Abstract
Pim-2 kinase is one of three highly conserved Pim family members which are known to be involved in cell survival and cell proliferation. Here we demonstrate that like Pim-1, Pim-2 also phosphorylates the cell cycle inhibitor p21Cip1/WAF1 (p21) on Thr145 in vitro and in vivo. Overexpression of Pim-2 in HCT116 cells leads to the increased stability of p21, and results in enhanced levels of both exogenous and endogenous p21 proteins. Knock-down of Pim-2 expression via siRNA results in reduced level of endogenous p21, indicating that like Pim-1, Pim-2 is another legitimate p21 kinase. However, Pim-2 has no influence on the nuclear localization of p21 in HCT116 cells. In addition, Pim-2 is able to arrest the cell cycle at G1/S phase and inhibit cell proliferation through phosphorylation of p21 in HCT116 cells. These data suggest that Pim-2 phosphorylation of p21 enhances p21’s stability and inhibits cell proliferation in HCT116 cells.
Keywords: Pim-2, Phosphorylation, p21Cip1/WAF1, protein stability, cell proliferation 3
Introduction
The proto-oncogene pim-2 was originally discovered as a result of proviral insertion discovered in a murine T cell lymphoma (Breuer et al., 1989). The gene encodes a serine/thronine kinase whose expression is regulated by hematopoietic cytokines (Fox et al., 2003). Pim-2 has a well-documented role in oncogenesis (for review see Allen and Bern (Allen and Berns, 1996)) and the co-expression of pim-2 and c-Myc transgenes induces rapid malignant transformation in vivo (Allen et al., 1997). Pim-2 transgenic mice have a low-rate of spontaneous tumor incidence, but are more susceptible to lymphomagenesis upon viral infection or chemical carcinogen exposure (Allen et al., 1997). In humans, dysregulated Pim-2 overexpression has been reported to occur in prostate cancer (Dhanasekaran et al., 2001;Dai et al., 2005), lymphoma (Yoshida et al., 1999), leukemia (Amson et al., 1989) and multiple myeloma (Claudio et al., 2002). These observations underscore Pim-2’s involvement in human tumor formation. One of the main functions attributed to Pim-2 is as a potent regulator of cell survival in response to growth factor signaling (Fox et al., 2003). Pim-2 is reported to inhibit apoptosis by phosphorylating BAD, thus interfering with BAD-induced cell death (Yan et al., 2003). Pim-2 has been found to promote cell survival by activating the NFκB survival pathway through both augmentation of IκB kinase activity and NFκB nuclear translocation (Hammerman et al., 2004). More recently, it is reported that Pim-2 is required to confer rapamycin resistance in hematopoietic cells (Fox et al., 2005), and Pim-2 is an independent regulator of chondrocyte survival and autophagy in the epiphyseal growth plate (Bohensky et al., 2007). Pim-2 kinase is also required for efficient pre-B-cell transfomation by v-Abl oncogene (Chen et al., 2008).
Pim-2 belongs to a serine/threonine kinase family which has three members: Pim-1, Pim-2 and Pim-3 (Wang et al., 2001). The Pim family kinases are highly homologous to each other and also share a unique hinge region sequence, but have < 30% sequence identity to other kinases (Mikkers et al., 2004). They also differ in their tissue expression (Eichmann et al., 2000). Yet, it is unknown to what extent the various family members differ in their biochemical effects. Functional similarity between Pim-1 and Pim-2 kinases has been inferred from the studies of transgenic mice. Both pim-1 and pim-2 genes induce lymphomas alone or in synergy with c-myc (van Lohuizen et al., 1989;van der Lugt et al., 1995). Furthermore, the relatively weak phenotype associated with disruption of the pim-1 gene (Laird et al., 1993) suggests that its functions may be assumed by other related molecules, such as pim-2, but what is not clear is whether the two kinases have functions that are unique to each kinase.
p21Cip1/WAF1(p21) is a well-known cyclin-dependent kinase inhibitor (CKI) and a prototypical member of the Cip/Kip family of CKIs (Gartel et al., 1996). It negatively modulates cell cycle progression by inhibiting the activity of cyclin/Cdk2 complexes (Harper et al., 1995) and it blocks DNA replication by binding to proliferating cell nuclear antigen (Waga et al., 1994). In addition to its role as an inhibitor of cell cycle progression, p21 may also function as an adaptor protein for cdk4/cyclin D complexes (LaBaer et al., 1997), thereby promoting cell cycle progression. Recent studies have revealed that p21 is involved in a variety of cellular events, such as differentiation (Asada et al., 1999), proliferation (Rossig et al., 2001), senescence (Brown et al., 1997), cell motility and tumor metastasis (Lee and Helfman, 2004), as well as cell survival (Mattiussi et al., 2004). The involvement of p21 in multiple cellular functions underscores its importance and that its precise regulation is crucial to the maintenance of the normal cellular function.
p21 is a key mediator of p53-dependent cell cycle arrest and may play the role of a tumor suppressor in cancer (Dotto, 2000). However, it has been shown that p21 may also act as an oncogene, because it inhibits apoptosis and may promote cell proliferation in some tumors (see reviews (Gartel and Tyner, 2002;Gartel, 2005;Cazzalini et al., 2010)). p21 is overexpressed in a variety of human tumors including prostate, cervical, breast and squamous cell carcinomas and, in many cases, p21 up-regulation correlates positively with tumor grade, invasiveness and aggressiveness and is poor prognostic indicator (see review (Abbas and Dutta, 2009)).
The regulation of p21 expression is mainly controlled at the transcriptional level by various transcription factors (Gartel and Radhakrishnan, 2005). This could occur in both p53 dependent and p53 independent manners (Gartel and Tyner, 1999). However, a series of new studies have shown that p21 can be also regulated by post-translational mechanisms. p21 is an unstable protein with the half-life of < 30 min and is degraded by the proteasome in a ubiquitin-independent manner (Sheaff et al., 2000). Over the past years, a number of phosphorylation sites in p21 have been identified, which are targeted by several important signaling protein kinases including Akt/PKB, GSK3, MAPK p38, PKC, PKA, and Pim-1 (Li et al., 2002;Rossig et al., 2001;Kim et al., 2002;Kashiwagi et al., 2000;Suzuki et al., 2000;Zhang et al., 2007;Rossig et al., 2001). The significance of some of the phosphorylation sites on p21 is that they lead to the alteration of its stability and/or subcellular localization.
