Abstract
Genes for tetrapod fibrillar procollagen chains can be divided into two clades, A and B, based on sequence homologies and differences in protein domain and gene structures. Although the major fibrillar collagen types I–III comprise only clade A chains, the minor fibrillar collagen types V and XI comprise both clade A chains and the clade B chains pro-α1(V), pro-α3(V), pro-α1(XI) and pro-α2(XI), in which defects can underlie various genetic connective tissue disorders. Here we characterize the clade B procollagen chains of zebrafish. We demonstrate that in contrast to the four tetrapod clade B chains, zebrafish have six clade B chains, designated here as pro-α1(V), proα3(V)a and b, pro-α1(XI)a and b, and pro-α2(XI), based on synteny, sequence homologies, and features of protein domain and gene structures. Spatiotemporal expression patterns are described, as are conserved and non-conserved features that provide insights into the function and evolution of the clade B chain types. Such features include differential alternative splicing of NH2-terminal globular sequences and the first case of a non-triple helical imperfection in the COL1 domain of a clade B, or clade A, fibrillar procollagen chain. Evidence is also provided for previously unknown and evolutionarily conserved alternative splicing within the pro-α1(V) C-propeptide, which may affect selectivity of collagen type V/XI chain associations in species ranging from zebrafish to human. Data presented herein provide insights into the nature of clade B procollagen chains and should facilitate their study in the zebrafish model system.
Keywords: Collagen type V/XI, Clade B procollagen chains, Spatiotemporal expression, Intron/exon organization, Alternative splicing, Zebrafish
1. Introduction
The 29 known tetrapod collagen types (Myllyharju and Kivirikko, 2004; Soderhall et al., 2007; Veit et al., 2006) can be divided into two major groups, based primarily on protein domain structure and types of macromolecular structures formed. Nonfibrillar collagens form varied macromolecular structures, have triple-helical regions differing greatly in length and containing numbers of non-triple helical interruptions, and are encoded by genes with limited conservation of intron/exon organization (Myllyharju and Kivirikko, 2004; Sandell, 1990). In contrast, fibrillar collagen types I–III, V, and XI, share a similar protein domain structure, comprising an uninterrupted triple-helical domain of ~1000 amino acids flanked by N- and C-terminal globular domains, and are encoded by genes with similar intron/exon organizations, denoting evolutionary kinship. The “major” fibrillar collagens, I–III, are among the most abundant ECM proteins, whereas collagens V and XI are designated “minor” fibrillar collagens, due to abundance levels markedly lower than those of I–III. Collagen V is widely distributed in tetrapod tissues as an α1(V)2α2(V) heterotrimer (Fessler, 1987), but is also found as an α1(V)α2(V)α3(V) heterotrimer in placenta, uterus, skin, synovial membranes, peripheral nerves (Abedin et al., 1982; Brown et al., 1978; Chernousov et al., 2000; Fessler, 1987; Sage and Bornstein, 1979), and perhaps in ligaments and skeletal muscle, in which expression of the α3(V) chain gene has been reported (Imamura et al., 2000). Collagen V can also be expressed as α1(V)3 homotrimers (Haralson et al., 1980). Collagen XI was originally characterized as an α1(XI)α2(XI)α3(XI) heterotrimer, constituting a minor component of cartilage ECM (Morris and Bachinger, 1987).
Collagen V α1(V)2α2(V) heterotrimers are incorporated into fibrils of the much more abundant collagen I, and regulate properties of the resultant heterotypic fibrils (Birk et al., 1990). Thus, defects in either α1(V) or α2(V) chain genes result in the genetic disorder classic Ehlers-Danlos syndrome, characterized by collagen I/V fibrils of abnormal geometry and diminished tensile strength (De Paepe et al., 1997; Nicholls et al., 1996; Richards et al., 1998; Toriello et al., 1996; Wenstrup et al., 1996). Collagen XI appears to interact with collagen II, the major fibrillar collagen of cartilage, in a manner similar to that in which collagens V and I interact in other tissues (Mendler et al., 1989), as evidenced by the abnormal collagen II fibrils and chondrodysplasia that can result from collagen XI defects (Li et al., 1995).
Although collagens V and XI were originally characterized as separate collagen types, findings of α1(XI) chains in bone (Niyibizi and Eyre, 1989), of α1(XI)α1(V)α3(XI) and α1(XI)2α2(V) heterotrimers in cartilage (Wu et al., 2009), and of heterotrimers comprising α2(V) and α1(XI) chains in non-cartilage tissues (Kleman et al., 1992; Mayne et al., 1993) have now made it apparent that collagen V and XI chains in fact constitute a single collagen type in which different combinations of chains can associate in a tissue-specific manner. In tetrapods, the pro-α1(V), pro-α3(V), proα1(XI), and pro-α2(XI) chains constitute a subgroup among fibrillar procollagen chains on the basis of sequence similarities, structures of cognate genes, size and configuration of N-propeptides, and modes of biosynthetic processing (Greenspan et al., 1991; Imamura et al., 1998; Kimura et al., 1989; Takahara et al., 1995; Takahara et al., 1991; Unsold et al., 2002; Vuristo et al., 1995; Yoshioka and Ramirez, 1990; Zhidkova et al., 1993). These chains have been designated the clade B fibrillar procollagen chains, whereas the related but distinct clade A fibrillar procollagen chains comprise the major fibrillar collagen type I–III chains, and the pro-α2(V) chain (Boot-Handford and Tuckwell, 2003). The pro-α3(XI) chain is also a clade A chain, as it is encoded by the same gene that encodes the pro-α1(II) chain of type II collagen, with the two chains differing only in post-translational modifications (Morris and Bachinger, 1987; Wu and Eyre, 1995).
Zebrafish (Danio rerio) are of great utility in studying functional roles of gene products, due to the almost ideal amenability of this species to combined embryological, molecular and genetic analyses (Kimmel et al., 1990; Mullins et al., 1994; Solnica-Krezel et al., 1994; Streisinger et al., 1981). Here we report that unlike tetrapods, which have four clade B procollagen chains, zebrafish possess six clade B chains, designated here pro-α1(V), pro-α3(V)a, pro-α3(V)b, pro-α1(XI)a, pro-α1(XI)b, and pro-α2(XI). Evidence is provided for proposed orthologous relationships between the 6 zebrafish and 4 tetrapod chains. Full-length sequences are provided for the zebrafish clade B chains, and intron-exon organizations are provided for the cognate genes, designated here col5a1, col5a3a, col5a3b, col11a1a, col11a1b, col11a2, respectively. Spatiotemporal gene expression is also provided. Importantly, evidence is provided of evolutionarily conserved alternative splicing within the pro-α1(V) C-propeptide, which may affect selectivity of collagen type V/XI chain associations in species ranging from zebrafish to humans. Data presented herein provide resources for furthering insights into roles of clade B fibrillar procollagen chains in normal biological and disease processes.
2. Results and Discussion
2.1. Identification/characterization of six zebrafish clade B type V/XI collagen genes
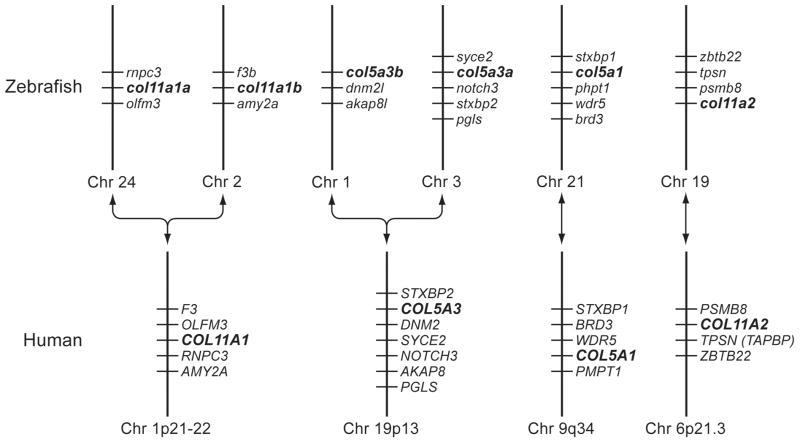
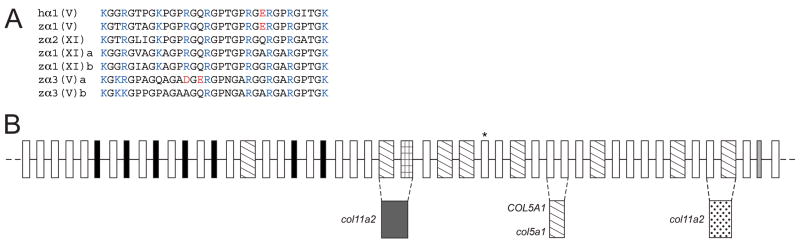
A protein BLAST search of zebrafish genome databases, using murine pro-α3(V) sequences (Imamura et al., 2000), identified 6 loci that seemed reasonable candidates for zebrafish clade B collagen genes, based on partial sequence homologies and intron/exon structures. Despite the numerous differential genomic rearrangements that have occurred in zebrafish and mammalian genomes since their divergence, attempts to establish synteny between the 6 zebrafish loci and mammalian type V/XI collagen genes were sufficiently successful (Fig. 1) to suggest correspondence of the human pro-α1(XI) gene COL11A1 with both loci NM_001083844 and XM_677653, human pro-α2(XI) gene COL11A2 with locus NM_001079992, human pro-α1(V) gene COL5A1 with XM_685787, and human pro-α3(V) gene COL5A3 with both XM_001921860 and XM_688785; leading to provisional designation of the zebrafish loci as col11a1a, col11a1b, col11a2, col5a1, col5a3a and col5a3b, respectively.
Fig. 1.
Apparent syntenic relationships between zebrafish and human clade B collagen genes. Apparent conserved synteny is dispayed for human gene COL11A1 with zebrafish loci NM_001083844 and XM_677653, of the human gene COL11A2 with locus NM_001079992, of the human gene COL5A1 with XM_685787, and of the human gene COL5A3 with both XM_001921860 and XM_688785; leading to provisional designation of the zebrafish loci as col11a1a, col11a1b, col11a2, col5a1, col5a3a and col5a3b, respectively. Changes in the order of genes in some instances between human and zebrafish is presumably due to differential chromosomal rearrangements during evolution. To determine conservation of synteny between zebrafish and human clade B procollagen chain genes, upstream and downstream genes on respective chromosomes in the NCBI ENTREZ D. rerio Tubingen and H. sapiens genome projects were manually compared.
Using the loci listed above as starting points, we confirmed, corrected, and completed coding sequences, UTRs and intron-exon structures of the zebrafish clade B procollagen chain genes. Sequences are available via GenBank accession numbers GQ485664, pro-α1(XI)a; GQ485665, pro-α1(XI)b; GQ485666, pro-α2(XI); GQ485668, pro-α3(V)a; GQ485669, pro-α3(V)b; and GQ485667, pro-α1(V). Only pro-α1(V) sequences are incomplete, lacking signal peptide and 5′-UTR sequences. The six cognate genes col11a1a, col11a1b, col11a2, col5a3a, col5a3b, and col5a1 have 67, 67, 66, 67, 65, and 65 exons, respectively. Comparison of coding sequences, intron-exon organization, and other features of the zebrafish genes (below) confirmed relationships with the 4 mammalian clade B collagen genes suggested by synteny.
2.2 NH2-terminal globular sequences and corresponding portions of cognate genes
Sequences NH2-terminal to the main collagenous (COL1) domain in mammalian clade B procollagen chains show similarities of size, sequence, and domain structure (Greenspan et al., 1991; Imamura et al., 2000; Tsumaki and Kimura, 1995; Zhidkova et al., 1993) that are conserved in the zebrafish clade B chains (Fig. 2). Immediately NH2-terminal to COL1 is the NC2 (noncollagenous 2) linker region, and immediately NH2-terminal to this is the short collagenous COL2 domain (Zhidkova et al., 1993). Although COL2 has been described as three small triple helical motifs divided by two short noncollagneous interruptions (Fichard et al., 1995), the central block of Gly-X-Y repeats, fixed at 17 repeats in all zebrafish and mammalian clade B chains, is probably the only portion capable of triple helix formation. In all mammalian and zebrafish clade B chain genes, COL2 is encoded by 42-, 63-, and 75-bp exons, with the exception of zebrafish Col5a3b (Fig. 3), in which the 42-bp exon has apparently fused with an upstream 51-bp exon to form a 93-bp exon. Col5a3a is the only clade B gene with a 51-bp exon adjacent to the 42-bp COL2 exon, further supporting relatedness of col5a3a and col5a3b.
