Abstract
Tsetse reproduction is unique among insects due to the small numbers of offspring the flies produce and because the female fly carries and nourishes her offspring for their entire immature development. Larval nourishment is supplied by the female as a “milk” substance synthesized by a specialized accessory gland. The milk consists of ~50% fat and ~50% protein. Two milk proteins were identified as the Major Milk gland Protein (GmmMGP) and Transferrin (GmmTsf). Here we describe the identification of two novel gene transcripts (gmmmgp2 and gmmmgp3) produced by the milk gland tissue. These putative secretory products bear no homology to known proteins in the NCBI nr database. Transcripts for these genes can only be detected in the milk gland and their temporal expression correlates with larval development. Functional analysis of these products by RNA interference (RNAi) knockdown analysis shows that GmmMGP2 is critical to reproductive function. The protein appears to affect ovulation, suggesting that it may play a regulatory role in the tsetse reproductive cycle. GmmMGP3 knockdown lacks a phenotype, suggesting its function as a milk protein is possibly redundant.
Keywords: Tsetse, larvigenesis, milk gland, fecundity
Introduction
Tsetse flies are the sole vectors of Human African Trypanosomiasis (HAT) in sub-Saharan Africa. The antigenic variation phenomenon the parasites display in the mammalian host has prevented the development of effective vaccines. Resistance emerging in parasites to the few drugs available for treatment further complicates disease control efforts. In contrast, reduction of tsetse fly populations in affected areas is a highly effective method due to tsetse’s low reproductive rate (Joja and Okoli 2001; Jordan 1986). Understanding of tsetse reproductive physiology can be instrumental in augmenting vector control techniques based on traps and targets to make them more effective and tsetse specific.
Most insects have a large reproductive capacity. Dipterans are capable of developing tens to over a hundred oocytes per gonotrophic cycle. The majority of Dipterans are oviparous (depositing eggs). Tsetse reproduction is unique among vector insects in that it is a viviparous system (intrauterine development and birth of live offspring). A number of fly species have developed various forms of viviparous reproduction. However, most of these species undergo facultative viviparity (deposition of developing embryos or live larvae that do not require maternal nourishment). Tsetse flies (Family: Glossinidae) are members of the Hippoboscoidea superfamily which also includes the Nycteribiidae (Bat flies) and Hippoboscidae (Louse flies) families. All the flies within the Hippoboscoidea superfamily have taken viviparity to the extreme of obligate viviparity. The mother develops a single oocyte per gonotrophic cycle and carries the resulting offspring in an intrauterine environment for the duration of embryonic and larval development. The mother undergoes parturition (gives birth) to a fully developed third instar larva that burrows into the ground and pupates within 30–60 minutes after deposition. Female tsetse flies live on average for ~90 days and are capable of generating an average of ~8 offspring during a lifetime (Langley and Clutton-Brock 1998).
A critical maternal process during larval development is the production and provisioning of nutrients for the larva. The nutrients are produced in a modified accessory gland (milk gland) that connects to the uterus and expands throughout the abdominal cavity of the fly as a series of bifurcating tubules (Tobe and Langley 1978).
Nutrient acquisition, metabolism and production for developing larvae require a large investment of maternal time, activity and nutritional resources. In the conditions associated with our colony, the first gonotrophic cycle in tsetse requires ~21 days from eclosion to parturition of a fully developed third instar larva. The following gonotrophic cycles average ~9 days per offspring (Attardo et al. 2006a). At the time of parturition a third instar larva weighs ~10 mg and is ~6 mM long. The nutrients are supplied in the form of a “milk” secretion that is generated by the milk gland, which empties its secretions into the uterus. During each gonotrophic cycle the mother produces ~30 mg (wet weight) of milk secretion. The milk is comprised of about 50% lipids and 50% protein (Denlinger and Ma 1974). Previous analysis of milk proteins by SDS-PAGE analysis indicates that multiple proteins are contained in the milk secretions and that the protein profile changes over the course of the pregnancy (Riddiford and Dhadialla 1990). Analysis of larval gut contents have also identified multiple proteins, the most predominant of which was sequenced resulting in its partial N-terminal sequence (Osir et al. 1991).
This abundant protein was recently characterized as Milk Gland Protein (GmmMGP) and appears to be necessary for larval development (Attardo et al. 2006a). Transferrin (GmmTsf) was also identified as an abundant milk protein synthesized in the milk gland tissue (Guz et al. 2007). Transferrin appears to undergo proteomic cleavage as it is mobilized into the offspring. Cleavage of transferrin has been associated with its activation for function in development and immunity (Harizanova et al. 2005; Stafford and Belosevic 2003).
Both GmmMGP and GmmTsf are expressed by the milk gland in tight coordination with the development of the larva in the uterus. A double stranded RNA (dsRNA) based RNA interference (RNAi) approach showed that reduction of GmmMGP resulted in reduction of host fecundity. The molecular mechanisms regulating the expression, production and secretion of milk protein genes are unknown. Identification of additional milk proteins will facilitate research into their transcriptional regulation and biochemical functions in the milk. Furthermore, the identification of multiple milk proteins will facilitate the characterization of regulatory factors responsible for the tissue, sex and development stage specific expression profile of these proteins.
Here we report on two novel putative milk associated proteins, identified via bioinformatic analysis of a fat body/milk gland Expressed Sequence Tag (EST) library. Molecular and functional characterization of these genes confirms their association with milk production by the milk gland and shows that at least one is essential for fecundity in female flies.