In this study, using p21 as a target, we found that p21 can be phosphorylated by Pim-2 kinase both in vitro and in vivo. This phosphorylation event increases p21 stability. Furthermore, the phosphorylation of p21 by Pim-2 does not change the nuclear localization of p21 in HCT116 cells and promotes the inhibition of cell cycle at G1/S phase. Knock-down of Pim-2 levels mediated by pim-2 siRNA results in decreased p21 levels in both HCT116 and DU145 cells. In addition, Pim-2 kinase inhibits cell proliferation in HCT116 cells via phosphorylation of p21. However, when compared side by side in the same cell line, both Pim-2 and Pim-1 have the same function indicating that it is the genetic background of the cells in which the kinases are expressed that dictate how their function will be manifested.
Materials and Methods
Cell culture and transfection
Human colorectal carcinoma HCT116 cells were cultured in McCoy’s 5A medium supplemented with 10% (v/v) fetal bovine serum (Atlanta biologicals), 100units/ml penicillin and 100μg/ml streptomycin. Human prostate carcinoma DU145 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 100units/ml penicillin and 100μg/ml streptomycin. Both cell lines were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2. Transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Silencer validated siRNA to Pim-2, Silencer GAPDH siRNA (positive control) and Silencer negative control siRNA( Ambion) were transfected into HCT116 and DU145 cells using SiPORT lipid siRNA transfection reagent (Ambion) and cells were cultured for 48h before harvesting for analysis. For cycloheximide treatment, cells were treated with a concentration of 25μg/ml for the indicated time.
Antibodies and reagents
Antibodies used included anti-p21, anti-p53 (BD Pharmingen); anti-actin, anti-HA, anti-FLAG (Sigma); anti-phospho-Ser146-p21, anti-phospho-Thr145-p21, anti-Pim-2 (Santa Cruz Biotech). Other reagents used included doxorubicin (Sigma) and cycloheximide (Sigma).
Plasmids and constructs
pBK/CMV-HA-p21 ( wild type(WT), T145D (T/D), T145A (T/A), S146D(S/D), S146A(S/A) T145D S146D(DD), T145A S146A(AA)), as well as pGEX-2T-p21 ( wild type(WT), T145A(T/A), S146A(S/A), T145A S146A(AA)) were used as described earlier (Zhang et al., 2007). To subclone Pim-2 (wild type (WT) and kinase dead (KD)) cDNA into pET30a, primers were designed to include a C-terminal 6X His-tag into mouse wild type Pim-2 and kinase dead Pim-2 (K61A) (cDNA for both were generous gifts from Dr. Michael Lilly, University of California at Irvine). PCR products and pET30a vector were then digested by NdeI/KpnI double enzymes and ligated. To subclone Pim-2 (WT and KD) into pBK/CMV, primers were designed to include a C-terminal FLAG tag into Pim-2. Pim-2 cDNA was then subcloned by double digestion of the PCR products into the vector with HindIII/XbaI and ligated. DNA sequence analysis was done to confirm the correct sequences.
Recombinant protein purification
GST-p21 (WT, T/A, S/A, AA) were used as described early (Zhang et al., 2007). To prepare recombinant Pim-2 kinases (WT and KD), pET30a-Pim-2 plasmid was transformed into E. coli strain BL-21pLysS (DE3), and 1mM IPTG was used to carry out the induction. The protein was affinity–purified with Ni-NTA beads, and eluted with 300 mM imidazole. Protein was subsequently dialyzed against Tris buffer at 4°C overnight.
In vitro kinase assay
For the kinase assay analyzed by 32P incorporation, each GST substrate protein (1μg) was incubated with 0.2 μg of Pim-2 (WT or KD) in 50 μl kinase buffer (20 mM MOPS, pH 7.4, 150 mM NaCl, 12.5 mM MgCl2, 1 mM MnCl2, 1 mM EGTA, 1 mM DTT, 10 μM ATP) containing 20 μCi γ-32P-ATP. The reactions were carried out at room temperature for 20 min. An appropriate amount of the reaction mixture was mixed with 2 X lammeli buffers and boiled for 5 min, then loaded onto SDS-PAGE and transferred to PVDF membrane until exposed to hyperfilm.
Cell lysates preparation and Western blotting
DNA plasmids were transfected using Invitrogen’s lipofectamine 2000 following the manufacturer’s instructions. Cells were trypsinized and harvested, washed with PBS once and resuspended in cell lysis buffer containing 25mM Tris-HCl (pH 7.5), 1% (w/v) NP-40, 1mM EDTA, protein phosphatase inhibitor cocktail and protease inhibitor cocktail set I (Calbiochem), as well as 150mM NaCl. After brief sonication, cell lysates were centrifuged at 13,000 rpm for 5 min. Protein concentration was determined so that equivalent amounts of lysate based on protein concentration was added to an equal volume of 2X Laemmli buffer and boiled for 10 min. For Western blot analysis, protein was separated by SDS-PAGE and transferred to PVDF membrane. Membranes were subsequently blocked with 5% non-fat dry milk-PBS-Tween for 1 hour at room temperature, incubated with primary antibody at optimized dilution overnight at 4°C. Membranes were then washed, incubated with HRP-conjugated secondary antibody (Santa Cruz Biotech) at 1:10,000 for 1 hour, washed, treated with ECR reagent (Pierce) and exposed to hyperfilm.
Cell cycle analysis
HCT116 cells were trypsinized and washed with PBS for once, cells were then resuspended into PBS buffer, −20°C absolute ethanol was then added dropwise while vortexing to make a final concentration of 80%. Cells were fixed overnight at −20°C and washed with ice cold PBS two times. Cells were then treated with RNase in the buffer consisting of 250mM Na2HPO4 and 250mM NaH2PO4, and then stained with propidium iodide until analysis by flow cytometry. The percentage of G1, S and G2/M phases was recorded.