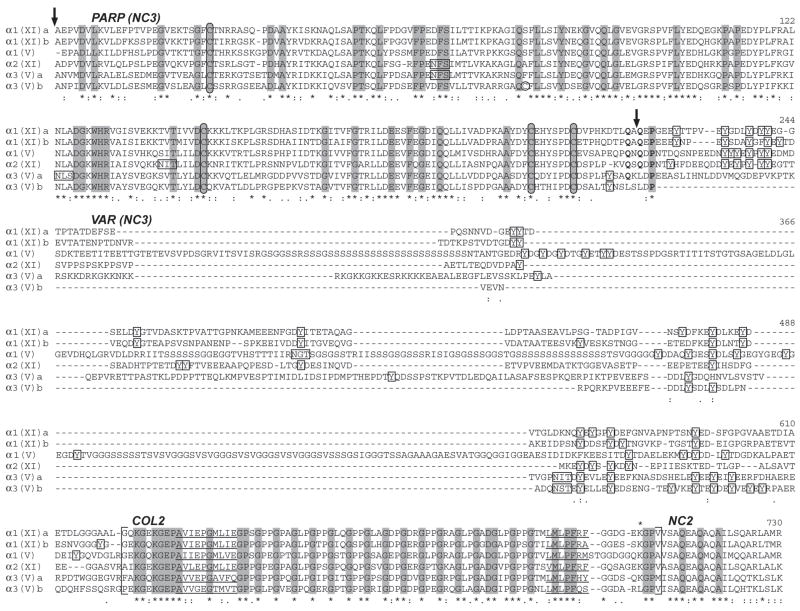
Fig. 2.
Alignment of NH2-terminal sequences of zebrafish clade B procollagen chains. NH2-terminal sequences were aligned using the EMBL-EBI ClustalW2 server. Dashes represent gaps introduced for optimal sequence alignment. Vertical arrows mark the approximate site of signal peptide cleavage and cleavage by BMP1-like proteinases, based on predicted and demonstrated sites in mammalian clade B chains (Gopalakrishnan et al., 2004; Greenspan et al., 1991; Imamura et al., 2000; Imamura et al., 1998; Unsold et al., 2002). PARP, and variable (VAR) subdomains of noncollagenous domain 3 (NC3) are labeled, as is collagenous domain 2 (COL2), and noncollagenous domain 2 (NC2). The extent of COL2 is marked by brackets. Noncollagenous interruptions in the COL2 domain are underlined. Cysteines are circled. Tyrosines between the PARP and COL2 domains are boxed, as are potential Asn-linked glycosylation sites. Residues found at the pro-α1(V) BMP1-cleavage site and conserved in zebrafish clade B chains are in boldface type. Asterisks and dots at the bottom of the alignment signify extent of similarity of aligned sequences, with asterisks denoting identity at a given position in all six zebrafish clade B chains. Residues identical in all six zebrafish and all reported mammalian clade B procollagen chains (Imamura et al., 2000) are shaded.
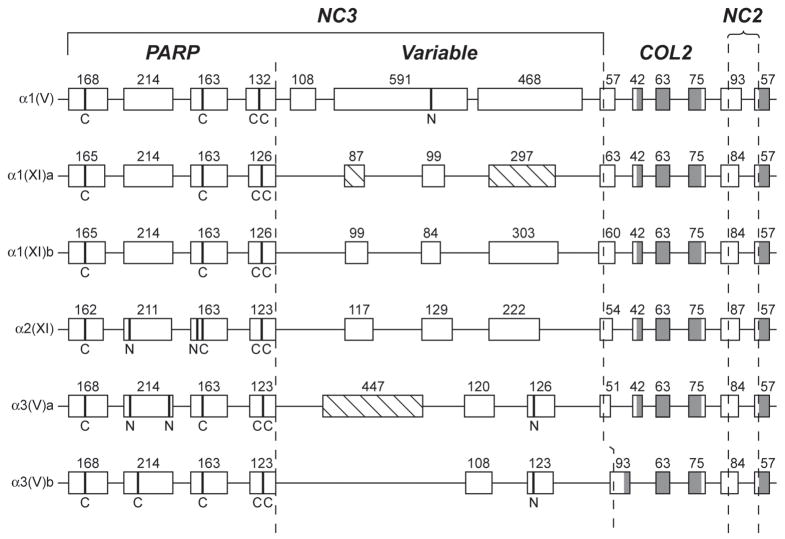
Fig. 3.
Comparison of the intron/exon organizations of NH2-terminal sequence-encoding regions of zebrafish clade B procollagen genes. Boxes represent exons. Numbers represent lengths in basepairs. Shaded and open regions of boxes represent triple-helix and non-triple helix coding sequences, respectively. Dashed lines demarcate PARP-, Variable-, COL2- and NC2-encoding regions. Hatched boxes represent alternatively spliced exons. Sequences encoding cysteines and potential Asn-linked glycosylation sites are marked by broad bands and the letters “C” and “N”, respectively.
Between COL2 and the signal peptide of all mammalian clade B chains is a large globular NC3 region that can be divided into two domains: an NH2-terminal “PARP” domain, with some sequence similarity with the thrombospondin heparin-binding domain (Bork, 1992) and similar domains in FACIT collagens, and a COOH-terminal “variable” domain, lacking discernable homology even between different clade B chains (Imamura et al., 2000; Zhidkova et al., 1993). The same domain structure is conserved in all zebrafish clade B chains (Fig. 2), each of which contains a PARP subdomain encoded by four exons similar in size to corresponding exons of the mammalian clade B chain genes (Takahara et al., 1995) (Fig. 3). As in mammals, each PARP domain has four invariant cysteines, the two most COOH-terminal of which form a cluster that approximately separates PARP and variable regions. These four cysteines were shown to form two intramolecular disulfide bonds in mammalian pro-α2(XI) chains (Neame et al., 1990). Notably, the zebrafish pro-α3(V)b PARP domain differs in having an extra cysteine (Figs. 2 and 3), the function of which remains to be determined. Residues perfectly conserved in PARP subdomains of all zebrafish and mammalian clade B chains (Fig. 2) may represent a consensus or signature clade B PARP domain sequence.
We previously noted that isoelectric points (pIs) of the human clade B PARP domains differ in ways that might reflect differences in physical and functional properties (Imamura et al., 2000). Interestingly, these differences are conserved in the zebrafish clade B chains: pro-α2(XI) has a basic pI (8.64), pro-α1(XI)a, pro-α1(XI)b, and proα1(V) have somewhat acidic pIs (5.31, 5.99, and 6.26, respectively), and pro-α3(V)a and pro-α3(V)b have more markedly acidic pIs (4.54, and 4.66, respectively). The characteristically different pIs may thus be important in providing distinct functional properties for each chain type across species.
Mammalian pro-α1(XI) and pro-α1(V) PARP domains are cleaved by bone morphogenetic protein-1 (BMP1) -like proteinases (Imamura et al., 1998; Pappano et al., 2003; Unsold et al., 2002), and some residues flanking the cleavage sites are conserved at corresponding positions in mammalian pro-α2(XI), but not pro-α3(V) chains (Imamura et al., 1998). The same residues are conserved in zebrafish pro-α1(XI)a, pro-α1(XI)b, pro-α1(V), and pro-α2(XI), but not pro-α3(V)a or pro-α3(V)b (Fig. 2), consistent with the possibility that cleavage by BMP1-like proteinases occurs at this site in all clade B chains except pro-α3(V), across species. The latter possibility is consistent with experimental evidence that mammalian pro-α3(V) chains are not cleaved at this site [(Gopalakrishnan et al., 2004), and unpublished data].
Comparison of PARP sequences shows zebrafish pro-α1(V) to be most homologous to mammalian pro-α1(V), and zebrafish pro-α1(XI)a and pro-α1(XI)b to be most homologous to mammalian pro-α1(XI), consistent with relationships derived from synteny (Table 1). However, zebrafish pro-α2(XI), pro-α3(V)a and pro-α3(V)b PARP domains are not highly homologous to any mammalian clade B chain (Table 1). Thus fixed PARP domain sequences may be less important to pro-α2(XI) and pro-α3(V) function, or functions of these chains may have diverged more than those of the other chains between zebrafish and mammals.
TABLE 1.
Amino Acid Sequence Comparisons of Mammalian and Zebrafish Clade B Protein Domains
| Synteny | PARP Sequence Homology* | Synteny | COL1 Sequence Homology | Synteny | C-prop. Sequence Homology |
|---|---|---|---|---|---|
| hα1(V) | |||||
| α1(V) | 80(91)** | α1(V) | 90(93) | α1(V) | 81(90) |
| α1(XI)b | 70(83) | α1(XI)a | 85(89) | α1(XI)a | 71(84) |
| α1(XI)a | 68(83) | α2(XI) | 80(86) | α1(XI)b | 65(81) |
| α2(XI) | 66(81) | α1(XI)b | 79(84) | α2(XI) | 60(76) |
| α3(V)a | 60(76) | α3(V)a | 73(78) | α3(V)b | 52(70) |
| α3(V)b | 53(72) | α3(V)b | 70(74) | α3(V)a | 50(69) |
| mα1(XI) | |||||
| α1(XI)b | 73(87) | α1(XI)a | 89(92) | α1(XI)a | 79(88) |
| α1(XI)a | 70(85) | α1(XI)b | 84(88) | α1(XI)b | 72(85) |
| α1(V) | 68(87) | α1(V) | 83(87) | α1(V) | 71(86) |
| α2(XI) | 61(79) | α2(XI) | 78(83) | α2(XI) | 58(76) |
| α3(V)a | 53(76) | α3(V)a | 75(79) | α3(V)a | 53(70) |
| α3(V)b | 52(73) | α3(V)b | 72(75) | α3(V)b | 49(69) |
| mα2(XI) | |||||
| α1(V) | 54(72) | α2(XI) | 84(88) | α2(XI) | 53(67) |
| α2(XI) | 52(75) | α1(V) | 80(85) | α1(XI)a | 45(66) |
| α1(XI)b | 52(74) | α1(XI)a | 79(84) | α1(V) | 44(64) |
| α1(XI)a | 50(73) | α1(XI)b | 75(81) | α3(V)b | 44(59) |
| α3(V)a | 48(66) | α3(V)a | 69(75) | α1(XI)b | 43(62) |
| α3(V)b | 47(63) | α3(V)b | 66(71) | α3(V)a | 40(58) |
| hα3(V) | |||||
| α1(XI)a | 44(62) | α1(XI)a | 72(77) | α3(V)a | 57(72) |
| α3(V)b | 43(61) | α1(V) | 70(77) | α3(V)b | 56(70) |
| α1(XI)b | 42(63) | α1(XI)b | 69(76) | α2(XI) | 56(68) |
| α3(V)a | 42(62) | α3(V)a | 68(74) | α1(V) | 52(68) |
| α2(XI) | 42(61) | α2(XI) | 65(72) | α1(XI)b | 48(68) |
| α1(V) | 41(58) | α3(V)b | 61(71) | α1(XI)a | 47(66) |
Percentages of identity and percentages of similarity (parentheses) were obtained by alignment using the BLAST Basic Local Alignment Search Tool.
Percentages of similarity and of identity of subject zebrafish sequences predicted by synteny to be orthologous to query mammalian sequences are given in boldface type.