Results
Characterization of novel milk gland specific proteins
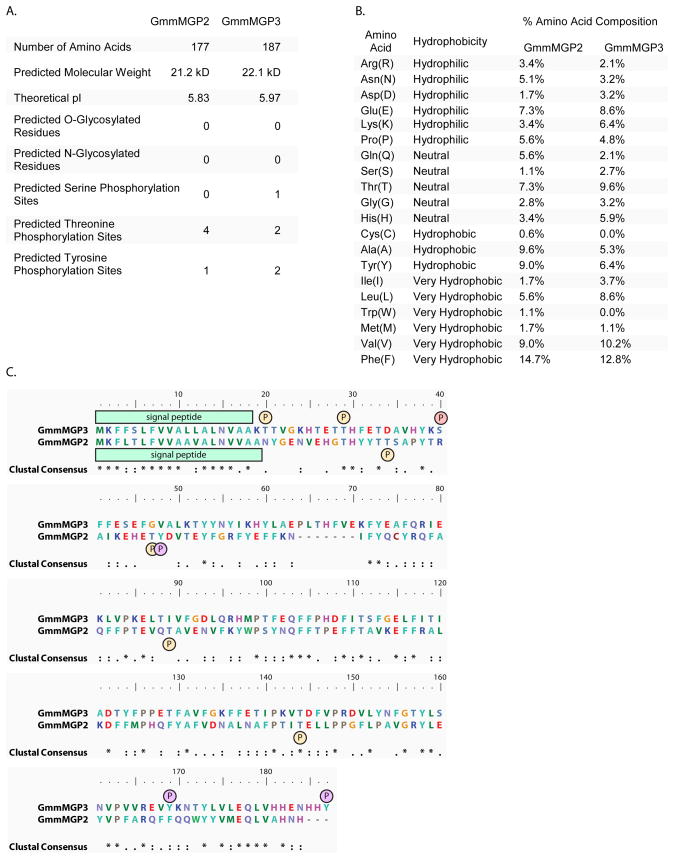
In silico analysis of the tsetse G. m. morsitans fatbody/milk gland EST library identified several small secretory proteins with no homologs in the NCBI NR database (Attardo et al. 2006b). Preliminary RT-PCR analysis showed two of these proteins to be fat body/milk gland specific. These two genes have been named milk gland protein 2 (gmmmgp2) and milk gland protein 3 (gmmmgp3). The 534 bp full-length cDNA for gmmmgp2 is predicted to code a 177-amino acid (aa) protein with a molecular mass of approximately 21 kDa and an estimated pI value of 5.83. The 564 bp full-length cDNA for gmmmgp3 encodes a predicted 187-aa mature protein (22 kDa). Neither protein contains predicted O or N glycosylation sites as predicted by NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) (Blom et al. 2004) and NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/) (Julenius et al. 2005). Potential Serine, threonine and tyrosine phosphorylation sites for these proteins were predicted by NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) (Blom et al. 1999) (Fig 1A). Analysis of the amino acid composition of these proteins reveals a large percentage of hydrophobic amino acids. In particular phenylalanine and valine were very high in both predicted proteins (Fig 1B). This suggests that these proteins could be a mechanism for mobilizing these amino acids which would be difficult to do in their free form. Alignment of these proteins shows high homology at the N-terminus which codes for a signal peptide in both sequences. There also appears to be some homology between the proteins as there are significant groupings of identical amino acids and conserved substitutions occurring throughout the sequence of the two proteins (Fig 1C).
Fig. 1. Comparative protein sequence analysis for gmmmgp2 and gmmmgp3 predicted proteins.
A. Physical characteristics, predicted O-, N- glycosylation sites and predicted serine, threonine and tyrosine phosphorylation sites.
B. Amino acid composition of predicted proteins sorted by hydrophobicity.
C. Clustalx alignment of gmmmgp2 + 3 with predicted signal peptide and phosphorylation sites marked.
Sex and tissue specificity of gmmmgp2 and gmmmgp3 expression
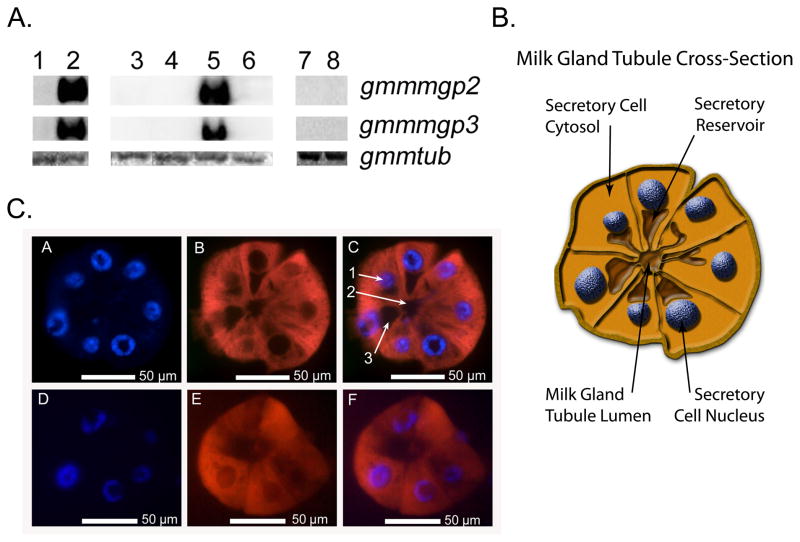
We evaluated the transcript abundance of gmmmgp2 and gmmmgp3 from whole adult males, females and from dissected female tissue including midgut, reproductive tract, and fat body/milk gland (Fig. 2A). Both genes are female specific (lane 2) and corresponding transcripts are only detectable in the fat body/milk gland tissue fractions (lane 5). Expression of both gene products was restricted to adult tissues and no corresponding transcripts could be detected in intrauterine larval or pupal developmental stages (lanes 7+8). Low levels of transcripts for both genes were also detected in the salivary glands by RT-PCR amplification but its significance is unknown (data not shown).