Cell proliferation assay
HCT116 cells were seeded in 24-well plate one day before transfection so that cells were at 30–40% confluence and transfected with indicated plasmids. Forty hours after transfection, cell proliferation was measured with CellTiter-Glo Luminescent Cell Viability assay kit (Promega) according to the manufacturer’s instructions. Relative cell proliferation rate was determined as a percentage of the sample with highest luminescence reading.
Confocol microscopy
Cells were grown on coverslips in 24-well plates and then transfected with the appropriate plasmids. Forty hours post transfection, cells were fixed in 4% paraformaldehyde for 20 minutes, washed in three changes of PBS and then permeabilized in 0.2% triton X-100 for 20 minutes followed by three changes of PBS. Cells were then blocked for 30 min with 3% BSA/PBS at room temperature and then incubated with primary antibody (1:100 anti-HA; 1:100 anti-FLAG) for two hours at room temperature. The coverslips were washed with three changes of PBS and then incubated with the secondary antibody (1:200 anti-mouse conjugated with Oregon green and Texas red, respectively) for 1 hour at room temperature in the dark. The cells were washed with three changes of PBS and then mounted onto coverglass using mounting medium with DAPI (4,6′-diamidino-2-phenylindole) and examined using confocal microscope after overnight staining.
Statistical analysis
Student’s t-test was performed to determine the statistical significance between unpaired values. Results were considered significant if P<0.05.
Results
Pim-2 kinase phosphorylates p21 in vitro and in vivo
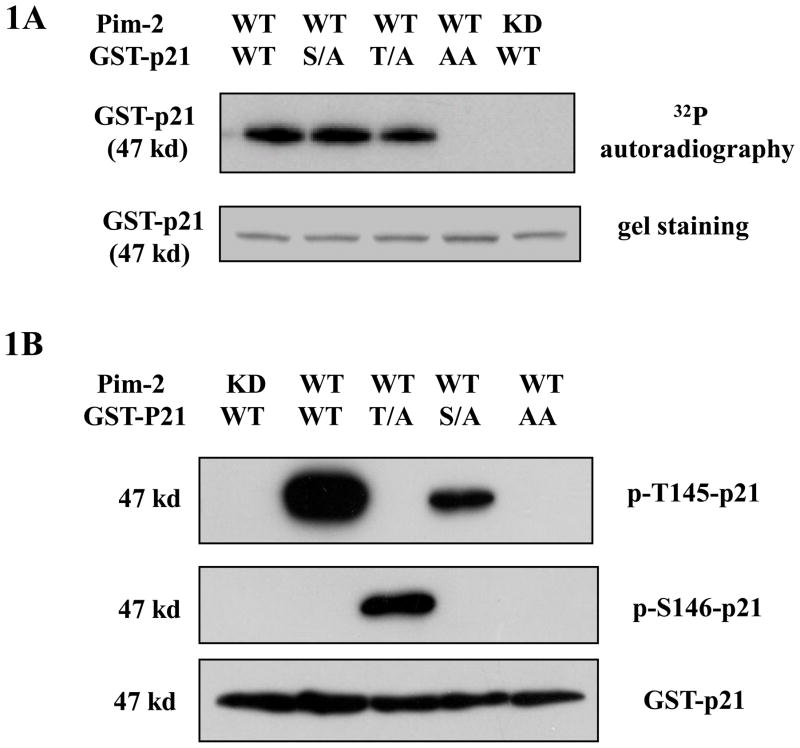
p21 has been reported previously to be phosphorylated on Thr 145 and Ser 146 by the PKB/Akt, PKC and Pim-1 kinases (Rossig et al., 2001;Li et al., 2002;Kashiwagi et al., 2000;Zhang et al., 2007). These two sites also sit in the predicted preferred phosphorylation consensus sequence for Pim-2 kinase (Peng et al., 2007). To determine on which site(s) Pim-2 kinase preferentially phophorylates p21, we first carried out an in vitro kinase assay with the purified recombinant Pim-2 and GST-p21 proteins. As shown in Figure 1A, Pim-2 kinase readily phosphorylates wild type p21. In addition, the incorporation of 32P into the p21 protein with a single mutation at either Thr145Ala (T/A) or Ser146Ala (S/A) indicates that both residues can be phosphorylated. The data also show that when either Thr145 or Ser146 residue cannot be phosphorylated, the other site becomes available for phosphorylation. Furthermore, when both residues are mutated to alanine, the phosphorylation is abolished indicating these two residues in p21 are the only sites phosphorylated by Pim-2. The question remains whether both sites are phosphorylated in vitro or is there one single site preferentially phosphorylated over the other as is observed for Pim-1 (Zhang et al., 2007). To answer this question, we utilized two phospho-specific antibodies for Ser146 and Thr145. As shown in Figure 1B, the Western blot result shows that Thr145 is phosphorylated in wild type p21, but not Ser146, indicating that only Thr145 is the preferred site for phosphorylation. This result is similar for what was found for Pim-1 indicating that with respect to p21, the substrate preferences for the two Pim kinases are very similar. Taken together, both Pim-2 and Pim-1 kinases are able to phosphorylate p21 on Thr145 in vitro. When Thr 145 site is not available for phosphorylation, both Pim-1 and Pim-2 can phosphorylate Ser146 site even though phosphorylation of Ser146 by Pim-1 is markedly less efficient compared to Pim-2.
Figure 1. Phosphorylation of p21 by Pim-2 kinase in vitro.
A) Pim-2 phosphorylation of p21 detected by an in vitro kinase assay. Recombinant GST-p21 proteins including wild type (WT), Thr145Ala (T/A), Ser146Ala (S/A) and Thr145Ala/Ser146Ala (AA) were used as substrates for Pim-2 kinase. The in vitro kinase assay was carried out as described in the Material and Methods. The upper panel shows the autoradiography for kinase reaction and the lower panel is the coomassie blue stained membrane for the same reaction. B) Western blot analysis shows the phosphorylation of p21 by Pim-2. The kinase assay was run on various GST-p21 proteins. However, there was no 32P-γ-ATP in the reaction. Reaction samples were separated by SDS-PAGE, transferred onto PVDF membrane and followed by Western blots with two phospho-specific antibodies (anti-phospho-Thr145 and anti-phospho Ser146 antibodies) and anti-p21 antibody.