NC3 variable domains are retained on the mature forms of clade B chains, subsequent to PARP cleavage (Broek et al., 1985; Imamura et al., 1998; Kumamoto and Fessler, 1981; Linsenmayer et al., 1993; Moradi-Ameli et al., 1994; Neame et al., 1990; Niyibizi and Eyre, 1993; Rousseau et al., 1996; Thom and Morris, 1991; Unsold et al., 2002; Zhidkova et al., 1993), and thus likely contribute to functional properties in the ECM. However, there is little homology between variable domains of different zebrafish clade B chains (Fig. 2), or between variable domains in the same chain type in zebrafish and mammals (not shown). Nevertheless, some features are retained. Mammalian proα1(V), pro-α1(XI) and pro-α2(XI) variable regions are highly acidic and sequence diversity is increased and pI modulated via alternative splicing of pro-α1(XI) and proα2(XI) variable region sequences (Greenspan et al., 1991; Oxford et al., 1995; Zhidkova et al., 1995). Similarly, the zebrafish pro-α1(XI)a variable domain is acidic (pI = 3.59), with evidence of alternative splicing yielding forms that contain both, one, or neither of 297- and 87-bp exons (Fig. 3). Zebrafish pro-α1(XI)b and pro-α2(XI) variable domains are also acidic (pIs = 3.73 and 3.78), but show no evidence of alternative splicing. There is no evidence for alternative splicing within mammalian (Greenspan et al., 1991; Zhidkova et al., 1995) or zebrafish pro-α1(V) regions. The zebrafish pro-α1(V) variable region is acidic (pI = 3.86) and, like the mammalian pro-α1(V) variable region and variable regions of zebrafish and mammalian pro-α1(XI) and pro-α2(XI) chains, is rich in tyrosines. Corresponding tyrosines in chick pro-α1(V) and pro-α1(XI) variable regions are sulfated (Fessler et al., 1986), further acidifying the domain. Interestingly, the zebrafish pro-α1(V) variable domain, encoded by large 591- and 468-bp exons, is considerably larger than, and contains large tracts of serines not found in its mammalian counterpart (Figs. 2 and 3). Functional significance of these serines is unknown, though post-translational modification by O-sulfonation (Medzihradszky et al., 2004), phosphorylation or glycosylation could affect the pI and/or other properties of this region.
There is no evidence of alternative splicing in mammalian pro-α3(V) variable regions which, unlike those of the other mammalian clade B chains, is highly basic and lacks tyrosines (Imamura et al., 2000). In contrast, zebrafish pro-α3(V)a and pro-α3(V)b variable regions are acidic (pIs = 4.78 and 3.97, respectively) and have tyrosines, indicating that basicity and lack of tyrosines in this region is not crucial to pro-α3(V) function in all vertebrates. However, the pro-α3(V)a variable region also contains an alternatively spliced 447-bp exon (Fig. 3) encoding basic (pI = 8.05) and highly charged (26 acidic and 28 basic) residues, such that domain properties would be quite different in the presence/absence of these sequences. The relatively short variable domain of proα3(V)b is encoded by an area of col5a3b apparently missing an exon, compared to corresponding regions of other zebrafish clade B chain genes (Fig. 3).
Lysines involved in homotypic covalent crosslinking are located 24 residues NH2-terminal of COL1 and at COL1 residue 924 in mammalian pro-α1(V), pro-α1(XI) and pro-α2(XI); whereas lysines involved in heterotypic crosslinking between types V and I and between types XI and II collagen are at pro-α1(V) and pro-α1(XI) COL1 residue 84 (Niyibizi and Eyre, 1994; Wu and Eyre, 1995). All six zebrafish clade B chains have lysines at COL1 positions 84 and 924, and all except pro-α3(V)b have lysines 24 residues NH2-terminal of COL1 (Fig. 2). Thus, although all zebrafish clade B chains appear capable of both homotypic and heterotypic crosslinking, pro-α3(V)b-containing heterotrimers should have decreased homotypic crosslinking, functional consequences of which are presently unclear.
A phylogram derived from comparison of sequences NH2-terminal to COL1 in the six zebrafish clade B chains supports a relatively close phylogenetic relatedness between pro-α1(XI)a, pro-α1(XI)b, and mammalian pro-α1(XI) chains; between proα3(V)a, pro-α3(V)b, and mammalian pro-α3(V) chains; between zebrafish and mammalian pro-α1(V) chains; and between zebrafish and mammalian pro-α2(XI) chains (Fig. 4A).
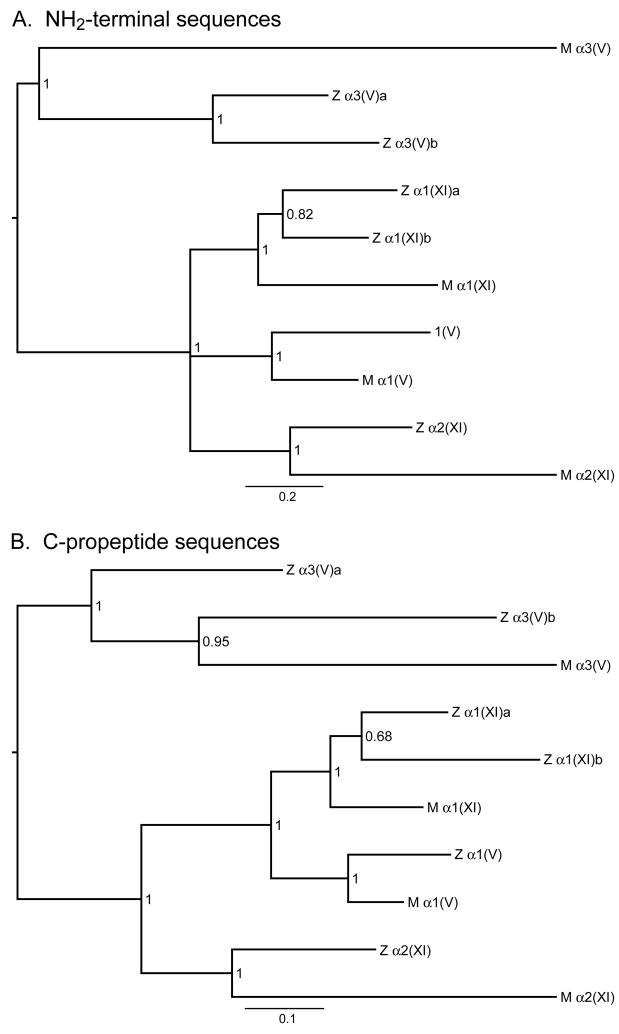
Fig. 4.
Bayesian majority-rule consensus phylograms for the NH2-terminal and C-propeptide regions of zebrafish and mammalian clade B procollagen chains. These midpoint rooted trees provide estimates of phylogenetic relationships among the N-terminal globular (A) and C-propeptide (B) amino acid sequences of these chains, with branch lengths drawn in proportion to the average number of substitutions per site on a given branch (scale bar provided). Numbers represent posterior probability values. Z and M designate zebrafish and mammalian chains, respectively. Mammalian sequences used were from human pro-α3(V) and pro-α1(V), and murine pro-α1(XI) and pro-α2(XI) chains. Both trees are consistent with hypothesized relatedness of the various genes hypothesized in the text. Agreement between the NH2-terminal and C-propeptide trees is very high, with identical topologies except for one polytomy in the NH2-terminal tree and one discrepancy in the relationships among pro-α3(V)a, pro-α3(V)b, and mammalian pro-α3(V). The NH2-terminal tree but not the C-propeptide tree is consistent with the hypothesis that the zebrafish pro-α3(V)a and pro-α3(V)b paralogs trace back to the whole genome duplication event that occurred early in the radiation of ray-finned fish. Although the optimal C-propeptide tree would seem to contradict this likely hypothesis, the posterior probability of the C-propeptide tree is low enough (0.95) that the hypothesis supported by the NH2-terminal region tree remains plausible. Comparison of branch lengths with associated scale bars for the two trees indicates different rates of evolutionary change, consistent with differences in the intensity of purifying selection for NH2-terminal and C-propeptide sequences.
2.3 COL1 domains
Mammalian pro-α3(V) COL1 domains are 3 residues shorter than the 1014-residue COL1 domains of the other mammalian clade B chains (Imamura et al., 2000). Similarly, zebrafish pro-α3(V)a and pro-α3(V)b COL1 domains are 1011 residues long, shorter at their C-termini by one Gly-X-Y triplet than the COL1 domains of the other zebrafish clade B chains, consistent with designation of pro-α3(V)a and pro-α3(V)b as homologues of mammalian pro-α3(V). A shortened mammalian pro-α3(V) COL1 domain and lower number of imino acids than in mammalian pro-α1(V) and pro-α2(V) COL1 domains may contribute to a lower melting temperature of α1(V)α2(V)α3(V) compared with α1(V)2α2(V) heterotrimers (Imamura et al., 2000). Consistent with conservation of lower α1(V)α2(V)α3(V) melting temperatures in zebrafish, numbers of imino acids in pro-α3(V)a and pro-α3(V)b COL1 domains (173 and 164 Pro codons, respectively) are reduced compared to those found in zebrafish pro-α1(V), pro-α1(XI)a, pro-α1(XI)b, and pro-α2(XI) (226, 221, 194, and 197 Pro codons, respectively).
Fibrillar collagen COL1 domains are characterized by uninterrupted Gly-X-Y repeats, a feature thought necessary for normal fibrillogenesis. Surprisingly, the zebrafish pro-α1(XI)a COL1 domain has Ala rather than Gly in what corresponds to the 205th Gly-X-Y COL1 triplet of other clade B chains (see Fig. 5B). Such an imperfection would be expected to destabilize the triple helix, and such Gly substitutions in type I–III collagen COL1 domains can result in severe and pathogenic effects (Myllyharju and Kivirikko, 2004). However, few disease-causing defects in mammalian clade B chain genes are COL1 Gly substitutions (Malfait and De Paepe, 2005), and it is unclear what effect this imperfection may have on ability of α1(XI) chains to regulate properties of type II/XI heterotypic fibrils. It should also be noted that the clade C pro-α1(XXVII) chain, with a domain structure similar to that of clade B chains, has multiple COL1 imperfections and yet can form fibrils (Boot-Handford et al., 2003; Plumb et al., 2007).
Fig. 5.
Features of the COL1-encoding regions of zebrafish clade B procollagen genes. (A) An alignment is shown of zebrafish clade B procollagen COL1 sequences corresponding to the human α1(V) heparin-binding domain. Basic and acidic residues are blue and red, respectively. (B) White, black, hatched, dark grey, light grey, checkered and stippled boxes represent 54-, 45-, 108-, 198-, 36-, 90-, and 162-bp exons, respectively. Dashed lines indicate fusion of 108- and 90-bp exons found in other clade B chain genes to form a 198-bp exon in zebrafish col11a2, fusion of two 54-bp exons found in other clade B chain genes to form a 108-bp exon in human COL5A1 and zebrafish col5a1, and fusion of 54- and 108-bp exons found in other clade B chain genes to form a 162-bp exon in zebrafish col11a2. An asterisk marks the exon encoding an imperfection of the triple helix in zebrafish col11a1a.
Mammalian collagen V has a heparin/heparan sulfate binding site localized to clustered basic amino acid residues in the α1(V) COL1 domain (Delacoux et al., 1998; LeBaron et al., 1989; Mizuno and Hayashi, 1996; Yaoi et al., 1990). Mammalian type XI collagen binds heparin, presumably due to high basicity in the corresponding region (Yaoi et al., 1990), but mammalian α3(V) chains do not bind heparin, perhaps due to less basic and more acidic residues in this region (Imamura et al., 2000). The zebrafish α1(V) chain is identical to human in numbers/placement of basic and acidic residues at this site (Fig. 5A) and zebrafish type XI clade B chains have similar numbers of basic residues, and no acidic residues, in the corresponding region. However, although mammalian α3(V) chains have 6 basic and 3 acidic residues at this site (compared with 9 basic and 0–2 acidic residues for other mammalian clade B chains) (Imamura et al., 2000), zebrafish α3(V)a has 8 basic and two acidic residues, and zebrafish α3(V)b chain has 8 basic, and no acidic residues (Fig. 5A). Thus, zebrafish and mammalian α3(V) COL1 domains may differ in ability to bind heparan sulfate proteoglycans.
