors contributing to these differences among the various groups may lead to improved clinical management and better risk assessment. Our study provides the initial characterization of 4 genes that might have a role in essential hypertension. In general, the data demonstrates greater genetic variation in the population of African descent in comparison to the other groups, as has been demonstrated in comparable studies [16, 17]. However, this study observed a greater than expected ethnic allele frequency difference in comparison to the Perlgen database. Fst scores, haplotype analysis and LD blocks have not been previously reported for these genes.
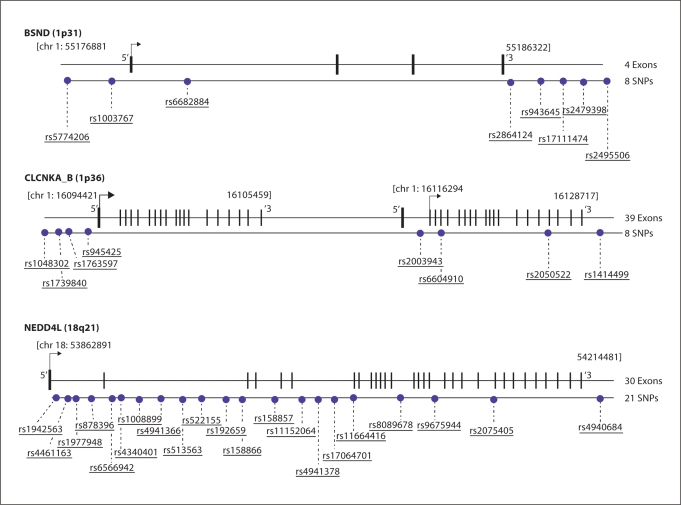
Fig. 1.
Schematic outline of the investigated SNPs in genes (BSND, CLCNKA_B and NEDD4L).
The four genes we chose to examine in this study are involved in salt reabsorption in the kidney. CLCNKA and CLCNKB are located within 11 kb of each other on chromosome 1p36. They are >95% identical, presumably due to a historic gene duplication event, therefore, we characterized them together in this study. BSND, an accessory subunit to both CLCNKA and CLCNKB, is located on chromosome 1p31 while E3 ubiquitin-protein ligase NEdd4-like protein (NEDD4L) which interacts with ClC-Ka (CLCNKA) through barttin (BSND) is on 18q21. Although all four genes have been proposed to be candidate genes for studying genetic variations in the hypertensive population, only two, CLCNKB and NEDD4L have been shown to have an association with essential hypertension. CLCNKB-T481S, a reported gain-of-function variant, was initially associated with hypertension in Ghanaians and White Germans, but other studies have not been able to replicate this association [18,19,20]. Interestingly, despite multiple attempts we were unable to design primers to examine CLCNKB-T481S; and this SNP has not been genotyped by the HapMap project. Russo et al. conducted a haplotype study and demonstrated an association of haplotypes in NEDD4L and hypertension in three populations: White Americans, African and Greek Americans [21]. To date there has not been any replication study of Russo et al. Aside from these two studies extensive examinations of allele, genotype frequencies and halpotype analysis have not been conducted to characterize CLCNKA, CLCNKB, BSND and NEED4L in distinct ethnic populations. Since the introduction of the HapMap, more studies including our group are now examining haplotypes as part of an association study; therefore our study will provide an initial characterization necessary for examining CLCNKA, CLCNKB, BSND and NEDD4Las part of a candidate gene, genome-wide or haplotype association study in various populations.
In this study, we defined haplotype blocks and noted that the number and diversity of haplotypes varied greatly from gene to gene, making generalizations about haplotypes across these candidate genes impossible. All four populations had consistently strong LD with one haplotype block in BSND with the same two SNPs (rs6682884 and rs2864124). These two SNPs clearly can be utilized as tag SNPs in candidate gene association studies in any of the four distinct ethnic groups we examined. This observation leads to the question: are tags SNPs population specific or is power comprised in haplotype association study when tagSNPs chosen from data in one population sample is examined in another sample. There are studies showing that tag SNPs chosen in one population (e.g. European-descent population) are often not appropriate for genotyping in a different population (e.g. African-descent population [22, 23]. Yet, others have shown that tag SNPs are transferable among multiple populations [24]. Our study was not designed to answer if tag SNPs are population specific, yet in these four selected genes there are unique haplotype block structures within each population that need to be considered when designing population association studies.
CLCNKA_B exhibited strong LD in all groups except for African-Americans. Similar to BSND, all groups had one haplotype block where there were two SNPs (rs1739840 and rs1763597) that was shared by all four groups. African-Americans had less LD reflecting evidence for historical recombination that has been noted in several other studies. We were unable to design primer and probes on the to ABI SNPlex platform to examine the only gain-of-function variant, CLCNKB-T481S (rs12140311), associated with hypertension. However, this marker is located 283 bp from rs2050522, making it a reasonable proxy for CLCNKB-T481S and surrounding markers. Marker rs2050522 is in strong LD with rs6604910 within both the African-American and Caucasian populations, suggesting that haplotype analyses using these markers and CLCNKB-T481S should be a focus of future studies.