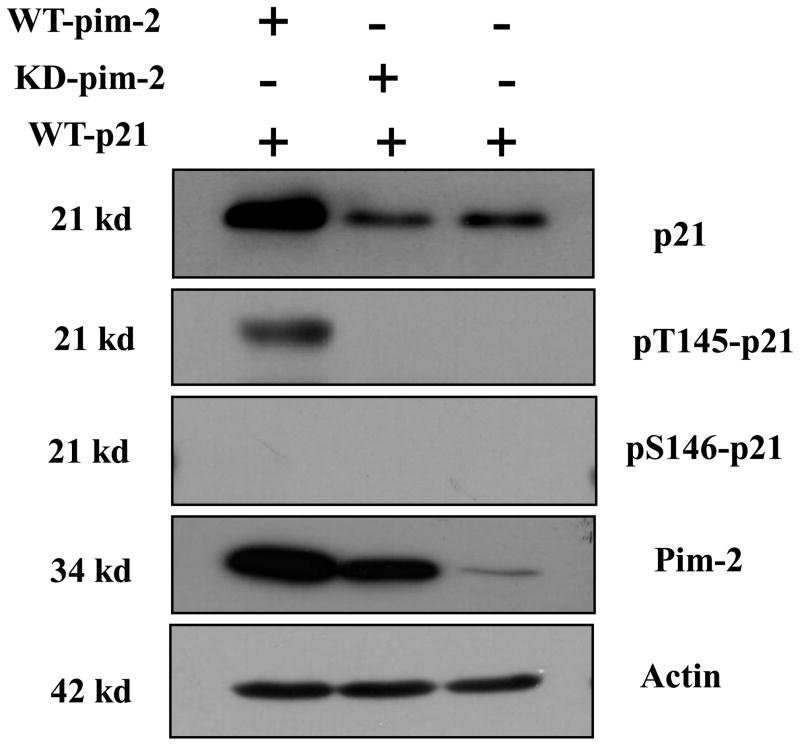
To study the phosphorylation site of p21 by Pim-2 in vivo, we performed a co-transfection of pBK/CMV-Pim-2-FLAG along with pBK/CMV-HA-p21 (wild type) into the colon cancer cell line HCT116. Forty hours post-transfection, cell lysates were prepared for Western blot analysis. As revealed by the data shown in Figure 2, only Thr145 is phosphorylated by Pim-2, while Ser146 is not. This is consistent with the in vitro results. By reprobing the blots with anti-p21 antibody to check the levels of p21 in each of the samples, we found that cells that are transfected with wild type Pim-2 appear to have an increase in the steady state level of p21, while the kinase dead Pim-2 seems to inhibit the accumulation of p21 as compared to the empty vector control in vivo which suggests that phosphorylation leads to stabilization of p21.
Figure 2. Pim-2 kinase phosphorylates p21 in vivo.
HCT116 cells were co-transfected with Pim-2 (either wild type (WT) or kinase dead (KD)) and p21. Cell lysates were made 40 h after transfection and analyzed by Western blotting with anti-p21, anti-phospho-Thr145, anti-phospho-Ser146, anti-Pim-2 and anti-actin antibodies.
Pim-2 kinase stabilizes p21 via phosphorylation on Thr145
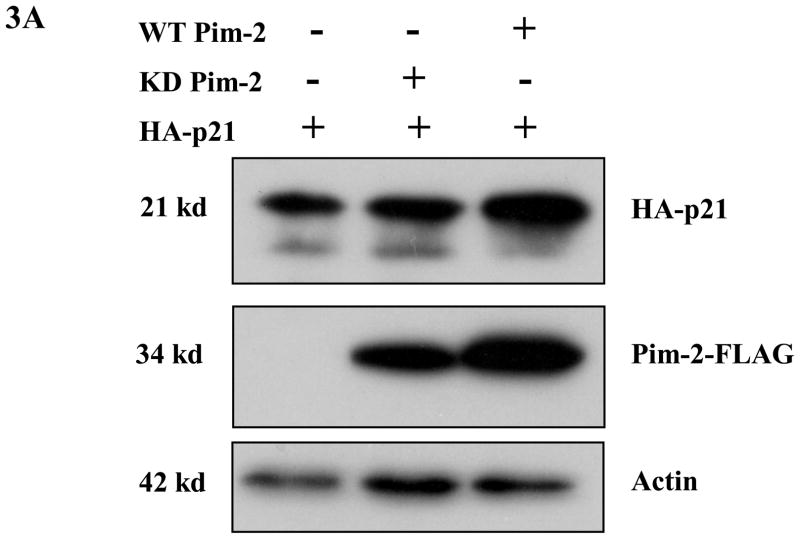
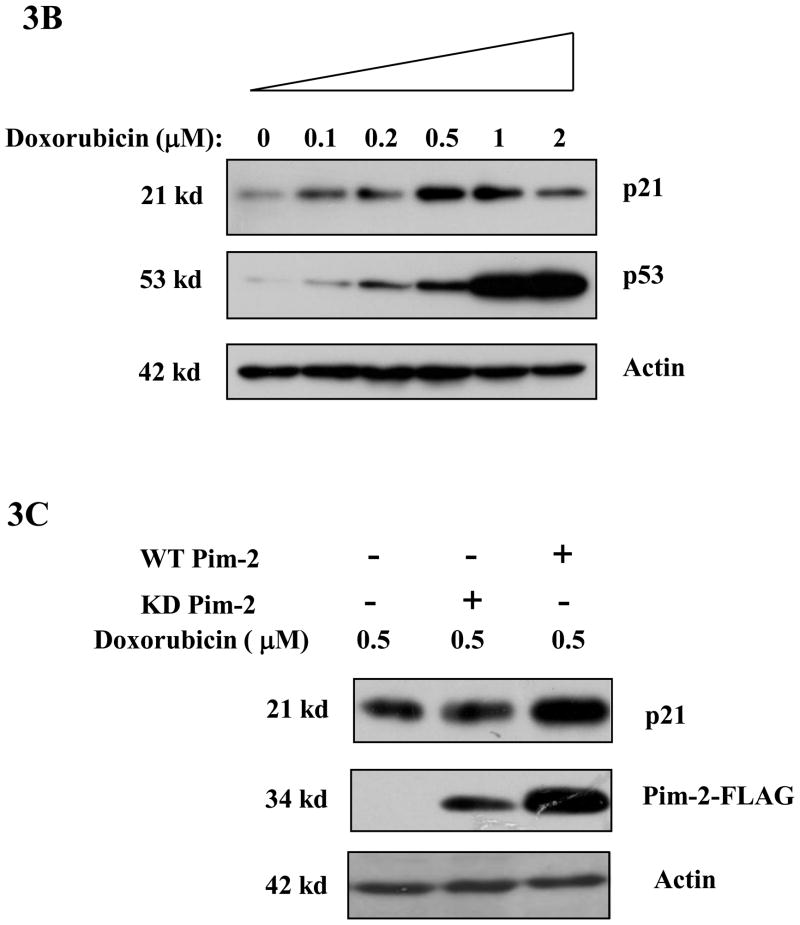
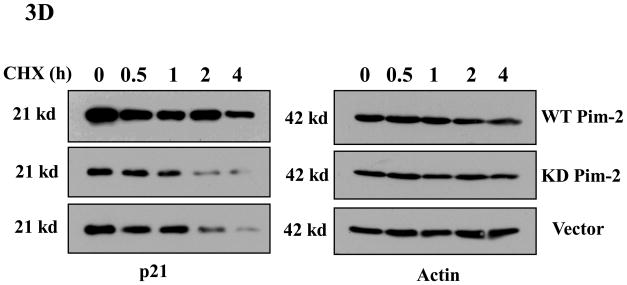
Phosphorylation of p21 by several kinases has been reported to regulate its protein stability (Li et al., 2002;Kim et al., 2002;Zhang et al., 2007). We had previously found that Pim-1 phosphorylates p21 on Thr145 which in the lung carcinoma cell line, H1299, leads to the stabilization of p21. Therefore, we predicted that Pim-2 should also cause p21 to be stabilized by phosphorylation on Thr145 in HCT116 cells. To determine if Pim-2 stabilizes p21, we co-transfected pBK/CMV-Pim-2-FLAG (wild type and kinase dead) with pBK/CMV-HA-p21 into HCT116 cells. As shown in Figure 3A, compared to the vector control, transfection of wild type Pim-2 leads to an increased level of exogenous p21, while kinase-dead Pim-2 has no effect on exogenous p21 levels in HCT116 cells. To determine whether Pim-2 also causes endogenous p21 to be stabilized, we first transfected pBK/CMV-Pim-2-FLAG (wild type and kinase dead) into HCT116 cells. Then endogenous