Intron-exon organization of COL1 regions of mammalian clade B and clade A chain genes are similar, in that early evolution of both appears to have involved tandem duplication of a 54-bp ancestral element (Takahara et al., 1995; Vuristo et al., 1995), but differ in ways that reflect divergence of the two clades (Takahara et al., 1991; Vuristo et al., 1995). COL1 intron-exon organization of mammalian and zebrafish clade B chain genes are generally conserved (Fig. 5B). Mammalian and zebrafish pro-α1(V) gene COL1 regions differ from other clade B genes in containing a 108-bp exon, apparently resulting from fusion of two 54-bp exons found in the other clade B genes [Fig. 5B and (Takahara et al., 1995)]. Zebrafish col11a2 has unique COL1 198-bp and 162-bp exons, apparently resulting from fusion of 108-bp and 90-bp exons, and of 54-bp and 108-bp exons, respectively, found in the other clade B genes (Fig. 5B).
Zebrafish pro-α1(V) COL1 amino acid sequences are most similar to mammalian pro-α1(V), zebrafish pro-α1(XI)a and pro-α1(XI)b sequences are most similar to mammalian pro-α1(XI), and zebrafish pro-α2(XI) is most similar to mammalian proα2(XI) (Table 1), consistent with synteny. However, zebrafish pro-α3(V)b and proα3(V)a COL1 sequences are less homologous to those of mammalian pro-α3(V) than are some other zebrafish clade B procollagen chains (Table 1). Thus, fixed COL1 sequences may not be as important to pro-α3(V) function as they are for other clade B chains, or pro-α3(V) function may have diverged more than functions of the other clade B chains.
2.4 C-propeptides
Fibrillar procollagen C-propeptides have 7 or 8 cysteines at fixed positions (Dion and Myers, 1987; Imamura et al., 2000). The most C-terminal four cysteines are invariant in number and are thought to form intrachain disulfide bonds, necessary to proper tertiary configuration and to trimer formation (Bernard et al., 1988; Dion and Myers, 1987). The more NH2-terminal 3 or 4 cysteines are thought to form interchain disulfide bonds (Bernard et al., 1988; Dion and Myers, 1987). Pro-α1(V) has eight C-propeptide cysteines in both zebrafish (Fig. 6) and higher vertebrates (Gordon et al., 1999; Greenspan et al., 1991; Takahara et al., 1991; Wu et al., 1998). Zebrafish proα1(XI)a and pro-α1(XI)b C-propeptides both contain seven cysteines, as in human, rat and mouse pro-α1(XI) (Bernard et al., 1988; Yoshioka et al., 1995), but differing from the 8 cysteines reported for chick (Nah et al., 1992). Interestingly, zebrafish pro-α3(V)a and pro-α3(V)b C-propeptides have 8 cysteines, in contrast to the 7 C-propeptide cysteines of human, mouse and rat pro-α3(V) (Chernousov et al., 2000; Imamura et al., 2000). Similarly, the zebrafish pro-α2(XI) C-propeptide contains 7 cysteines, unlike the 8 cysteines of mammalian pro-α2(XI) (Kimura et al., 1989; Lui et al., 1996). Thus, either the changed numbers of cysteines change the functions of zebrafish pro-α3(V) and pro-α2(XI) C-propeptides compared to mammalian counterparts, or these changes do not have marked functional consequences. It has been speculated that a full complement of 8 C-propeptide cysteines enables procollagen chains to form both homo- and hetero-trimers, whereas procollagen chains with 7 C-propeptide cysteines are restricted to incorporation into heterotrimers (Bernard et al., 1988). However, although this correlation appears to hold for major fibrillar collagen types I–III, and for mammalian type V collagen chains (Bernard et al., 1988; Fichard et al., 1997; Gopalakrishnan et al., 2004; Imamura et al., 2000; Imamura et al., 1998), mammalian pro-α2(XI) chains, with 8 C-propeptide cysteines (Kimura et al., 1989), are not known to form homotrimers, and addition of an eighth cysteine to the proper position (position 2) in the pro-α2(I) chain is not sufficient for homotrimer formation (Lees and Bulleid, 1994). It thus remains to be determined the extent to which differing numbers of C-propeptide cysteines may affect function in pro-α3(V), pro-α1(XI), and pro-α2(XI) chains of different species.
Fig. 6.
Alignment of C-propeptide sequences of zebrafish clade B procollagen chains. C-propeptide sequences were aligned using the EMBL-EBI ClustalW2 server. Dashes represent gaps introduced for optimal alignment. Ends of COL1 domains are marked by brackets. Consensus sites for cleavage by proprotein convertases are underlined, and a vertical arrow marks the predicted site for cleavage. Cysteines are circled and potential Asn-linked glycosylation sites are boxed. Asterisks and dots at the bottom of the alignment signify extent of similarity of aligned sequences, with asterisks denoting identity in all six zebrafish clade B chains. Residues identical in all six zebrafish and all reported mammalian clade B procollagen chains (Imamura et al., 2000) are shaded.
In contrast to the major fibrillar procollagens, in which C-propeptides are cleaved by BMP1-like proteinases (Kessler et al., 1996; Scott et al., 1999), mammalian proα1(V), pro-α3(V), and pro-α1(XI) C-propeptides are cleaved by furin-like proprotein convertases (PCs) at canonical RX(K/R)R sites (Gopalakrishnan et al., 2004; Imamura et al., 1998; Pappano et al., 2003; Unsold et al., 2002). Similar sites in mammalian proα1(XI) and pro-α2(XI) chains align with the pro-α1(V) and pro-α3(V) PC cleavage sites (Imamura et al., 2000). All zebrafish clade B chains have sequences that fit the necessary requirements for cleavage by PCs (Fig. 6); an invariant Arg in the P1 position, and basic residues at the P2 and P4 positions (Nakayama, 1997), and all of these sites align with the demonstrated and putative PC cleavage sites of the mammalian clade B chains (not shown). Thus, it seems likely that zebrafish clade B procollagen C-propeptides are cleaved by PCs, the major proprotein processing enzymes of the constitutive secretory pathway. Human, pro-α1(V) C-propeptides can be cleaved in vitro with BMP1 (Kessler et al., 2001). However, the site at which such cleavage occurs does not align with potential BMP1 cleavage sites in zebrafish pro-α1(V), consistent with the probability that in vivo cleavage of clade B procollagen C-propeptides is via PCs across species.
A potential Asn-linked glycosylation site preceding cysteine 6 is the only such site in clade A C-propeptides, and has been suggested to be of functional significance (Dion and Myers, 1987). This site is also conserved in mammalian pro-α1(V) and proα1(XI) (Bernard et al., 1988; Greenspan et al., 1991; Takahara et al., 1991), and in zebrafish pro-α1(V), pro-α1(XI)a and pro-α1(XI)b (Fig. 6), suggesting functional significance in these chains. However, the site is absent in mammalian pro-α2(XI) (Kimura et al., 1989), but present in zebrafish pro-α2(XI), and is in human, but not mouse pro-α3(V) (Imamura et al., 2000) or zebrafish pro-α3(V)a or pro-α3(V)b. Thus the site does not appear to be either necessary or detrimental to pro-α2(XI) or pro-α3(V) function. A potential glycosylation site (NQT) conserved between cysteines 6 and 7 in human and mouse pro-α3(V) C-propeptides, and not found in any other fibrillar procollagen C-propeptide, is absent in zebrafish pro-α3(V)a and pro-α3(V)b, indicating that glycosylation at this site is not necessary to pro-α3(V) function in zebrafish. A potential NFT glycosylation site conserved immediately downstream of cysteine 4 in all mammalian clade B procollagen C-propeptides (Imamura et al., 2000), but missing from all clade A chains, is conserved in all zebrafish clade B chains (Fig. 6), consistent with the possibility that glycosylation at this site marks a fundamental structural/functional difference between clade A and B fibrillar procollagen chains.
The mammalian pro-α2(XI) C-propeptide is shortened between cysteines 5 and 6, relative to other mammalian clade B chains (Imamura et al., 2000; Kimura et al., 1989), due a shortened penultimate exon of the cognate gene and absence of a 69 bp exon found in mammalian pro-α1(V) and pro-α1(XI) genes [(Takahara et al., 1995), NT_039240]. Interestingly, zebrafish pro-α2(XI) does not a shortened C-propeptide (Fig. 6), as the penultimate 237-bp col11a2 exon is not reduced in size compared to the corresponding 234-bp exons in other zebrafish clade B genes (Fig. 8B); and because the 69-bp exon missing from mammalian pro-α2(XI) genes is present in col11a2, as in all other zebrafish clade B collagen genes (e.g. Fig. 8B). Thus, absence of sequences missing in mammalian pro-α2(XI) chains is not necessary to pro-α2(XI) function in zebrafish.
Fig. 8.
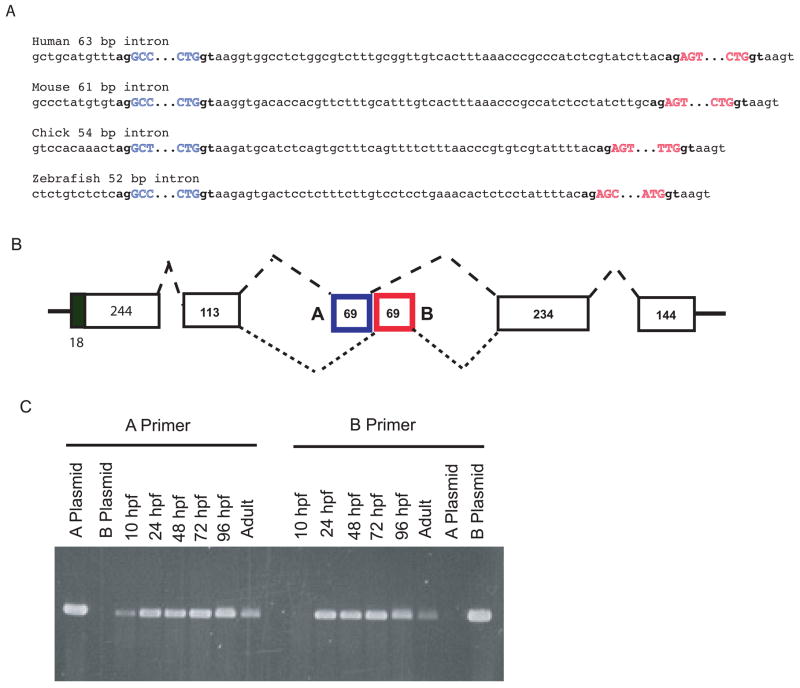
(A) Mutually exclusive alternative splicing of exons A and B in pro-α1(V) genes of zebrafish and other species. Sequences are shown of splice junctions and introns separating exons A (blue) and B (red) in the C-propeptide regions of pro-α1(V) genes of human, mouse, chick, and zebrafish. Exonic sequences are uppercase and intronic sequences are lower case. Invariable 5′ gt and 3′ ag ends of introns are in boldface type. (B) A model is shown for the proposed mutually exclusive alternative splicing of exons A and B. (C) RT-PCR with exon A- and B-specific primers demonstrates expression of both exons in col5a1 transcripts of zebrafish embryos and adults. PCR of plasmids containing cloned zebrafish exon A and B sequences (A plasmid and B plasmid, respectively) demonstrates specificity of the primers.
Zebrafish pro-α1(V) C-propeptide amino acid sequences are most similar to mammalian pro-α1(V); pro-α1(XI)a and pro-α1(XI)b are most similar to mammalian pro-α1(XI); zebrafish pro-α2(XI) is most similar to mammalian pro-α2(XI); and proα3(V)b and pro-α3(V)a are most similar to mammalian pro-α3(V) (Table 1). Thus, C-propeptide sequence homologies are in perfect agreement with predictions of relatedness based on synteny. As shown for NH2-terminal sequences (Fig. 4A), a phylogram comparison of C-propeptide sequences supports close phylogenetic relatedness between pro-α1(XI)a, pro-α1(XI)b, and mammalian pro-α1(XI) chains; between pro-α3(V)a, proα3(V)b, and mammalian pro-α3(V) chains; between zebrafish and mammalian proα1(V) chains; and between zebrafish and mammalian pro-α2(XI) chains (Fig. 4B).