Fig. 2. Tissue and sex specific gene expression profile for gmmmgp2 and gmmmgp3.
A. For Northern blot analysis samples were analyzed from 15-day old flies and 15-day old female tissues. Lane 1: whole males, 2: whole mated females, 3: midguts, 4: reproductive tracts, 5: milk gland and fat body, 6: remaining female carcass. Developmental analysis included: 7: larvae, 8: pupae. Fifteen μg of total RNA collected from 5 individual flies was loaded per lane. Gmmtub-β was used as a loading control.
B. Physiological schematic of a milk gland tubule cross section for reference with 2C.
C. Fluorescent in situ hybridization (FISH) analysis in pregnant female milk gland organ. Tissue sections were stained with DAPI, DIG-labeled antisense gmmmgp2 or gmmmgp3 RNA probe, and anti-DIG antisera. A and D: DAPI staining shows the milk gland cell nuclei; B: In situ staining with gmmmgp2; E: In situ staining with gmmmgp3; C and F : merged images. The corresponding transcripts are localized in the cytoplasm of the secretory cells. 1 = nucleus; 2 = milk gland tubule lumen; 3 = secretory reservoir.
To visualize the cellular localization of gmmmgp2 and gmmmgp3 expression within the fatbody/milk gland tissue fraction, we used in situ hybridization. The structure of the milk gland tubule is displayed in (Fig. 2B). Hybridization with digoxigenin labeled antisense probes showed uniform distribution of transcripts for both genes throughout the cytoplasm of the milk gland secretory cells (Fig 2C). Staining for gmmmgp2 or gmmmgp3 transcripts in the surrounding fat body cells was negative, suggesting these genes undergo milk gland specific expression.
Expression of gmmmgp2 and gmmmgp3 by adult females during larval development
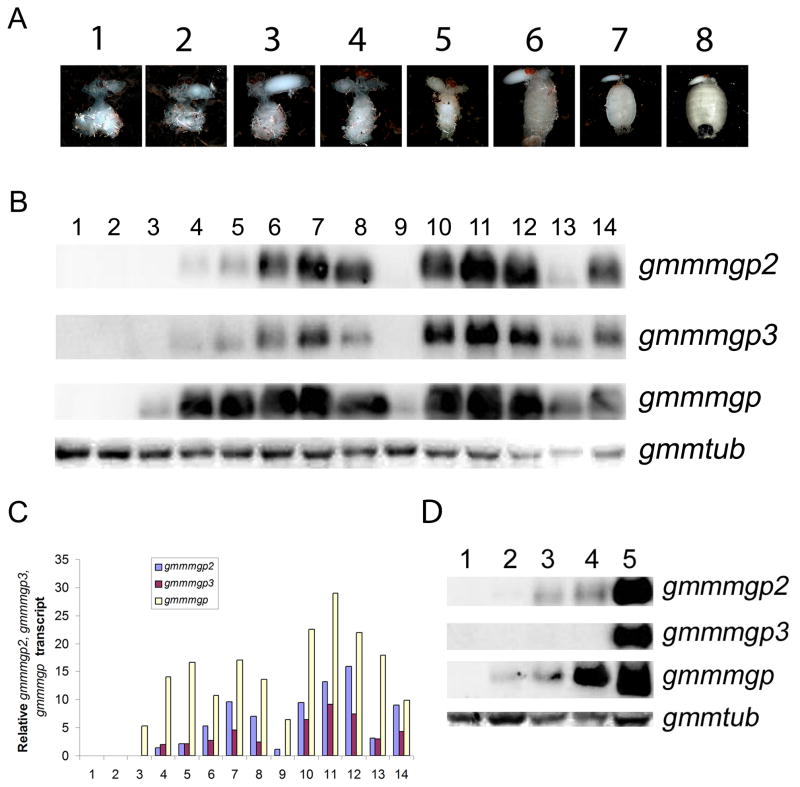
A time course based upon tsetse reproductive physiology was performed to measure gmmmgp2 and 3 expression in whole flies during the first two gonotrophic cycles. The stages analyzed are based upon the physiological state of the female reproductive tract. Fertile females were dissected during the first 2 gonotrophic cycles and the pregnancy status of each female was evaluated by examining the yolk content of the developing oocyte (stages: 1 <25% yolk; 2 >25% and <75% yolk; 3 >75% yolk), the ovary in which oocyte development is occurring (left or right), the presence of an embryo or larva in the uterus and the instar of the larva in the uterus (1st instar, 2nd instar, 3rd instar). The representative physiological states associated with each time point during a gonotrophic cycle were imaged by dissection and microscopy (Fig. 3A).