p21 levels were induced by treating the cells with 0.5 μM doxorubicin 24 hours post-transfection. As shown in Figure 3B, the induced expression of endogenous p21 and p53 correspond well with the increased concentration of doxorubicin. Figure 3C shows that the presence of wild type Pim-2 increases the levels of endogenous p21 as compared to kinase-dead Pim-2 or vector control. The half-life of p21 was determined by treating cells with cycloheximide and collecting cells at the indicated time points for measurement of protein levels as shown in Figure 3D. This result confirms that wild type Pim-2 markedly increases the half-life of p21, whereas in cells expressing the kinase-dead form of Pim-2, the half life of p21 is even short compared to the vector control.
Figure 3. Pim-2 phosphorylation of p21 increases the stability of p21 proteins.
A) Pim-2 kinase activity increases the protein level of p21. HCT116 cells were co-transfected with HA-tagged p21 and either wild-type (WT) Pim-2-FLAG, Kinase dead (KD) Pim-2-FLAG or control vector. Forty hours after transfection, the protein levels of p21 were assayed by Western blotting with anti-HA antibody. Exogenous Pim-2 levels and control actin level were measured with anti-FLAG antibody and anti-actin antibody, respectively. B) Induction of p21 and p53 by increasing doses of doxorubicin in HCT116 cells. HCT116 cells were treated with the indicated amounts of doxorubicin for 24h. Induced p21 and p53 proteins were analyzed by Western blot with the designated antibodies. C) Exogenous Pim-2 kinase stabilizes p21 protein in doxorubicin-treated HCT116 cells. Either wild type (WT) Pim-2-FLAG, kinase dead (KD) Pim-2-FLAG or control vector was transfected into HCT116 cells. Doxorubicin was added at a final concentration of 0.5 μM to induce endogenous p21 after transfection. Cell lysates were then analyzed via Western blot for the expression of p21, Pim-2 and actin. D) Pim-2 kinase stabilizes p21 proteins. HCT116 cells were co-transfected with p21 and either WT Pim-2, KD Pim-2 or control vector. Forty hours post transfection, cells were treated with 25 μg/ml cycloheximide (CHX) and harvested at the indicated time points. The amount of degradation of p21 and actin proteins was analyzed by Western blot with anti-p21 and anti-actin antibodies.