In all fibrillar collagen genes a junctional exon encodes the end of COL1 and the beginning of the C-propeptide (Takahara et al., 1991) (Fig. 8B). The relative shortness of the zebrafish pro-α3(V)a and pro-α3(V)b COL1 domains is reflected, as in mammalian pro-α3(V) chains, in the presence of only 9 bp of COL1-encoding sequences in the cognate junctional exons, compared to 18 bp in the junctional exons of all other mammalian and zebrafish clade B collagen genes, again consistent with relatedness of pro-α3(V)a, pro-α3(V)b, and mammalian pro-α3(V) chains.
2.5 Alternative splicing in the pro-α1(V) C-propeptide
In previous RT-PCR screens of dermal fibroblasts from human subjects (unpublished data), we identified a subset of cloned pro-α1(V) cDNAs in which the reported 69 bp sequence of COL5A1 exon 64 (Greenspan et al., 1991; Takahara et al., 1995; Takahara et al., 1991) is replaced by an alternate 69 bp sequence (Fig. 7A, exon B). To assay for possible alternative splicing in COL5A1 at this site, direct sequencing was performed on cDNA synthesized from human heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas RNA. In each human tissue, superimposed sequences of both exons A and B, flanked by single sequences from upstream and downstream exons provided clear evidence for alternative splicing in all tissues examined, with some differences in ratios of A and B sequences suggesting a degree of tissue-specificity (not shown). Restriction with EcoRI, which cleaves within human exon A but not B sequences, and with SexAI, which cleaves within exon B but not A, demonstrates different ratios of exons A and B sequences in human placenta and liver (Fig. 7B).
Fig. 7.
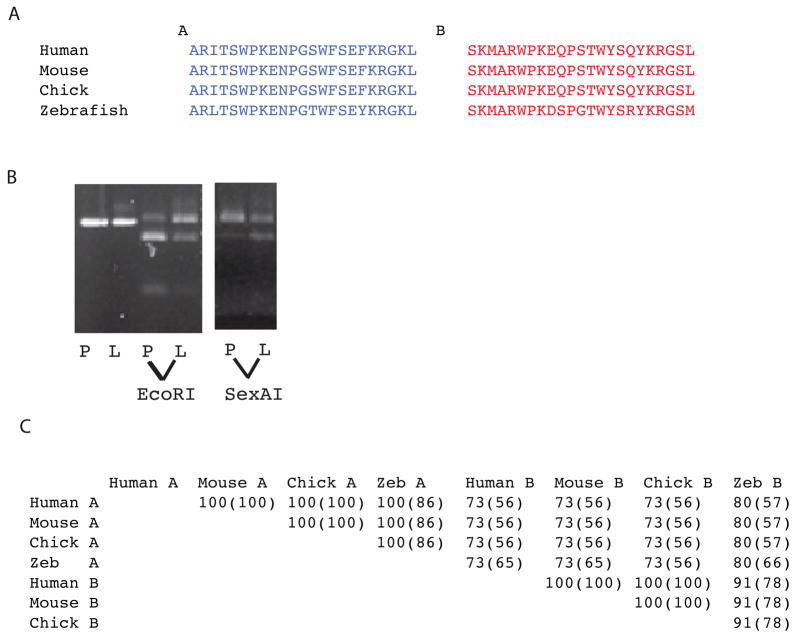
Alternative splicing in the COL5A1 C-propeptide region and conservation of alternatively spliced exon sequences across species. (A) Amino acid sequences are shown for exons A and B for human mouse, chick and zebrafish. (B) Restriction with EcoRI, which cuts within exon A, but not exon B sequences, or SexAI, which cuts within exon B but not exon A sequences, demonstrates varying ratios of exon A and B sequences in human placenta (P) and liver (L). The first two lanes are uncut cDNA. (C) Percentages of similarity and identity (parentheses) between the amino acid sequences of exons A and B in the pro-α1(V) genes of human, mouse, chick, and zebrafish were obtained by alignment using the BLAST Basic Local Alignment Search Tool.
Only 63 bp separate COL5A1 exon 64A from exon 64B (Fig. 8A), suggesting a mechanism of mutually exclusive use of one or the other exon in a given mRNA (Fig. 8B), as a minimum intron length of 70–80 nucleotides is thought necessary for splicing in higher eukaryotes (Ruskin et al., 1985; Wieringa et al., 1984). To determine whether similar alternative splicing might be conserved across species, we examined equivalent areas in the pro-α1(V) genes of zebrafish and other species. Potential alternative exons are found a short distance downstream of the exon corresponding to COL5A1 exon 64 in all species examined (Figs. 7A and 8A). In addition, in all species examined intervening sequences between exons A and B are shorter than the ~70–80 nucleotides thought necessary for splicing (Fig. 8A), consistent with conservation of the mechanism for alternative splicing first suggested by COL5A1 sequences. Interestingly, some sequence homology exists between exons A and B in the various species (Fig. 7C), suggesting derivation of one exon from the other during evolution, perhaps via exon duplication. Importantly, amino acid sequences are highly conserved across species not only between the various exons A, but between the exons B as well (Fig. 7C), consistent with functional significance for sequences encoded by both exons in each species.
To assay for alternative splicing in zebrafish col5a1, direct sequencing was performed on cDNA synthesized from RNA of 24, 48, 72, and 96 hpf embryos, and adults. Sequences from embryos of all stages showed superimposed exon A and B sequences, flanked by single sequences from upstream and downstream exons (not shown), providing clear evidence for alternative splicing. A and B sequence ratios were similar in 24–72 hpf embryos, with a slightly increased ratio of A to B at 96 hpf, and a more obvious increase of A to B sequences in adults (not shown). PCR amplification with primers specific for exon A or exon B sequences shows the presence of both A and B sequences in 24–96 hpf embryos and adults (Fig. 8C). At tailbud stage (10 hpf), PCR detected exon A, but not B sequences (Fig. 8C), suggesting that exon A may be of more importance at earlier developmental stages, when col5a1 expession is at considerably lower levels than at later stages (Fig. 9).
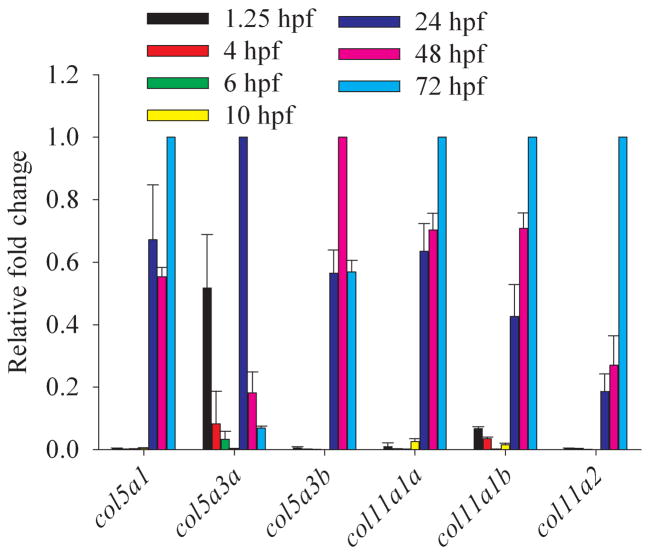
Fig. 9.
Temporal distribution of clade B procollagen gene expression throughout embryogenesis. Real time quantitative RT-PCR analysis of RNA levels of all zebrafish clade B procollagen genes was performed for harvested embryos from 1.25 hpf (8-cell), 4 hpf (sphere), 6 hpf (shield), 10 hpf (bud), 24 hpf (prim-5), 48 hpf (long-pec), and 72 hpf (protruding mouth) stages. Levels of expression of each gene are given, relative to the stage at which they are maximally expressed, and normalized to expression of the β-actin housekeeping gene.
Potential exons separated by small introns were not observed 5′ or 3′ of exons corresponding to exon A in any other zebrafish clade B chain gene, suggesting alternative splicing of C-propeptide sequences to be unique to pro-α1(V). Alternative splicing has not previously been reported for fibrillar procollagen C-propeptides, but such could have profound effects upon biosynthesis. Procollagen chains first associate into trimers via their C-propeptides, which provide the selectivity necessary to ensure that only correct combinations of chains bind each other, and C-propeptides then guide correct alignment and registration of chains, resulting in correct C- to N-terminal triple helix propagation (Lees et al., 1997; Prockop, 1990). Sequences encoded by exon A or B lie between proα1(V) C-propeptide cysteines 5 and 6 (Fig. 6), which are conserved in all fibrillar procollagen chains, and appear to be involved in intrachain disulfide bonding important to correct folding and C-propeptide function (Olsen, 1982). Also between these two cysteines and only 4 amino acids C-terminal to exon A or B sequences are the molecular recognition sequences thought to be involved in fibrillar procollagen chain selection (Lees et al., 1997). Thus, alternative splicing of exons A and B may influence pro-α1(V) function by affecting presentation of the molecular recognition sequence, perhaps via local effects on tertiary structure, thus affecting chain selectivity. In mammals, α1(V) chains can be found in α1(V)2α2(V), α1(V)α2(V)α3(V), and α1(XI)α1(V)α3(XI) heterotrimers, or in α1(V)3 homotrimers (Fichard et al., 1995; Haralson et al., 1980; Wu et al., 2009). It will be of interest to determine whether alternative splicing of exons A and B influences preferential incorporation of pro-α1(V) chains into one type of trimeric association or another, across species.
2.6 Spatiotemporal expression patterns of zebrafish clade B procollagen genes
Temporal expression of zebrafish clade B chain genes was examined via quantitative real time RT-PCR, using RNA from embryos of developmental stages ranging from 1.25 to 72 hpf (Fig. 9). In early embryos, col5a3 is unique in having levels of expression that approach those seen later in development, while readily detectable transcripts are also detected for Col11a1b. Thus, the clade B chains encoded by these genes may play early roles in embryogenesis. In fact, RNA transcripts from these two genes found at 1.25 hpf (8-cell stage) are likely to be maternally transmitted, as most zygotic transcription does not commence until the midblastula transition (MBT) at ~2.75 hpf (Kane and Kimmel, 1993; Mathavan et al., 2005). During the segmentation period (10–24 hpf), during which the embryo elongates and somites, muscle cells, notochord, central nervous system primordium, and rudiments of primary organs develop, expression of all six clade B chain genes increases to maximal or near maximal levels, with robust detection of all genes by 24 hpf (Fig. 9). Relatively high levels continue from 24 through 72 hpf (Fig. 9), during which time the fins and circulatory system form. All clade B chain genes are thus at relatively high levels during formation of definitive cartilage (stainable with Alcian blue), as earliest formation of such cartilage begins in cranial cartilage during the late pharyngula period (~45 hpf), with most cartilage differentiation (fins, jaws, and chondrocranium) occurring during the 48 to 72 hpf “hatching” period (Kimmel et al., 1995; Schilling and Kimmel, 1997). Interestingly, all type XI clade B chains are at maximal levels towards the end of this time interval, during the period at which most cartilage is formed. In contrast, col5a3 expression actually declines from 24 to 72 hpf, consistent with the probability that this gene does not play a major role in cartilage formation.