Fig. 3. Northern blot analysis of gmmmgp, gmmmgp2 and gmmmgp3 expression in Virgins and during the first and second gonotrophic cycle.
A. Images of reproductive tracts from the first 8 developmental stages represented in B. Stages 9–14 are physiologically equivalent to Stages 3–8 due to the beginning of a new gonotrophic cycle. Physiological status was evaluated by measuring yolk content of developing oocytes (Static = no development, Stage 1 = <25% yolk, stage 2 = >25% and <75% yolk, stage 3 = > 75% yolk), the ovary in which oocyte development is occurring (left or right) and the instar of the larva in the uterus (embryo, 1st instar, 2nd instar, 3rd instar). Stage number = 1: Left ovary static, empty uterus and right ovary stage 1; 2: Left ovary static, empty uterus and right ovary stage 2; 3: Left ovary static, empty uterus and right ovary stage 3; 4: Left ovary stage 1, uterus embryo, right ovary static; 5: Left ovary stage 2, uterus embryo or 1st instar, right ovary static; 6: Left ovary stage 3, uterus 1nd instar, right ovary static; 7: Left ovary stage 3, uterus 2nd instar, right ovary static; 8: Left ovary stage 3, uterus 3rd instar, right ovary static.
B. Developmental expression analysis of gmmmgp2, gmmmgp3 and gmmmgp. Each point in the time course is based upon the pregnancy status of the female during the pregnancy cycle as described in A. Each lane represents 15 μg of total RNA from three individual flies for each time point. gmmtub-β was used as a loading control.
C. Quantitated data from B measured by chemoluminescent detection and normalized to gmmtub-β levels.
D. Expression analysis of gmmmgp2, gmmmgp3 and gmmmgp in unmated and mated females, 15 μg of total RNA was collected from three individual flies. Lanes 1–4 correspond to 1 day old, 10 days old, 12 days old and 15 days old virgin females, respectively. Lane 5 shows 15 days old mated females. Gmmtub-β was used as a loading control.
Northern blot analysis of gmmmgp 2 + 3 was performed for 2 gonotrophic cycles and the previously characterized Gmmmgp was included as a comparative control (Fig. 3B). Gmmmgp was included as it expressed in a female and milk gland specific pattern that correlates with larval development (Attardo et al. 2006a). Transcript abundance was measured and normalized using the house keeping gene β-tubulin. The expression profiles of gmmmgp2 and 3 are very similar to gmmmgp, which is a well characterized milk protein (Fig 3C). Transcripts for gmmmgp2 and 3 were absent in young females immediately post eclosion and expression correlated with intrauterine embryonic and larval development and increased until parturition (birth) of the first offspring. Immediately after deposition of the larva, a rapid reduction in transcript abundance was observed. However, transcripts for both genes increased again as the second offspring underwent embryonic and larval development. The expression patterns observed indicate that these two genes are associated with the larval developmental cycle and with milk production.
We also evaluated the expression patterns of both genes in mated and virgin females as a function of female age (Fig. 3D). Transcripts for gmmmgp2 were detectable at 12 days post eclosion in virgin females, while transcripts for gmmmgp3 were undetectable in virgin females by Northern analysis. Comparative analysis of gmmmgp2 levels in mated 15 day old females versus their virgin counterparts showed much higher transcript levels in the mated flies. The same analysis of gmmmgp3 shows expression in mated 15 day old females and no expression in 15 day old virgin females.
Impact of reduced gmmmgp2 and gmmmgp3 functions on host fecundity
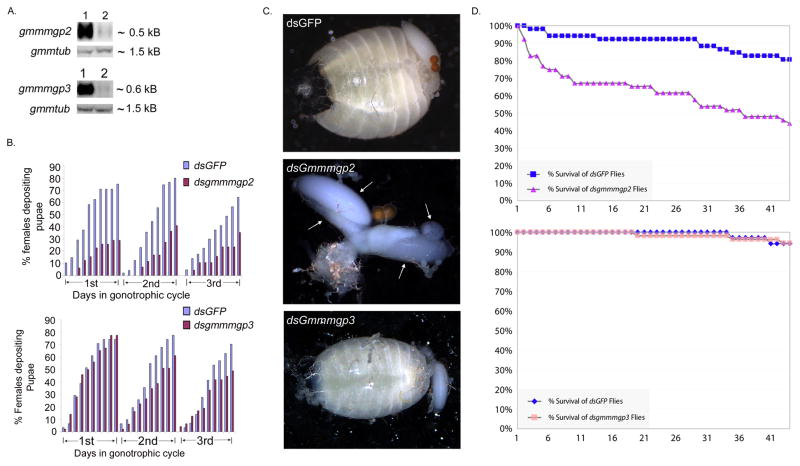
Functional roles of GmmMGP2 and GmmMGP3 were investigated by dsRNA-mediated gene knockdown experiments. Groups of fertile females were treated with dsRNAs corresponding to gmmmgp2 and gmmmgp3 or control gfp, respectively. Knockdown efficacy was determined by northern blot analysis of dsRNA treated flies (Fig. 4A). Mortality of dsRNA treated flies and larval depositions were monitored daily over the first 3 gonotrophic cycles. The cumulative percentage of females depositing pupae relative to the control group was significantly reduced in the dsmgp2 treated group during three gonotrophic cycles. In contrast, the dsmgp3 knockdown group showed no significant difference in pupal deposition from the dsGFP control group (Fig. 4B). The reproductive defects associated with loss of fecundity in the gmmmgp2 knockdown flies were examined by dissection and examination of the reproductive tract. All flies were confirmed to have motile sperm in their spermatheca by microscopic analysis indicating that matings were successful. The ovaries of the gmmmgp2 knockdown flies contained multiple oocytes, some of which were undergoing resorption (Fig. 4C). In a normal fly, only one developing oocyte is present in the ovaries in addition to a maturing larva in the uterus as was observed in the dsgmmmgp3 and dsgfp treated flies. Loss of fecundity in the gmmmgp2 knockdown flies appears to result from a defect in ovulation. The mortality data for these experiments indicates that the dsgmmmgp2 treatment may have had an effect upon fly mortality as well as by the end of the experiment almost ½ of the treated flies had died. The gmmmgp3 treated flies did not show any significant signs of mortality associated with the treatment.