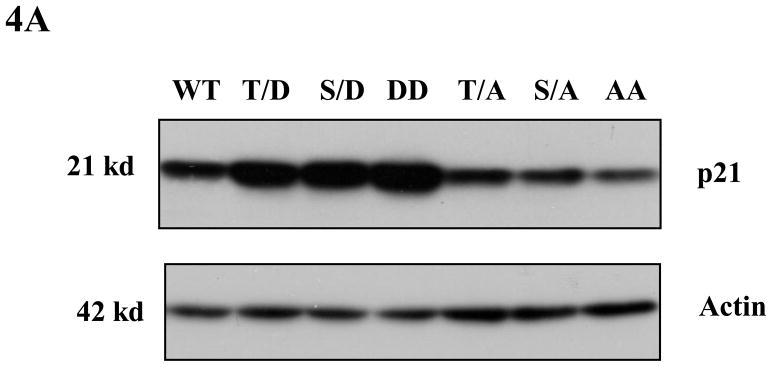
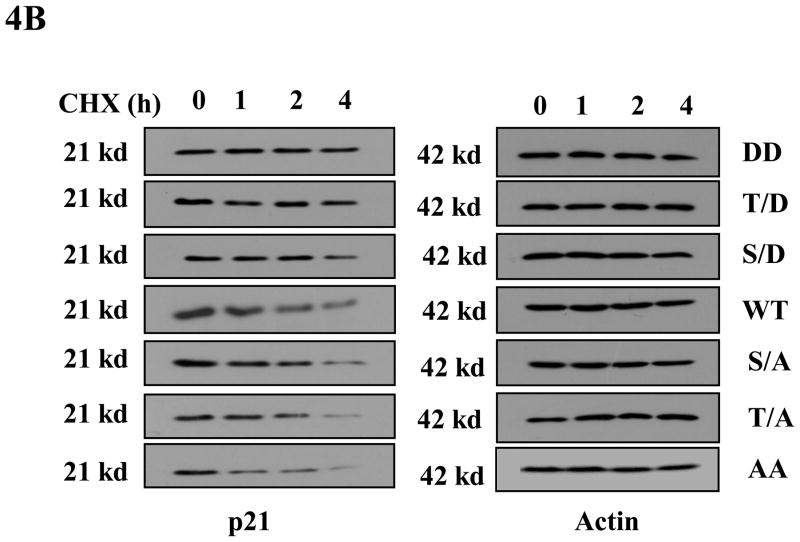
Phosphorylation mimics have been widely used in studies to determine the effect of p21 phosphorylation on its stability (reviewed in (Child and Mann, 2006)). In order to assess whether the phosphorylation of p21 on Thr145 is important for its stability, we used six different phospho-mimic mutants of HA-p21 plus wild type HA-p21, and transfected equal amounts of each construct into HCT116 cells. Western blot was used to determine the steady-state levels of p21 for each of these mutants. As shown in Figure 4A, phosphomimic on Thr145 or Ser146 (T/D, S/D and DD mutants) increases the steady-state level of transfected p21, while cells transfected with non-phosphomimic (T/A, S/A and AA mutants) have less p21 protein compared to wild type p21. When the half-lives of these p21 mutants were examined with cycloheximide treatment, we found that the phosphomimic mutants DD, T/D and S/D are more stable than wild type, while mutants T/A, S/A and AA are far less stable compared with the phosphomimic mutants (Figure 4B). It is also noticeable that the phosphomimic double mutant DD has the highest steady-state level of p21 and a longer half-life compared to other p21 mutants.
Figure 4. Phosphorylation of p21 increases its stability.
A) Phospho mimics of p21 increase steady levels of p21 in HCT116 cells. HCT116 cells were transfected with equal amounts of one of the HA-tagged p21 plasmids (WT, T/D, S/D, T/A, S/A, DD or AA). Forty hours post transfection, cells were harvested and the steady level of the p21 proteins were evaluated by Western blot with anti-HA antibody. The membrane was reprobed with anti-actin antibody to verify equal loading. B) Stability assay of various p21 proteins. HCT116 cells were transfected with one of the HA-tagged p21 plasmids (DD, T/D, S/D, WT, S/A, T/A, or AA). Thirty-six hours after transfection, cells were treated with 25μg/ml cycloheximide (CHX) and harvested at indicated time points. The stabilities of p21 and actin proteins were analyzed by Western blot with anti-p21 and anti-actin antibodies.
Endogenous p21 level can be modulated by Pim-2 kinase
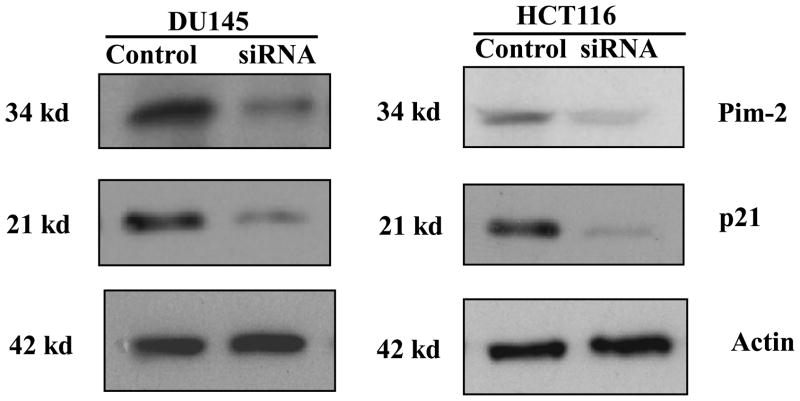
To further confirm Pim-2 kinase can regulate p21 stability endogenously, we knocked down endogenous Pim-2 with Pim-2 siRNA in HCT116 cells and also in DU145 cells for a comparison. As shown in Figure 5, corresponding with the decreased levels of Pim-2 by the transfection of Pim-2 siRNA, endogenous levels of p21 protein in both cell lines was reduced significantly. These results are consistent with the Pim-2 overexpression studies and indicate that Pim-2 kinase is capable of regulating p21 stability.
Figure 5. Pim-2 knockdown reduces endogenous level of p21.
HCT116 and DU145 cells were transfected with siRNA for pim-2. Two days after transfection, cell lysates were prepared and analyzed by the indicated antibodies to determine Pim-2 and p21 protein levels. Actin was used as a protein loading control.