Similar to the findings with CLCNKA_B,African-Americans have less LD in haplotype block analysis of NEED4L. Caucasians, Chinese and Mexicans had longer haplotype blocks (4–6) for the same region. Russo et al. reported that one SNP (rs513563) in NEDD4L was associated with hypertension in both African and Caucasian Whites. In our study we found that this variant is present in high frequency in all four populations (MAF = 0.34–0.048). In addition, it exhibits strong LD in all four populations; therefore, this SNP should be utilized as a tag SNP in any haplotype analysis that examines blood pressure and NEDD4L not only in the African and Caucasian samples but also in the Han Chinese and Mexican populations.
Overall African-Americans had weaker LD structure than the other groups most notably within the NEDD4L gene. In NEDD4L (table 4) African-Americans have several haplotypes with less than 5% frequency that together account for 59% of the haplotypes therefore making it difficult to use these cut-offs for African-Americans. Common BSND and CLCNKA_B haplotype frequencies in African-Americans were consistently less frequent but not significantly different than the other groups. However, haplotype frequencies for NEDD4L demonstrated significant differences with haplotype >5% in African-Americans accounting for insignificantly lower proportions of all haplotypes than in the other groups (table 4). This could be a function of having more markers in NEDD4L, but more importantly this indicates that haplotype variation makes haplotype association studies more difficult especially in African Americans. Lastly, we note that the Han Chinese had no variation at four markers, three in NEDD4L and one in CLCNKA_B.
In this study we observed ‘yin yang’ haplotype pairs for all four genes (table 5) [14]. There are several explanations for the yin-yang phenomenon observed in this study. First, other characteristics being equal, yin-yang reflects the differences in genes with regards to lower LD, mutation and recombination rates. This observation in this study might be affected by the limited number of SNPs and genes examined but does reflect a substantially lower frequency of haplotypes compared to the number expected (2 vs. 4 or 2 vs. 8) [15]. Understanding yin-yang phenomenon of a gene especially when considering studies involving complex diseases allows insight into the gene's LD, recombination and mutation rate.
Genes responsible for phenotypes that differ greatly between populations are expected to show large allele frequency differences between these populations and thus also demonstrate high Fst. In this study we calculated Fst for each SNP common across all populations. CLCNKA_B had 8/8 SNPs with Fst scores >0.05, 5/8 markers in BSND and 15/21 in NEDD4L had Fst scores >0.05. These genes have been proposed to be associated with hypertension, and they have moderately high Fst which may account for unusually large between-population differences in blood pressure and other intermediate phenotypes surrounding blood pressure. The extent of the differences was also seen in the comparisons of allele frequencies between populations, where all genes differed at more than 50% of the SNPs assayed except BSND between AA and CAU. The differences observed in this study were much greater than expected based on comparisons we have made using the entire Perlegen database (1,585,674 SNPs). In our previous analysis African Americans differed from Caucasians in 38.78% of the markers assayed, African Americans from Han Chinese in 45.01%, Caucasians from Han Chinese at 34.08% (personal communication, unpublished data, Velez, DR et al). These data support the argument that the genes we have studied are more different between populations than the average across the genome. There were no data for Mexican-Americans.
Our study demonstrated genes with high Fst that can be identified as candidate SNPs responsible for yet unidentified intermediate phenotypes affecting blood pressure that differ greatly between populations and thus novel genotype-phenotype relationships may be exposed in further exploring SNPs with high Fst. High Fst values can suggest either local positive selection or drift driving changes in allele frequencies. Although, one of the markers (rs2050522) in CLCNKA_B had an unusually high Fst (>0.5), it is not possible to definitely differentiate which factor is operating based on our data. In addition, markers or SNPs with low (0–0.05) Fst scores were further examined for conservation among species using ensembl genome browser, a comprehensive suite of programs and databases for comparative analysis of genomic sequences (http://www.ensembl.org/index.html). Figure 5 shows SNPs in circles denoting low Fst scores and >50% conservation across species results. Conservation was examined by sequence alignment where >50% of bases in alignments match across species. We observed that 36% of low Fst SNPs are also >50% conserved. SNPs with low Fst scores and conservation across species suggest that perhaps selection constraint may be a factor.
In summary, our data demonstrates initial characterization of four proposed hypertension candidate genes. Allelic frequency data and Fst values indicate the large variation that exists between the various ethnic populations. This study will serve as a starting tool for examining these genes and blood pressure phenotypes in distinct ethnic groups.
Acknowledgements
This work was supported by the National Institute of Health (DK071742, S.S.) and The Robert Wood Johnson Foundation (Amos Medical Faculty Development Program, S.S.).