Fig. 4. Functional role of GmmMGP2 in tsetse fecundity.
A. Silencing of gmmmgp2 and gmmmgp3 in mated female flies. Northern analysis of RNA from 15 days old mated females treated with either dsGfp (Top blot: (Lane 1) or dsGmmmgp2 (Lane 2)); and (Bottom blot: dsGfp (Lane 1) and dsGmmmgp3 (Lane 2)).
B. Schematic graphs showing the daily cumulative percentage of females depositing pupae over the three gonotrophic cycles for flies treated with dsGmmmgp2 and dsGmmmgp3 compared with control dsGFP injected flies.
C. Morphological impairment effects on egg development following dsGmmmgp2, dsGmmmgp3 and dsGFP treatment. Arrows indicate the 4 ovarian follicles showing oogenesis without ovulation as well as oocyte degradation and reabsorbtion.
D. Mortality statistics for dsGmmmgp2 and dsGmmmgp3 treatments. Graphs illustrate the percentage of surviving flies over the time in days.
Discussion
In this work, we used an in silico approach to search for unique milk proteins in a tsetse cDNA database and describe functional aspects of two novel milk genes resulting from that search (gmmmgp2 and gmmmgp3). The predicted open reading frames of both genes contain a signal peptide at the N-terminus suggesting that these proteins are secretory products. The expression profiles for both of these genes is female and milk gland tubule specific and expression levels correlate with intrauterine embryonic and larval development events during the gonotrophic cycle. Knockdown of gmmmgp2 by RNAi results in reduced fecundity suggesting that this protein component is required for ovulation and intrauterine larval development. Flies in the gmmmgp2 knockdown group also appeared to have higher mortality levels than the controls. Knockdown of gmmmgp3 does not result in an effect upon fecundity suggesting that this protein may be non-essential or redundant.
Previous work on the tsetse milk proteins identified a number of unknown proteins produced by the milk gland tubules over the course of a pregnancy (Riddiford and Dhadialla 1990). Data mining and analysis of the fat body/milk gland specific cDNA library facilitated the analysis of these two novel proteins that appear to function either as nutrient proteins, regulatory/signaling proteins or both. The phenotype associated with the gmmmgp2 knockdown suggests that this protein may have regulatory properties. Female gmmmgp2 knockdown flies undergo oogenesis, however the fully developed oocytes are never ovulated into the uterus and appear to be broken down and absorbed back into the ovary. These flies also appear to have a higher mortality rate. The cause of this increase is not apparent and further research will be required to understand the physiological consequences of losing the function of this protein. The mode and site of action for this protein remains unknown and bears further analysis to understand its role in the reproductive cycle and tsetse physiology in general. It is possible that GmmMGP2 is provided to the larva as a component of the milk. However, another possibility is that it functions as a regulatory protein that is secreted from the milk gland tubule into the hemolymph rather than the milk gland lumen where it effects ovulation by acting on the uterus/ovaries. Alternatively, the protein could be a milk component that acts as a signal for ovulation from within the uterus. In contrast, knockdown of the second protein, GmmMGP3, resulted in no apparent negative effects upon larval development and deposition. This suggests that this protein may be a non-essential or redundant component of the milk the loss of which is compensated for by other milk proteins.
The development of milk production by females to nourish offspring has evolved multiple times. Mammals, marsupials and a small number of insect species have developed lactation in a convergent manner. While lactation has developed in independent systems, there are interesting parallels in the type of proteins expressed in the milk secretions produced by these dramatically different organisms. The major milk protein in tsetse GmmMGP is in the lipocalin family of proteins (Attardo et al. 2006a). Lipocalins bind small hydrophobic molecules and are thought to act as transporters for insoluble moieties (Flower et al. 2000). Lipocalins are an important component in the milk secretions of viviparous cockroaches (Williford et al. 2004), marsupials (Piotte et al. 1998; Trott et al. 2002) and mammals (Kontopidis et al. 2004). Proteins with iron binding capability such as transferrin and lactoferrin are another common component of milk secretions and are associated with immune function through the sequestration of iron from pathogenic bacteria in the digestive tract of the offspring (Guz et al. 2007; Raiha 1985). The novel proteins identified here may not have structural orthologs in other lactation systems, but they may be representative of orthologous functions in other species. Further functional characterization of these proteins may reveal a unique solution to a common need in lactation or they may represent a specific function required by tsetse’s physiology.