Pim-2-stabilized p21 localizes in the nucleus
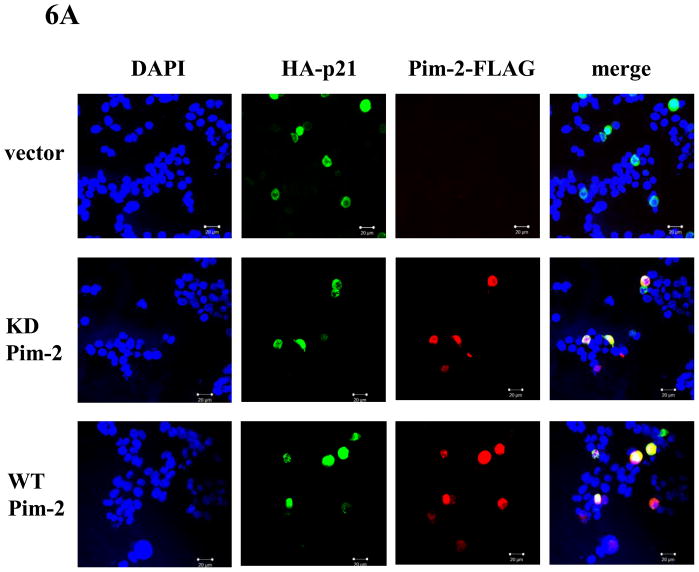
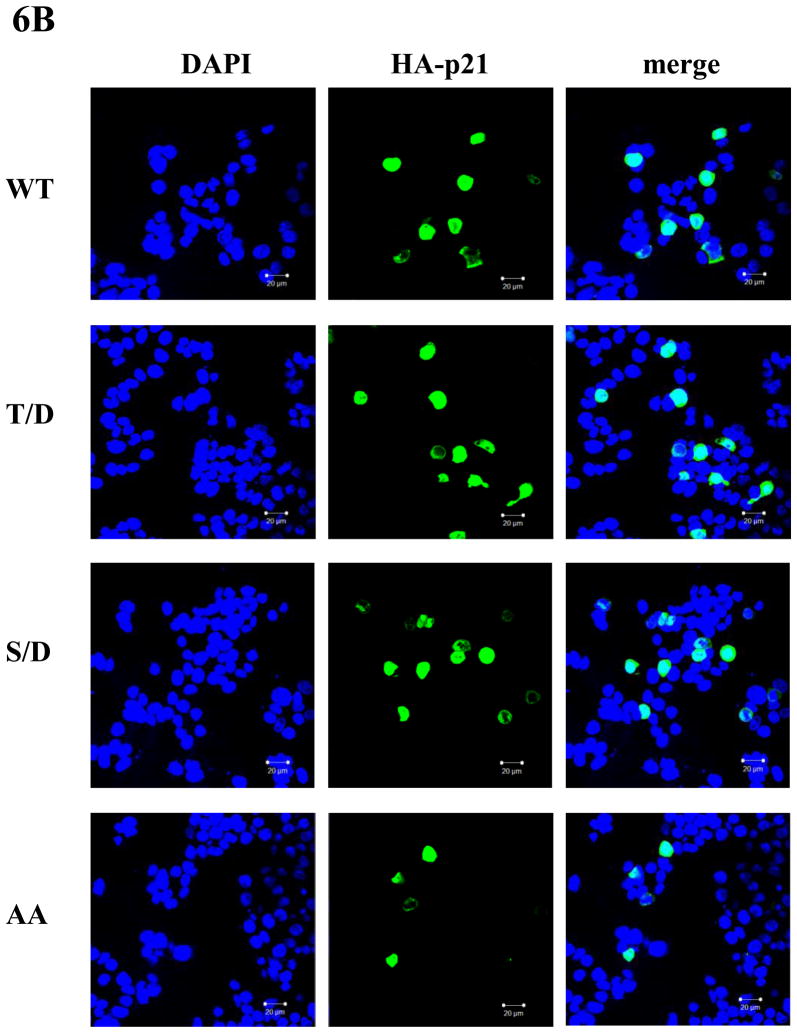
In addition to enhancing its stability, phosphorylation of p21 by various kinases can also regulate its subcellular localization (reviewed in (Child and Mann, 2006)). However, where p21 localizes after phosphorylation by these kinases is not consistent. For example, phosphorylation of p21 on Thr145 has been reported to be either translocated to the cytoplasm or remain in the nucleus. This outcome seemed to depend on the experimental cellular systems used although this was not suspected at the time (Rossig et al., 2001;Zhou et al., 2001). In order to determine the fate of phosphorylated p21 in the HCT116 cells, we used confocol microscopy to determine the subcellular localization of p21 that has been phosphorylated by Pim-2. After co-transfecting pBK/CMV-Pim-2-FLAG (either WT or KD) and pBK/CMV-HA-p21, HCT116 cells were stained with anti-HA and anti-FLAG antibodies. Anti-FLAG antibody was used to detect localization of Pim-2 while anti-HA antibody was used to detect that of p21. As shown in Figure 6A, compared to the vector control, neither wild type nor kinase dead Pim-2 changes the nuclear localization of HA-p21. However, it was previously found that Pim-1 phosphorylation of p21 causes p21 to be translocated from the nucleus into the cytoplasm in H1299 cells (Zhang et al., 2007). To compare the function of Pim-1 and Pim-2 under an identical cellular background, we repeated the co-transfection experiments with Pim-1 and p21. The results showed that like Pim-2, Pim-1 does not change the nuclear localization of p21 in HCT116 cells (See supplementary data). This observation suggests that phosphorylation of p21 by either Pim-1 or Pim-2 does not influence its cellular localization in HCT116 cells. As evidence for supporting this notion, we compared the localization of wild type p21 to the phosphomimic mutants T/D, S/D and AA by confocal microscopy. As shown in Figure 6B, all of the HA-p21 proteins were found to be localized in the nucleus. This indicates that phosphorylation on Thr145 or Ser146 does not change the subcellular localization of p21 in HCT116 cells, which is in contrast to what we have observed for other cell lines such as the lung carcinoma cell line, H1299 (Zhang et al., 2007).
Figure 6. Pim-2 does not change the nuclear localization of p21 in HCT116 cells.
A) Pim-2 has no effect on p21 cellular localization. HCT116 cells were co-transfected with HA-p21 and either WT Pim-2-FLAG, KD Pim-2-FLAG or control vector. Thirty-six hours later, cells were stained with anti-HA and anti-FLAG antibodies and visualized by confocal microscopy. Blue color depicts the nucleus stained with DAPI. Red color represents the localization of Pim-2-FLAG while green color depicts the localization of HA-p21. B) Phospho mimics of p21 have no effect on p21 localization. HCT116 cells were transfected with one of the p21 plasmids (WT, T/D, S/D or AA) and localization of p21 proteins were determined as above.