References
- 1.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 2.Lane DA, Lip GY. Ethnic differences in hypertension and blood pressure control in the UK. QJM. 2001;94:391–396. doi: 10.1093/qjmed/94.7.391. [DOI] [PubMed] [Google Scholar]
- 3.Nesbitt S, Victor RG. Pathogenesis of hypertension in African Americans. Congest Heart Fail. 2004;10:24–29. doi: 10.1111/j.1527-5299.2004.02021.x. [DOI] [PubMed] [Google Scholar]
- 4.Nesbitt SD. Hypertension in black patients: special issues and considerations. Curr Cardiol Rep. 2004;6:416–420. doi: 10.1007/s11886-004-0048-1. [DOI] [PubMed] [Google Scholar]
- 5.Birkenhager R, Otto E, Schurmann MJ, Vollmer M, Ruf EM, Maier-Lutz I, et al. Mutation of BSND causes Bartter syndrome with sensorineural deafness and kidney failure. Nat Genet. 2001;29:310–314. doi: 10.1038/ng752. [DOI] [PubMed] [Google Scholar]
- 6.Schlingmann KP, Konrad M, Jeck N, Waldegger P, Reinalter SC, Holder M, et al. Salt wasting and deafness resulting from mutations in two chloride channels. N Engl J Med. 2004;350:1314–1319. doi: 10.1056/NEJMoa032843. [DOI] [PubMed] [Google Scholar]
- 7.Simon DB, Bindra RS, Mansfield TA, Nelson-Williams C, Mendonca E, Stone R, et al. Mutations in the chloride channel gene, CLCNKB, cause Bartter's syndrome type III. Nat Genet. 1997;17:171–178. doi: 10.1038/ng1097-171. [DOI] [PubMed] [Google Scholar]
- 8.Liu K, Muse SV. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 9.Selander RK. Behavior and genetic variation in natural populations. Am Zool. 1970;10:53–66. doi: 10.1093/icb/10.1.53. [DOI] [PubMed] [Google Scholar]
- 10.Weir B, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 11.Zaykin DV, Westfall PY, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous triats in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 13.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 14.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Rowe WL, Clark AG, Buetow KH. Genomewide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet. 2003;73:1073–1081. doi: 10.1086/379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmueller KE, Wong LJ, Mauney MM, Jiang L, Felder RA, Jose PA, et al. Patterns of genetic variation in the hypertension candidate gene GRK4: Ethnic variation and haplotype structure. Ann Hum Genet. 2006;70(Pt 1):27–41. doi: 10.1111/j.1529-8817.2005.00197.x. [DOI] [PubMed] [Google Scholar]
- 17.Lonjou C, Zhang W, Collins A, Tapper WJ, Elahi E, Maniatis N, et al. Linkage disequilibrium in human populations. Proc Natl Acad Sci USA. 2003;100:6069–6074. doi: 10.1073/pnas.1031521100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeck N, Waldegger S, Lampert A, Boehmer C, Waldegger P, Lang PA, et al. Activating mutation of the renal epithelial chloride channel ClC-Kb predisposing to hypertension. Hypertension. 2004;43:1175–1181. doi: 10.1161/01.HYP.0000129824.12959.f0. [DOI] [PubMed] [Google Scholar]
- 19.Kokubo Y, Iwai N, Tago N, Inamoto N, Okayama A, Yamawaki H, et al. Association analysis between hypertension and, CYBA, CLCNKB: KCNMB1 functional polymorphisms in the Japanese population–the Suita Study. Circ J. 2005;69:138–142. doi: 10.1253/circj.69.138. [DOI] [PubMed] [Google Scholar]
- 20.Speirs HJ, Wang WY, Benjafield AV, Morris BJ. No association with hypertension of CLCNKB and TNFRSF1B polymorphisms at a hypertension locus on chromosome 1p36. J Hypertens. 2005;23:1491–1496. doi: 10.1097/01.hjh.0000174300.73992.cc. [DOI] [PubMed] [Google Scholar]
- 21.Russo CJ, Melista E, Cui J, DeStefano AL, Bakris GL, Manolis AJ, et al. Association of NEDD4L ubiquitin ligase with essential hypertension. Hypertension. 2005;46:488–491. doi: 10.1161/01.HYP.0000178594.63193.c0. [DOI] [PubMed] [Google Scholar]
- 22.Crawford DC, Nickerson DA. Definition and clinical importance of haplotypes. Annu Rev Med. 2005;56:303–320. doi: 10.1146/annurev.med.56.082103.104540. [DOI] [PubMed] [Google Scholar]
- 23.Fullerton SM, Buchanan AV, Sonpar VA, Taylor SL, Smith JD, Carlson CS, et al. The effects of scale: Variation in the APOA1/C3/A4/A5 gene cluster. Hum Genet. 2004;115:36–56. doi: 10.1007/s00439-004-1106-x. [DOI] [PubMed] [Google Scholar]
- 24.De Bakker PI, Graham RR, Altshuler D, Henderson BE, Haiman CA. Transferability of tag SNPs to capture common genetic variation in DNA repair genes across multiple populations. Pac Symp Biocomput. 2006:478–486. [PubMed] [Google Scholar]