Understanding of the transcriptional regulation of these proteins is important for the identification of the signals and factors regulating lactation in tsetse. The reduced (gmmmgp2) or absent (gmmmgp3) expression of these genes in virgin flies suggests that their regulation is associated with a mating stimulus (such as the transfer of male accessory gland proteins to the female) or a developmental stimulus resulting from larvigenesis.
The role of mating stimuli on female Diptera mating behavior and reproductive physiology is well documented. After mating Drosophila enter a phase called the “Long-Term Post Mating Response” (LTR). During this period female flies are non-receptive to mating advances from males and the rate of egg production and deposition is increased (Manning 1967). These changes in behavior and physiology are activated by the transfer of accessory gland proteins and sperm from the male to the female during copulation. One of the accessory proteins, “Sex Peptide or Acp70A”, stimulates the LTR response temporarily (1–2 days) if injected alone (Chen et al. 1988). However, accessory gland proteins appear to require sperm for the effect to last as females mating with males that secrete accessory gland proteins but not sperm also show a temporary LTR response (Xue and Noll 2000).
Recent work, demonstrates that accessory gland protein function requires a complex network of interactions amongst the proteins as well as physical association with sperm for mobilization to the spermathica which is required for longevity of the LTR response (Ram and Wolfner 2009). This system appears to be conserved as orthologus proteins are present in Aedes aegypti (Sirot et al. 2008) which undergo behavioral and physiological changes including suppression of host seeking behavior, stimulation of vitellogenesis, oviposition and decreased mating receptivity (Klowden 1999). While we do see transcription of some milk proteins in unmated tsetse (gmmmgp1, gmmtsf, and gmmmgp2) the level of transcription appears to be significantly lower than mated and pregnant females. Also gmmmgp3 does not appear to be transcribed at all in unmated females suggesting that mating status could play a larger role in its expression.
Another possible mechanism of milk protein activation is via stimuli related to larvigenesis. As is seen in Fig. 3B, gmmmgp transcription increases over time in correlation with the size and demand for nutrients by the larvae. Transcriptional activation of milk-protein genes could be associated with feedback from stimulation associated with larval development (such as uterine stretch receptors) and/or the volume of milk in the storage reservoirs of the secretory cells lining the milk gland tubules. Transcriptional behavior of the GmmMGP2 and GmmMPG3 proteins can be studied in association with GmmMGP and GmmTsf to build a consensus of genes regulated by the pregnancy cycle in tsetse to understand the signals and factors regulating milk gland protein transcription and translation processes.
Comparative analysis of the regulatory regions of these and other genes will be important for identifying key transcription factors responsible for milk gland tissue specificity and for regulating expression levels during larval development to compensate for larval growth and demand for nutrients. Identification of the regulatory mechanisms controlling milk production in tsetse is essential to development of novel tsetse specific population control strategies.
Experimental Procedures
Tsetse Fly rearing
The Glossina morsitans morsitans colony maintained in the insectary at Yale University was established from puparia obtained from the Tsetse Research Laboratory (Bristol University, Bristol, UK). The original colony was established from puparia collected in Zimbabwe. Flies were maintained at 24±1°C with 50–55% relative humidity and received defibrinated bovine blood every other day by using an artificial membrane system (Moloo 1971). Females were mated at day 3 post-emergence, males were removed after 5 days.
PCR amplification and cloning of gmmmgp2 and gmmmgp3
The cDNA for gmmmgp2 and gmmmgp3 was amplified by PCR utilizing the primer pairs for gmmmgp2: 5′ACTTTGTTCGTCGTAGCCG; 5′ GTTGGAAGAATTGTCTGGCG and for gmmmgp3: 5′ ACCACCCATTTTGAGACC; 5′ ACGGGAACATTGGACAAG, respectively. The PCR amplification conditions were 94°C for 3 min, followed by 30 cycles of 95°C for 50 s, 60°C for 30 s and 72°C for 45 s and by 1 cycle at 72°C for 10 min. Amplification products were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) prior to ligating into the T-vector plasmid (Invitrogen, Carlsbad, CA).
Northern blot analysis
Three flies were collected for each time point/developmental stage and snap frozen in liquid nitrogen. Total RNA was isolated from individual flies using TRIzol®Reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Equal amounts of RNA from the three corresponding flies were combined to provide a single sample. Fifteen micrograms of RNA from each sample was analyzed on a 2.0% agarose/formaldehyde gel. Northern blot analysis was performed following standard protocols (Sambrook and Russell 2001). Digoxigenin labeled antisense RNA probes were generated using the MAXIscript In vitro transcription kit following the manufacturer’s protocol (Ambion, Austin, TX). For probe synthesis, linearized T-vector plasmids containing the open reading frame for either gmmmgp2 or gmmmgp3 were used (Invitrogen, Carlsbad, CA). Chemiluminescent detection of blots was performed with Dig High Prime DNA Labeling and Detection Starter Kit II (Roche Applied Science) according to manufacturer’s instructions. All blots were stripped and rehybridized with gmmtub probe as an internal loading control and hybridization signals for gmmmgp 2 and gmmmgp3 were normalized to the GmmTub signal using Kodak 1D 3. 6. 1. Imaging Software.