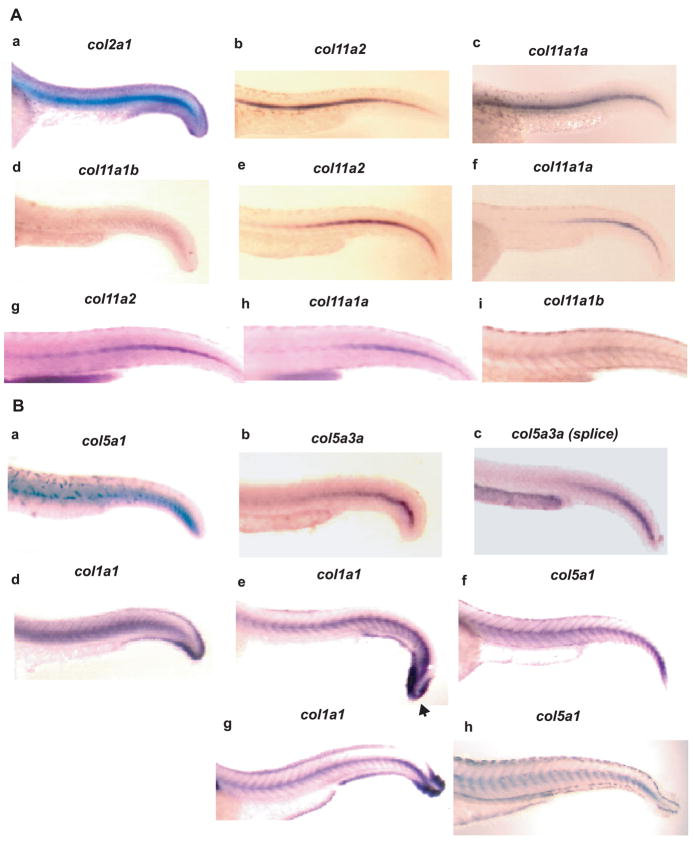
Spatial expression of col11a2, col11a1a, and col11a1b was analyzed by whole mount in situ hybridization of 30, 48, and 72 hpf embryos, using probes from low homology sequences from within each gene (see Experimental Procedures), to avoid cross-hybridization. Col11a2 and col11a1a were both readily detected at 30 hpf, with strong expression evident throughout the notochord, similar to that seen for the type II collagen gene col2a1, whereas signal for Col11a1b was less readily detected, more diffuse, and confined to the caudal notochord, thus showing divergence of spatial expression in notochord for this chain, compared to the other two collagen XI chains (Fig. 10A). At 48 hpf, expression in the notochord persisted for both col11a2 and col11a1a. However, although col11a2 expression persisted throughout the entire length of the notochord, Col11a1a expression seemed sequestered to the caudal notochord at this stage. Col11a1b expression was not readily detected in the notochord at 48 hpf (not shown). At 72 hpf, expression of all three genes was detected in notochord, with col11a1b and coll11a2 expression persisting throughout the notochord, and col11a1a expression remaining confined to the caudal notochord. Expression of col11a1a, col11a1b, and col11a2 in notochord is consistent with their identification as collagen type XI genes, as collagen type II is similarly expressed in zebrafish notochord, as shown in Fig. 10A and as reported (Yan et al., 1995), and as collagens II and XI are thought to interact and form heterotypic fibrils (Mendler et al., 1989). Also consistent with identification of col11a1a, col11a1b, and col11a2 as collagen XI genes was their expression in cartilaginous structures of the head and fins at 48 and 72 hpf, and in the otic vesicle, in which type II collagen is similarly expressed (Supplemental Fig. 1) (Yan et al., 1995).
Fig. 10.
Spatial distribution of clade B procollagen gene expression in caudal regions of 30, 48 and 72 hpf embryos. (A) Whole mount in situ hybridization shows expression of col11a1b, col11a1a and col11a2, and col2a1 as a control, in the notochord of 30 (a-d), 48 (e and f), and 72 (g-i) hpf embryos. (B) Whole mount in situ hybridization shows expression of col5a1, col5a3a and col1a1 as a control, in the notochord of 30 (a-d), 48 (e and f) and 72 (g and h) hpf embryos. Note similar localization of col5a1 and col1a1 expression surrounding the notochord and between myotomes at 48 and 72 hpf (panels eh). Col1a1 (arrowhead), but not col5a1, expression, is strong in the caudal fin fold. A probe corresponding to Col5a3a exons 1-5 and 7 sequences detected expression at 30 hpf throughout the length of the notochord (panel b), whereas a probe corresponding to alternatively spliced Col5a3a exon 6 (splice) only detected expression in the caudal notochord (panel c) at this time.
While in higher vertebrates collagen XI is, in large part, co-expressed with collagen II in cartilage (Mendler et al., 1989; Morris and Bachinger, 1987), collagen V is more broadly expressed, co-localizing in various tissues with collagen I, with which it interacts (Birk et al., 1990; Fessler, 1987). The zebrafish type I collagen gene col1a1 has been shown to be expressed in cells surrounding the notochord and between the myotomes at 72 hpf (Fisher et al., 2003). Thus, it is consistent with designation of col5a1 as a collagen V gene that strong col5a1 expression co-localizes with that of col1a1 in cells surrounding the notochord and between myotomes at 48 and 72 hpf (Fig. 10B), a pattern not seen with zebrafish collagen XI genes (Fig. 10A). Col5a3a expression was not detected at 48 or 72 hpf. At 30 hpf, col5a1 expression seemed limited to the caudal portion of the notochord, whereas col1a1 was expressed throughout the length of the notochord (Fig. 10B, panels a and d). At this time col5a3a expression was strongest in the caudal notochord, but was detectable throughout the length of the notochord (Fig. 10B, panel b), as detected by a probe corresponding to exons 1-5 and 7, sequences found in all col5a3a transcripts. In contrast, col5a3a expression detected by a probe specific for alternatively spliced exon 6, found in only some col5a3a transcripts, was, like col5a1 expression, limited to caudal notochord (Fig. 10B, panel c). Thus, it is possible that forms of pro-α3(V)a containing exon 6 sequences co-distribute with pro-α1(V), whereas forms of pro-α3(V)a lacking exon 6 sequences associate with chains other than proα1(V). Interestingly, although col1a1 expression is high in the caudal fin fold (Fisher et al., 2003) (Fig. 10B, panels e and g), neither col5a1 nor col5a3a expression was observed in this structure (Fig. 10B, panels a–c, f and h), suggesting that a postulated function of type V collagen in providing a nucleation event for collagen I fibril formation (Wenstrup et al., 2004) may not occur in this structure. In the head region at 48 hpf, col5a1 and col1a1 are similarly expressed in mesenchyme investing neural tissue and in the hyoid process (Supplemental Fig. 1A). However, by 72 hpf, col5a1 expression overlapped that of the type XI collagen genes (Supplemental Fig. 1B), perhaps reflecting ability of collagen V and XI chains to form cross-type heterotrimers in cartilaginous and non-cartilaginous structures (Niyibizi and Eyre, 1989; Wu et al., 2009), or perhaps reflecting expression of type V and XI collagen chains in ossified and non-ossified portions of the same skeletal structures. Col5a3b expression was not readily detected at 30, 48, or 72 hpf, despite attempts with four different probes directed against UTR, NH2-terminal globular, and variable domain sequences.
2.7 Summary/perspectives
Here we have shown zebrafish to have 6 clade B collagen chain genes, in contrast to the 4 clade B chain genes possessed by tetrapods. Full-length coding sequences and intron-exon organizations were determined for each zebrafish gene. Moreover, probable syntenic relationships, sequence homologies, features of protein domain and gene structures, and spatiotemporal expression patterns were used to determine phylogenetic relationships between tetrapod and zebrafish clade B collagen chain genes. Data presented here will be of value in determining relationships between structural features and functions of the different clade B procollagen chains across species. In addition, evidence is presented for evolutionarily conserved alternative splicing in pro-α1(V) C-propeptide sequences, with the potential to affect selectivity of collagen type V/XI chain associations. Recently, Baas et al reported phenotypic effects of down-regulating a zebrafish gene identified by them as col11a1 (Baas et al., 2009). However, data presented herein show the down-regulated gene to be col11a2. Thus, the current report can represent a useful guide to researchers interested in studying clade B procollagen biology in zebrafish, as it not only characterizes many features of these procollagen chains and their cognate genes, but also clearly defines phylogenetic relationships between zebrafish and tetrapod clade B procollagen chains and their cognate genes.
3. Experimental procedures
3.1. Determination of Full-length Zebrafish Clade B Procollagen Sequences
See supplementary data for detailed methodology for how full-length cDNA sequences were obtained for zebrafish loci XM_001921860, XM_685787, XM_688785, NM_001083844, NM_001079992, and XM_677653.
3.2 Phylogenetic analysis
Aligned propeptide sequences were subjected to Bayesian phylogenetic analysis using MrBayes 3.1 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) implemented through the CIPRES portal (Miller, 2009). We ran two MCMC runs each comprising four linked chains (heat=0.2) of one million generations, with sampling every thousand generations. We used the WAG model of amino acid evolution and modeled site-to-site rate heterogeneity using a discrete approximation to a gamma distribution with four rate categories. The first 250 trees from the posterior sample of each run were discarded as burn-in, and the remaining trees were used to generate a majority rule consensus tree with posterior probabilities.
3.3 Restriction and RT-PCR examination of alternative splicing in COL5A1 and col5a1
Total RNA was extracted with Trizol (Invitrogen), and SuperScriptIII reverse transcriptase (Invitrogen) was used to synthesize cDNA. For restriction analysis of COL5A1 sequences, PCR was with forward primer 5′-CAAGGATGCTCCAGGGATTCCTTCAAGGTTTAC-3′, corresponding to exon 63 sequences, and reverse primer 5′-CATGCTGAGGTACGAGGTTGCTCT-3′, corresponding to 3′-UTR sequences, to produce a ~600 bp product. This was gel purified and restricted with EcoRI or SexAI. For analyzing alternative splicing in zebrafish, PCR employed a common forward primer from exon 54 (5′-AGGGAGAGAAGGGAGACCGAGGCT-3′) and a reverse primer specific for exon 64A (5′-GTTTACCACGTTTGTATTCACTGA-3′) or 64B (5′-TTGACCCTCTCTTGTAGCGACTAT-3′). In both cases, a ~650-bp product was obtained.
3.4 Examination of spatiotemporal expression patterns
In situ hybridization probes for clade B chain genes were made by PCR, using an adult zebrafish cDNA library as template. For col11a1a, a 299-bp 3′-UTR probe was made using primers 5′-ATACCAATAACTTGGCTGGGTAGGA-3′ (forward) and 5′-GCACTGCACCGTTGAGAGGTCCT-3′ (reverse); for col11a1b, a 610-bp 3′-UTR probe was made using primers 5′-TCATTCCAAAAGCCAAGGAGTGCG-3′ (F) and 5′-GTTTCTCAGTGCATGTTCAGAACA-3′ (R); for col11a2, a 656-bp 3′-UTR probe was made using primers 5′-AGTTGCACACGAACTACACACCCA-3′ (forward) and 5′-GTACAGCGAGTGTTGGTTGTTTTC-3′ (reverse); for col5a1, an 850-bp probe corresponding to sequences from exons 7, 8, and part of 9 (variable subdomain sequences), using vector primer 1644 and reverse primer 5′-TTTCTCTCCTCTCAGACCATCCA-3′; for col5A3a, a 447-bp probe corresponding to exon 6 (variable subdomain) sequences, was made using primers 5′-GCCAGTCTCATCCATAACCTTGAT-3′ (forward) and 5′-CTTTGGGGACTCAGAAAATGCAG-3′ (reverse), and a 891-bp probe corresponding to exons 1-5 and 7 (but lacking exon 6 sequences) was made using primers 5′-GGCTTGTGGTGTTTGTGAGTGGTA-3′ (forward) and 5′-GCACATTGTGCTGATCGCCGTAAA-3′ (reverse). For each anti-sense probe, a corresponding sense probe served as a control for specificity. Col1a1 and col2a1 probes were as previously described (Fisher et al., 2003; Yan et al., 1995). In situ hybridization was performed as previously described (Pelegri and Maischein, 1998).
For quantitative real-time RT-PCR, embryos were collected from various stages (1.25 to 72 hpf) followed by total RNA extraction and reverse transcription, as described above. Real-time PCR was performed using Power SYBR Green Master Mix (Applied Biosystems) on an ABI 7300 instrument, under default cycling conditions (95°C 15 s followed by 60°C 1 min for 45°C cycles). Relative expression levels were determined by real-time RT-PCR from a standard curve of serial dilutions of cDNA samples and were normalized to β-actin expression. Primer sets used for real-time PCR are described in Supplemental materials.