Immunohistochemistry/Fluorescent in situ hybridization (FISH) and microscopy
Fat body and milk gland organs from pregnant females were detached from the abdomen and reproductive tract and placed directly into 4% paraformaldehyde for fixation for one week. Paraformaldehyde was replaced with several changes of phosphate buffered saline (PBS) over 2–3 hours. The tissue was then dehydrated through a graded ethanol series: 70% ethanol 2 times for 15 minutes, 95% ethanol 2 times for 30 minutes, 100% ethanol 2 times for 30 minutes. Samples were rinsed in 100% butanol 2 times for 10 minutes and stored in butanol at 4°C for one week. Tissues were embedded in paraffin and sectioned on a rotary microtome at a thickness of 5 μm. The sections were placed on poly-L-Lysine glass slides, left at room temperature until dried and kept at 4°C until used.
For hybridization studies, slides were dewaxed by incubation in methylcyclohexane and ethanol as described in (Attardo et al. 2008). Samples were air dried and prehybridized in hybridization buffer (200 mM NaCl, 8.9 mM TrisHCl, 1.1 mM Tris Base, 6.5 mM NaH2PO4, 5 mM Na2HPO4, 5 mM EDTA, 50% formamide, 10% dextrane sulphate, 1 mg/ml tRNA, 1X Denhardts solution (.02% bovine serum albumin, .02% ficoll, .02% PVP)) for 30 minutes at 65°C. Digoxigenin labeled antisense RNA Probes were diluted in hybridization buffer to a final concentration of 1 ng/μl. Slides were incubated overnight in a humidified chamber at 65°C. Following hybridization, slides were washed in wash buffer (1X SCC, 50% formamide and 0.1% Tween 20) at 65°C and in PBST (1X PBS and 0.1% Tween 20). Slides were blocked in 1X blocking solution (Roche, Indianapolis, IN) for 1 h at room temperature. Anti-DIG-rhodamine Fab fragments (1:200 dilution, Roche) were added to the slides and incubated overnight in the dark at 4°C.
Slides were washed 3 times in PBST and once for 2 minutes in ddH2O. Slides were mounted using GelMountTM medium (Biomedia, Foster City, CA) with 4′,6-diamidino-2-phenylindole (DAPI). Samples were analyzed using a Zeiss Axioskop2 microscope (Zeiss, Thornwood, NY) equipped with a fluorescent filter set with fluorescein, rhodamine and DAPI specificity at magnifications noted. Images were captured using an Infinity1 USB 2.0 camera and software (Lumenera Corporation, Ottawa, Ontario, Canda).
Preparation of double strand RNA
PCR amplicons tailed with T7 promoter sequences were used to synthesize dsRNAs with the Ambion T7 MEGAscript kit (Austin, TX; Cat No. 1334) according to the manufacturer’s instruction. The cDNA clones gmmmgp2, gmmmgp3 and plasmid eGFP (BD Bioscience) served as templates for PCR amplification using the following primers:
dsMGP2: 5′ TAATACGACTCACTATAGGGAGACTTTGTTCGTCGTAGCCG 3′
5′ TAATACGACTCACTATAGGGAGTTGGAAGAATTGTCTGGCG 3′
dsMGP3: 5′ TAATACGACTCACTATAGGGAGACCACCCATTTTGAGACC 3′
5′ TAATACGACTCACTATAGGGAGACGGGAACATTGGACAAG 3′
dsGFP : 5′ TAATACGACTCACTATAGGGTCAGTGGAGAGGGTGAAG 3′
5′ TAATACGACTCACTATAGGCTAGTTGAACGGATCCATC 3′.
The PCR amplification conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 50 s, 60°C for 30 s and 72°C for 50 s and by 1 cycle at 72°C for 10 min in a MJ Research (PTC-200) thermal cycler. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) prior to use for in vitro transcription. Sense and anti-sense RNAs were synthesized using the MEGAscript® RNAi Kit (Ambion, Austin, TX) following the manufacturer’s protocol. The transcripts were precipitated using isopropanol, resuspended in RNase-free water and annealed by incubation at 65°C for 30 min followed by gradual cooling to room temperature. The concentration of dsRNA was determined by spectrophotometry.
RNA interference
Flies for the knockdown experiment were collected within a 24 hour eclosion period to synchronize their reproductive status. The flies were mated at day 3 post eclosion. On day 6 post-eclosion, three groups of 50 flies each received 4 μg of dsGFP, dsMGP2 and dsMGP3 injections, respectively. Cages were monitored daily for fly mortality and larval deposition. At the end of the experiment at day 51, all flies were dissected to confirm mating status by visual examination of spermathica for the presence of sperm and for the accumulation of unovulated oocytes (virgin flies do not ovulate).