Supplementary Material
Acknowledgments
We thank Dr. David Baum (University of Wisconsin, Madison, USA) for help with the phylogenetic analyses. This work was supported by grants AR047746 and AR53815-12S1 from the National Institutes of Health (to D.S.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abedin MZ, Ayad S, Weiss JB. Isolation and native characterization of cysteine-rich collagens from bovine placental tissues and uterus and their relationship to types IV and V collagens. Biosci Rep. 1982;2:493–502. doi: 10.1007/BF01115247. [DOI] [PubMed] [Google Scholar]
- Baas D, Malbouyres M, Haftek-Terreau Z, Le Guellec D, Ruggiero F. Craniofacial cartilage morphogenesis requires zebrafish col11a1 activity. Matrix Biol. 2009 doi: 10.1016/j.matbio.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Bernard M, Yoshioka H, Rodriguez E, Van der Rest M, Kimura T, Ninomiya Y, Olsen BR, Ramirez F. Cloning and sequencing of pro-alpha 1 (XI) collagen cDNA demonstrates that type XI belongs to the fibrillar class of collagens and reveals that the expression of the gene is not restricted to cartilagenous tissue. J Biol Chem. 1988;263:17159–66. [PubMed] [Google Scholar]
- Birk DE, Fitch JM, Babiarz JP, Doane KJ, Linsenmayer TF. Collagen fibrillogenesis in vitro: interaction of types I and V collagen regulates fibril diameter. J Cell Sci. 1990;95(Pt 4):649–57. doi: 10.1242/jcs.95.4.649. [DOI] [PubMed] [Google Scholar]
- Boot-Handford RP, Tuckwell DS. Fibrillar collagen: the key to vertebrate evolution? A tale of molecular incest. Bioessays. 2003;25:142–51. doi: 10.1002/bies.10230. [DOI] [PubMed] [Google Scholar]
- Boot-Handford RP, Tuckwell DS, Plumb DA, Rock CF, Poulsom R. A novel and highly conserved collagen (pro(alpha)1(XXVII)) with a unique expression pattern and unusual molecular characteristics establishes a new clade within the vertebrate fibrillar collagen family. J Biol Chem. 2003;278:31067–77. doi: 10.1074/jbc.M212889200. [DOI] [PubMed] [Google Scholar]
- Bork P. The modular architecture of vertebrate collagens. FEBS Lett. 1992;307:49–54. doi: 10.1016/0014-5793(92)80900-2. [DOI] [PubMed] [Google Scholar]
- Broek DL, Madri J, Eikenberry EF, Brodsky B. Characterization of the tissue form of type V collagen from chick bone. J Biol Chem. 1985;260:555–62. [PubMed] [Google Scholar]
- Brown RA, Shuttleworth CA, Weiss JB. Three new alpha-chains of collagen from a non-basement membrane source. Biochem Biophys Res Commun. 1978;80:866–72. doi: 10.1016/0006-291x(78)91324-4. [DOI] [PubMed] [Google Scholar]
- Chernousov MA, Rothblum K, Tyler WA, Stahl RC, Carey DJ. Schwann cells synthesize type V collagen that contains a novel alpha 4 chain. Molecular cloning, biochemical characterization, and high affinity heparin binding of alpha 4(V) collagen. J Biol Chem. 2000;275:28208–15. doi: 10.1074/jbc.M003922200. [DOI] [PubMed] [Google Scholar]
- De Paepe A, Nuytinck L, Hausser I, Anton-Lamprecht I, Naeyaert JM. Mutations in the COL5A1 gene are causal in the Ehlers-Danlos syndromes I and II. Am J Hum Genet. 1997;60:547–54. [PMC free article] [PubMed] [Google Scholar]
- Delacoux F, Fichard A, Geourjon C, Garrone R, Ruggiero F. Molecular features of the collagen V heparin binding site. J Biol Chem. 1998;273:15069–76. [PubMed] [Google Scholar]
- Dion AS, Myers JC. COOH-terminal propeptides of the major human procollagens. Structural, functional and genetic comparisons. J Mol Biol. 1987;193:127–43. doi: 10.1016/0022-2836(87)90632-2. [DOI] [PubMed] [Google Scholar]
- Fessler JHaFLI. Type V collagen. In: Mayne R, Burgeson RE, editors. Structure and Function of Collagen Types. Academic Press; Orlando, FL: 1987. pp. 81–103. [Google Scholar]
- Fessler LI, Brosh S, Chapin S, Fessler JH. Tyrosine sulfation in precursors of collagen V. J Biol Chem. 1986;261:5034–40. [PubMed] [Google Scholar]
- Fichard A, Kleman JP, Ruggiero F. Another look at collagen V and XI molecules. Matrix Biol. 1995;14:515–31. doi: 10.1016/s0945-053x(05)80001-0. [DOI] [PubMed] [Google Scholar]
- Fichard A, Tillet E, Delacoux F, Garrone R, Ruggiero F. Human recombinant alpha1(V) collagen chain. Homotrimeric assembly and subsequent processing. J Biol Chem. 1997;272:30083–7. doi: 10.1074/jbc.272.48.30083. [DOI] [PubMed] [Google Scholar]
- Fisher S, Jagadeeswaran P, Halpern ME. Radiographic analysis of zebrafish skeletal defects. Dev Biol. 2003;264:64–76. doi: 10.1016/s0012-1606(03)00399-3. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan B, Wang WM, Greenspan DS. Biosynthetic processing of the Pro-alpha1(V)Pro-alpha2(V)Pro-alpha3(V) procollagen heterotrimer. J Biol Chem. 2004;279:30904–12. doi: 10.1074/jbc.M402252200. [DOI] [PubMed] [Google Scholar]
- Gordon MK, Marchant JK, Foley JW, Igoe F, Gibney EP, Nah HD, Barembaum M, Myers JC, Rodriguez E, Dublet B, van der Rest M, Linsenmayer TF, Upholt WB, Birk DE. Complete primary structure of the chicken alpha1(V) collagen chain. Matrix Biol. 1999;18:481–6. doi: 10.1016/s0945-053x(99)00037-2. [DOI] [PubMed] [Google Scholar]
- Greenspan DS, Cheng W, Hoffman GG. The pro-alpha 1(V) collagen chain. Complete primary structure, distribution of expression, and comparison with the pro-alpha 1(XI) collagen chain. J Biol Chem. 1991;266:24727–33. [PubMed] [Google Scholar]
- Haralson MA, Mitchell WM, Rhodes RK, Kresina TF, Gay R, Miller EJ. Chinese hamster lung cells synthesize and confine to the cellular domain a collagen composed solely of B chains. Proc Natl Acad Sci U S A. 1980;77:5206–10. doi: 10.1073/pnas.77.9.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Scott IC, Greenspan DS. The pro-alpha3(V) collagen chain. Complete primary structure, expression domains in adult and developing tissues, and comparison to the structures and expression domains of the other types V and XI procollagen chains. J Biol Chem. 2000;275:8749–59. doi: 10.1074/jbc.275.12.8749. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Steiglitz BM, Greenspan DS. Bone morphogenetic protein-1 processes the NH2-terminal propeptide, and a furin-like proprotein convertase processes the COOH-terminal propeptide of pro-alpha1(V) collagen. J Biol Chem. 1998;273:27511–7. doi: 10.1074/jbc.273.42.27511. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kessler E, Fichard A, Chanut-Delalande H, Brusel M, Ruggiero F. Bone morphogenetic protein-1 (BMP-1) mediates C-terminal processing of procollagen V homotrimer. J Biol Chem. 2001;276:27051–7. doi: 10.1074/jbc.M102921200. [DOI] [PubMed] [Google Scholar]
- Kessler E, Takahara K, Biniaminov L, Brusel M, Greenspan DS. Bone morphogenetic protein-1: the type I procollagen C-proteinase. Science. 1996;271:360–2. doi: 10.1126/science.271.5247.360. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–94. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Kimura T, Cheah KS, Chan SD, Lui VC, Mattei MG, van der Rest M, Ono K, Solomon E, Ninomiya Y, Olsen BR. The human alpha 2(XI) collagen (COL11A2) chain. Molecular cloning of cDNA and genomic DNA reveals characteristics of a fibrillar collagen with differences in genomic organization. J Biol Chem. 1989;264:13910–6. [PubMed] [Google Scholar]
- Kleman JP, Hartmann DJ, Ramirez F, van der Rest M. The human rhabdomyosarcoma cell line A204 lays down a highly insoluble matrix composed mainly of alpha 1 type-XI and alpha 2 type-V collagen chains. Eur J Biochem. 1992;210:329–35. doi: 10.1111/j.1432-1033.1992.tb17425.x. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA, Fessler JH. Propeptides of procollagen V (A,B) in chick embryo crop. J Biol Chem. 1981;256:7053–8. [PubMed] [Google Scholar]
- LeBaron RG, Hook A, Esko JD, Gay S, Hook M. Binding of heparan sulfate to type V collagen. A mechanism of cell-substrate adhesion. J Biol Chem. 1989;264:7950–6. [PubMed] [Google Scholar]
- Lees JF, Bulleid NJ. The role of cysteine residues in the folding and association of the COOH-terminal propeptide of types I and III procollagen. J Biol Chem. 1994;269:24354–60. [PubMed] [Google Scholar]
- Lees JF, Tasab M, Bulleid NJ. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. Embo J. 1997;16:908–16. doi: 10.1093/emboj/16.5.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lacerda DA, Warman ML, Beier DR, Yoshioka H, Ninomiya Y, Oxford JT, Morris NP, Andrikopoulos K, Ramirez F, et al. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell. 1995;80:423–30. doi: 10.1016/0092-8674(95)90492-1. [DOI] [PubMed] [Google Scholar]
- Linsenmayer TF, Gibney E, Igoe F, Gordon MK, Fitch JM, Fessler LI, Birk DE. Type V collagen: molecular structure and fibrillar organization of the chicken alpha 1(V) NH2-terminal domain, a putative regulator of corneal fibrillogenesis. J Cell Biol. 1993;121:1181–9. doi: 10.1083/jcb.121.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui VC, Ng LJ, Sat EW, Cheah KS. The human alpha 2(XI) collagen gene (COL11A2): completion of coding information, identification of the promoter sequence, and precise localization within the major histocompatibility complex reveal overlap with the KE5 gene. Genomics. 1996;32:401–12. doi: 10.1006/geno.1996.0135. [DOI] [PubMed] [Google Scholar]
- Malfait F, De Paepe A. Molecular genetics in classic Ehlers-Danlos syndrome. Am J Med Genet C Semin Med Genet. 2005;139C:17–23. doi: 10.1002/ajmg.c.30070. [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, Ruan Y, Korzh V, Gong Z, Liu ET, Lufkin T. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–76. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne R, Brewton RG, Mayne PM, Baker JR. Isolation and characterization of the chains of type V/type XI collagen present in bovine vitreous. J Biol Chem. 1993;268:9381–6. [PubMed] [Google Scholar]
- Medzihradszky KF, Darula Z, Perlson E, Fainzilber M, Chalkley RJ, Ball H, Greenbaum D, Bogyo M, Tyson DR, Bradshaw RA, Burlingame AL. O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes. Mol Cell Proteomics. 2004;3:429–40. doi: 10.1074/mcp.M300140-MCP200. [DOI] [PubMed] [Google Scholar]
- Mendler M, Eich-Bender SG, Vaughan L, Winterhalter KH, Bruckner P. Cartilage contains mixed fibrils of collagen types II, IX, and XI. J Cell Biol. 1989;108:191–7. doi: 10.1083/jcb.108.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T. The CIPRES Portals. [Accessed: 2009-08-04];CIPRES. 2009 2009-08-04. URL: http://www.phylo.org/subsections/portal. Archived by WebCite(r) at http://www.webcitation.org/5imQlJeQa)
- Mizuno K, Hayashi T. Separation of the subtypes of type V collagen molecules, [alpha 1(V)]2 alpha 2(V) and alpha 1(V) alpha 2(V) alpha 3(V), by chain composition-dependent affinity for heparin: single alpha 1(V) chain shows intermediate heparin affinity between those of the type V collagen subtypes composed of [alpha 1(V)]2 alpha 2(V) and of alpha 1(V) alpha 2(V) alpha 3(V) J Biochem. 1996;120:934–9. doi: 10.1093/oxfordjournals.jbchem.a021509. [DOI] [PubMed] [Google Scholar]
- Moradi-Ameli M, Rousseau JC, Kleman JP, Champliaud MF, Boutillon MM, Bernillon J, Wallach J, Van der Rest M. Diversity in the processing events at the N-terminus of type-V collagen. Eur J Biochem. 1994;221:987–95. doi: 10.1111/j.1432-1033.1994.tb18815.x. [DOI] [PubMed] [Google Scholar]
- Morris NP, Bachinger HP. Type XI collagen is a heterotrimer with the composition (1 alpha, 2 alpha, 3 alpha) retaining non-triple-helical domains. J Biol Chem. 1987;262:11345–50. [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Nah HD, Barembaum M, Upholt WB. The chicken alpha 1 (XI) collagen gene is widely expressed in embryonic tissues. J Biol Chem. 1992;267:22581–6. [PubMed] [Google Scholar]
- Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327(Pt 3):625–35. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neame PJ, Young CN, Treep JT. Isolation and primary structure of PARP, a 24-kDa proline- and arginine-rich protein from bovine cartilage closely related to the NH2-terminal domain in collagen alpha 1 (XI) J Biol Chem. 1990;265:20401–8. [PubMed] [Google Scholar]
- Nicholls AC, Oliver JE, McCarron S, Harrison JB, Greenspan DS, Pope FM. An exon skipping mutation of a type V collagen gene (COL5A1) in Ehlers-Danlos syndrome. J Med Genet. 1996;33:940–6. doi: 10.1136/jmg.33.11.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyibizi C, Eyre DR. Identification of the cartilage alpha 1(XI) chain in type V collagen from bovine bone. FEBS Lett. 1989;242:314–8. doi: 10.1016/0014-5793(89)80492-2. [DOI] [PubMed] [Google Scholar]
- Niyibizi C, Eyre DR. Structural analysis of the extension peptides on matrix forms of type V collagen in fetal calf bone and skin. Biochim Biophys Acta. 1993;1203:304–9. doi: 10.1016/0167-4838(93)90099-d. [DOI] [PubMed] [Google Scholar]
- Niyibizi C, Eyre DR. Structural characteristics of cross-linking sites in type V collagen of bone. Chain specificities and heterotypic links to type I collagen. Eur J Biochem. 1994;224:943–50. doi: 10.1111/j.1432-1033.1994.00943.x. [DOI] [PubMed] [Google Scholar]
- Olsen BR. In: New Trends in Basement Membrane Research. Kuhn K, Schoene H, Timpl R, editors. Raven Press; New York: 1982. pp. 225–236. [Google Scholar]
- Oxford JT, Doege KJ, Morris NP. Alternative exon splicing within the amino-terminal nontriple-helical domain of the rat pro-alpha 1(XI) collagen chain generates multiple forms of the mRNA transcript which exhibit tissue-dependent variation. J Biol Chem. 1995;270:9478–85. doi: 10.1074/jbc.270.16.9478. [DOI] [PubMed] [Google Scholar]
- Pappano WN, Steiglitz BM, Scott IC, Keene DR, Greenspan DS. Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases. Mol Cell Biol. 2003;23:4428–38. doi: 10.1128/MCB.23.13.4428-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegri F, Maischein HM. Function of zebrafish beta-catenin and TCF-3 in dorsoventral patterning. Mech Dev. 1998;77:63–74. doi: 10.1016/s0925-4773(98)00132-4. [DOI] [PubMed] [Google Scholar]
- Plumb DA, Dhir V, Mironov A, Ferrara L, Poulsom R, Kadler KE, Thornton DJ, Briggs MD, Boot-Handford RP. Collagen XXVII is developmentally regulated and forms thin fibrillar structures distinct from those of classical vertebrate fibrillar collagens. J Biol Chem. 2007;282:12791–5. doi: 10.1074/jbc.C700021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prockop DJ. Mutations that alter the primary structure of type I collagen. The perils of a system for generating large structures by the principle of nucleated growth. J Biol Chem. 1990;265:15349–52. [PubMed] [Google Scholar]
- Richards AJ, Martin S, Nicholls AC, Harrison JB, Pope FM, Burrows NP. A single base mutation in COL5A2 causes Ehlers-Danlos syndrome type II. J Med Genet. 1998;35:846–8. doi: 10.1136/jmg.35.10.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rousseau JC, Farjanel J, Boutillon MM, Hartmann DJ, van der Rest M, Moradi-Ameli M. Processing of type XI collagen. Determination of the matrix forms of the alpha1(XI) chain. J Biol Chem. 1996;271:23743–8. doi: 10.1074/jbc.271.39.23743. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Greene JM, Green MR. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985;41:833–44. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Sage H, Bornstein P. Characterization of a novel collagen chain in human placenta and its relation to AB collagen. Biochemistry. 1979;18:3815–22. doi: 10.1021/bi00584a027. [DOI] [PubMed] [Google Scholar]
- Sandell LJ, Boyd CD. Conserved and divergent sequence and functional elements witin collagen genes. In: Sandell LJ, Boyd CD, editors. Extracellular Matrix Genes. Academic Press; San Diego, CA: 1990. pp. 1–56. [Google Scholar]
- Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–60. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Scott IC, Blitz IL, Pappano WN, Imamura Y, Clark TG, Steiglitz BM, Thomas CL, Maas SA, Takahara K, Cho KW, Greenspan DS. Mammalian BMP-1/Tolloid-related metalloproteinases, including novel family member mammalian Tolloid-like 2, have differential enzymatic activities and distributions of expression relevant to patterning and skeletogenesis. Dev Biol. 1999;213:283–300. doi: 10.1006/dbio.1999.9383. [DOI] [PubMed] [Google Scholar]
- Soderhall C, Marenholz I, Kerscher T, Ruschendorf F, Esparza-Gordillo J, Worm M, Gruber C, Mayr G, Albrecht M, Rohde K, Schulz H, Wahn U, Hubner N, Lee YA. Variants in a novel epidermal collagen gene (COL29A1) are associated with atopic dermatitis. PLoS Biol. 2007;5:e242. doi: 10.1371/journal.pbio.0050242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel L, Schier AF, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136:1401–20. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–6. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- Takahara K, Hoffman GG, Greenspan DS. Complete structural organization of the human alpha 1 (V) collagen gene (COL5A1): divergence from the conserved organization of other characterized fibrillar collagen genes. Genomics. 1995;29:588–97. doi: 10.1006/geno.1995.9961. [DOI] [PubMed] [Google Scholar]
- Takahara K, Sato Y, Okazawa K, Okamoto N, Noda A, Yaoi Y, Kato I. Complete primary structure of human collagen alpha 1 (V) chain. J Biol Chem. 1991;266:13124–9. [PubMed] [Google Scholar]
- Thom JR, Morris NP. Biosynthesis and proteolytic processing of type XI collagen in embryonic chick sterna. J Biol Chem. 1991;266:7262–9. [PubMed] [Google Scholar]
- Toriello HV, Glover TW, Takahara K, Byers PH, Miller DE, Higgins JV, Greenspan DS. A translocation interrupts the COL5A1 gene in a patient with Ehlers-Danlos syndrome and hypomelanosis of Ito. Nat Genet. 1996;13:361–5. doi: 10.1038/ng0796-361. [DOI] [PubMed] [Google Scholar]
- Tsumaki N, Kimura T. Differential expression of an acidic domain in the amino-terminal propeptide of mouse pro-alpha 2(XI) collagen by complex alternative splicing. J Biol Chem. 1995;270:2372–8. doi: 10.1074/jbc.270.5.2372. [DOI] [PubMed] [Google Scholar]
- Unsold C, Pappano WN, Imamura Y, Steiglitz BM, Greenspan DS. Biosynthetic processing of the pro-alpha 1(V)2pro-alpha 2(V) collagen heterotrimer by bone morphogenetic protein-1 and furin-like proprotein convertases. J Biol Chem. 2002;277:5596–602. doi: 10.1074/jbc.M110003200. [DOI] [PubMed] [Google Scholar]
- Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R. Collagen XXVIII, a novel von Willebrand factor A domain-containing protein with many imperfections in the collagenous domain. J Biol Chem. 2006;281:3494–504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- Vuristo MM, Pihlajamaa T, Vandenberg P, Prockop DJ, Ala-Kokko L. The human COL11A2 gene structure indicates that the gene has not evolved with the genes for the major fibrillar collagens. J Biol Chem. 1995;270:22873–81. doi: 10.1074/jbc.270.39.22873. [DOI] [PubMed] [Google Scholar]
- Wenstrup RJ, Florer JB, Brunskill EW, Bell SM, Chervoneva I, Birk DE. Type V collagen controls the initiation of collagen fibril assembly. J Biol Chem. 2004;279:53331–7. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- Wenstrup RJ, Langland GT, Willing MC, D’Souza VN, Cole WG. A splice-junction mutation in the region of COL5A1 that codes for the carboxyl propeptide of pro alpha 1(V) chains results in the gravis form of the Ehlers-Danlos syndrome (type I) Hum Mol Genet. 1996;5:1733–6. doi: 10.1093/hmg/5.11.1733. [DOI] [PubMed] [Google Scholar]
- Wieringa B, Hofer E, Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984;37:915–25. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Eyre DR. Structural analysis of cross-linking domains in cartilage type XI collagen. Insights on polymeric assembly. J Biol Chem. 1995;270:18865–70. doi: 10.1074/jbc.270.32.18865. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Weis MA, Kim LS, Carter BG, Eyre DR. Differences in chain usage and cross-linking specificities of cartilage type V/XI collagen isoforms with age and tissue. J Biol Chem. 2009;284:5539–45. doi: 10.1074/jbc.M806369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YL, Sumiyoshi H, Khaleduzzaman M, Ninomiya Y, Yoshioka H. cDNA sequence and expression of the mouse alpha1(V) collagen gene (Col5a1) Biochim Biophys Acta. 1998;1397:275–84. doi: 10.1016/s0167-4781(98)00016-5. [DOI] [PubMed] [Google Scholar]
- Yan YL, Hatta K, Riggleman B, Postlethwait JH. Expression of a type II collagen gene in the zebrafish embryonic axis. Dev Dyn. 1995;203:363–76. doi: 10.1002/aja.1002030308. [DOI] [PubMed] [Google Scholar]
- Yaoi Y, Hashimoto K, Koitabashi H, Takahara K, Ito M, Kato I. Primary structure of the heparin-binding site of type V collagen. Biochim Biophys Acta. 1990;1035:139–45. doi: 10.1016/0304-4165(90)90108-9. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Inoguchi K, Khaleduzzaman M, Ninomiya Y, Andrikopoulos K, Ramirez F. Coding sequence and alternative splicing of the mouse alpha 1(XI) collagen gene (Col11a1) Genomics. 1995;28:337–40. doi: 10.1006/geno.1995.1151. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Ramirez F. Pro-alpha 1(XI) collagen. Structure of the amino-terminal propeptide and expression of the gene in tumor cell lines. J Biol Chem. 1990;265:6423–6. [PubMed] [Google Scholar]
- Zhidkova NI, Brewton RG, Mayne R. Molecular cloning of PARP (proline/arginine-rich protein) from human cartilage and subsequent demonstration that PARP is a fragment of the NH2-terminal domain of the collagen alpha 2(XI) chain. FEBS Lett. 1993;326:25–8. doi: 10.1016/0014-5793(93)81753-m. [DOI] [PubMed] [Google Scholar]
- Zhidkova NI, Justice SK, Mayne R. Alternative mRNA processing occurs in the variable region of the pro-alpha 1(XI) and pro-alpha 2(XI) collagen chains. J Biol Chem. 1995;270:9486–93. doi: 10.1074/jbc.270.16.9486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.