Acknowledgments
This work was generously funded by grants to S.A. from the NIAID (AI51584), and Ambrose Monell Foundation and G.M.A from the NIH Ruth Kirshstein Postdoctoral Training Award F32 GM077964. Guangxiao Yang was supported by a Li Foundation grant to S.A
Reference List
- Attardo GM, Guz N, Strickler-Dinglasan P, Aksoy S. Molecular aspects of viviparous reproductive biology of the tsetse fly (Glossina morsitans morsitans): Regulation of yolk and milk gland protein synthesis. Journal of Insect Physiology. 2006a;52:1128–1136. doi: 10.1016/j.jinsphys.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. Analysis of milk gland structure and function in Glossina morsitans: Milk protein production, symbiont populations and fecundity. Journal of Insect Physiology. 2008;54:1236–42. doi: 10.1016/j.jinsphys.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo GM, Strickler-Dinglasan P, Perkin SA, Caler E, Bonaldo MF, Soares MB, El-Sayeed N, Aksoy S. Analysis of fat body transcriptome from the adult tsetse fly, Glossina morsitans morsitans. Insect Molecular Biology. 2006b;15:411–424. doi: 10.1111/j.1365-2583.2006.00649.x. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. Journal of Molecular Biology. 1999;294:1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Chen PS, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, Bohlen P. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell. 1988;54:291–8. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- Denlinger DL, Ma WC. Dynamics of the pregnancy cycle in the tsetse Glossina morsitans. Journal of Insect Physiology. 1974;20:1015–26. doi: 10.1016/0022-1910(74)90143-7. [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochimica et Biophysica Acta: Protein Structure and Molecular Enzymology. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Guz N, Attardo GM, Wu Y, Aksoy S. Molecular aspects of transferrin expression in the tsetse fly (Glossina morsitans morsitans) Journal of Insect Physiology. 2007;53:715–23. doi: 10.1016/j.jinsphys.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harizanova N, Georgieva T, Dunkov BC, Yoshiga T, Law JH. Aedes aegypti transferrin. Gene structure, expression pattern, and regulation. Insect Molecular Biology. 2005;14:79–88. doi: 10.1111/j.1365-2583.2004.00533.x. [DOI] [PubMed] [Google Scholar]
- Joja LL, Okoli UA. Trapping the vector: community action to curb sleeping sickness in southern Sudan. American Journal of Public Health. 2001;91:1583–5. doi: 10.2105/ajph.91.10.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan AM. Trypanosomiasis control and African rural development. Longman; London: 1986. [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Klowden MJ. The check is in the male: male mosquitoes affect female physiology and behavior. Journal of the American Mosquito Control Association. 1999;15:213–20. [PubMed] [Google Scholar]
- Kontopidis G, Holt C, Sawyer L. Invited review: beta-lactoglobulin: binding properties, structure, and function. Journal of Dairy Science. 2004;87:785–96. doi: 10.3168/jds.S0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- Langley PA, Clutton-Brock TH. Does reproductive investment change with age in tsetse flies, Glossina morsitans morsitans (Diptera : Glossinidae)? Functional Ecology. 1998;12:866–870. [Google Scholar]
- Manning A. The control of sexual receptivity in female Drosophila. Animal Behaviour. 1967;15:239–50. doi: 10.1016/0003-3472(67)90006-1. [DOI] [PubMed] [Google Scholar]
- Moloo SK. An artificial feeding technique for Glossina. Parasitology. 1971;63:507–12. doi: 10.1017/s0031182000080021. [DOI] [PubMed] [Google Scholar]
- Osir EO, Kotengo M, Chaudhury MF, Otieno LH. Structural studies on the major milk gland protein of the tsetse fly, Glossina morsitans morsitans. Comparative Biochemistry and Physiology, Part B: Biochemistry and Molecular Biology. 1991;99:803–9. doi: 10.1016/0305-0491(91)90145-4. [DOI] [PubMed] [Google Scholar]
- Piotte CP, Hunter AK, Marshall CJ, Grigor MR. Phylogenetic analysis of three lipocalin-like proteins present in the milk of Trichosurus vulpecula (Phalangeridae, Marsupialia) Journal of Molecular Evolution. 1998;46:361–9. doi: 10.1007/pl00006313. [DOI] [PubMed] [Google Scholar]
- Raiha NC. Nutritional proteins in milk and the protein requirement of normal infants. Pediatrics. 1985;75:136–41. [PubMed] [Google Scholar]
- Ram KR, Wolfner MF. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proceedings of the National Academy of Sciences; USA. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, Dhadialla TS. Protein-Synthesis by the Milk Gland and Fat-Body of the Tsetse-Fly, Glossina-Pallidipes. Insect Biochemistry. 1990;20:493–500. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Sirot LK, Poulson RL, McKenna MC, Girnary H, Wolfner MF, Harrington LC. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect Biochemistry and Molecular Biology. 2008;38:176–89. doi: 10.1016/j.ibmb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JL, Belosevic M. Transferrin and the innate immune response of fish: identification of a novel mechanism of macrophage activation. Developmental & Comparative Immunology. 2003;27:539–54. doi: 10.1016/s0145-305x(02)00138-6. [DOI] [PubMed] [Google Scholar]
- Tobe SS, Langley PA. Reproductive physiology of Glossina. Annual Review of Entomology. 1978;23:283–307. doi: 10.1146/annurev.en.23.010178.001435. [DOI] [PubMed] [Google Scholar]
- Trott JF, Wilson MJ, Hovey RC, Shaw DC, Nicholas KR. Expression of novel lipocalin-like milk protein gene is developmentally-regulated during lactation in the tammar wallaby, Macropus eugenii. Gene. 2002;283:287–297. doi: 10.1016/s0378-1119(01)00883-6. [DOI] [PubMed] [Google Scholar]
- Williford A, Stay B, Bhattacharya D. Evolution of a novel function: nutritive milk in the viviparous cockroach, Diploptera punctata. Evolution and Development. 2004;6:67–77. doi: 10.1111/j.1525-142x.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Xue L, Noll M. Drosophila female sexual behavior induced by sterile males showing copulation complementation. Proceedings of the National Academy of Sciences, USA. 2000;97:3272–5. doi: 10.1073/pnas.060018897. [DOI] [PMC free article] [PubMed] [Google Scholar]