Pim-2 kinase causes cell cycle arrest at G1/S phase
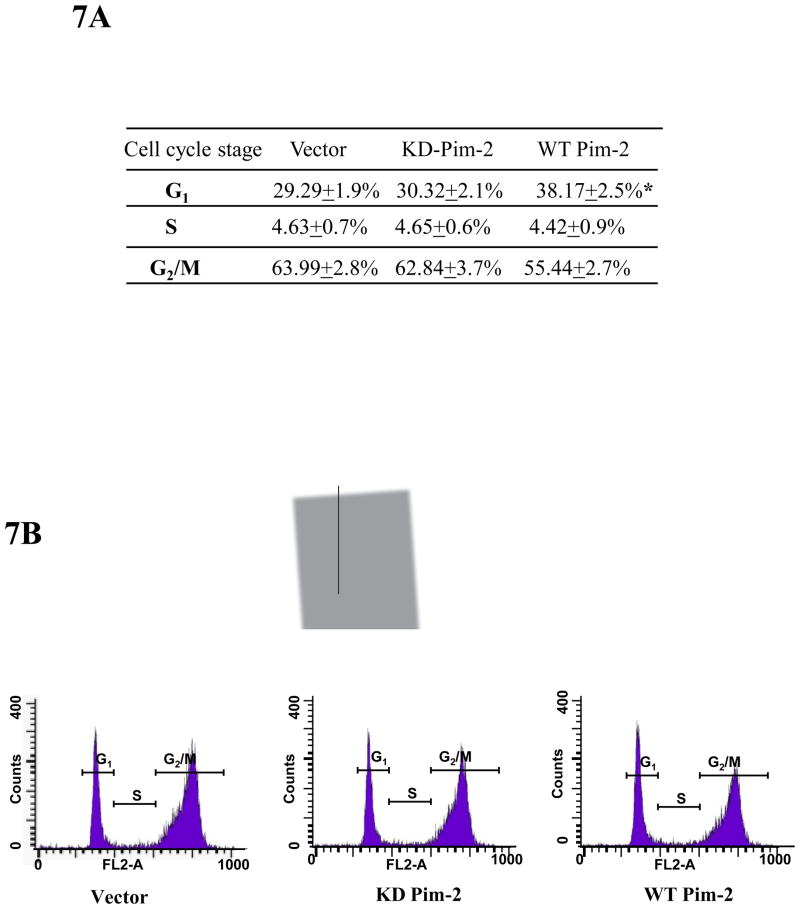
Nuclear localization is important for p21 to arrest cell cycle at G1/S. The increased p21 in the nucleus should potentially lead to cell cycle arrest as p21 is recognized as an important cyclin-dependent kinase inhibitor. As shown in Figure 3B and 3C, overexpression of Pim-2 kinase leads to stabilization of endogenous p21 induced by doxorubicin treatment. In order to study the functional consequences of Pim-2 phosphorylation of p21, we performed flow cytometric analysis to evaluate the cell cycle distribution of HCT116 cells. As shown in Figure 7A and 7B, we found that more of the cells transfected with the wild type Pim-2 kinase appear to be distributed in the G1 phase of the cell cycle as compared to the controls. The statistical analysis indicates that there is significant difference between the wild type Pim-2 and the controls. This indicates that by stabilizing endogenous p21 levels in doxorubicin-treated cells, Pim-2 kinase promotes cell cycle arrest at the G1/S transition.
Figure 7. Pim-2 causes cell cycle arrest at G1/S phase.
A) HCT116 cells were transfected with WT Pim-2, KD Pim-2 or control vector. Twenty four hours after transfection, 0.5 μM doxorubicin was added to induce endogenous p21. Cell cycle analysis was carried out as described in the Material and Methods. A persistent higher distribution in G1 phase was observed in cells that express WT Pim-2. Data represent means ± SD for three independent experiments, P< 0.01 as determined by student’s t-test. B) A typical histogram for the cell cycle analysis is illustrated for each sample. Gated portions represent G1, S and G2/M phases.
Pim-2 kinase inhibits cell proliferation through phosphorylation of p21
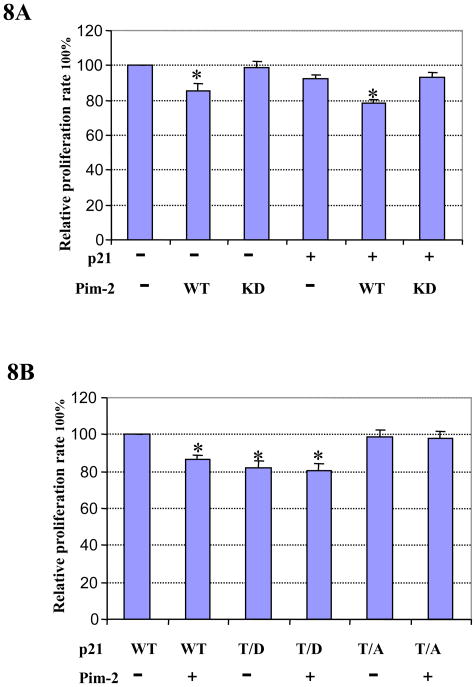
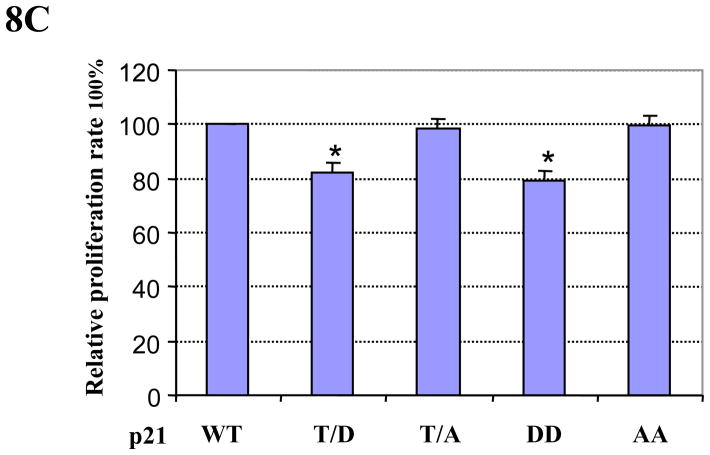
We found that Pim-2 kinase phosphorylates p21 on Thr145 and thereby increases its stability in the nucleus. Cell cycle arrest exerted by Pim-2 kinase at G1/S transition indicates that Pim-2 could in fact have a negative effect on cell proliferation. To test this hypothesis, HCT116 cells were co-transfected with p21 and Pim-2 (either WT or KD) plasmids and assayed for cell proliferation. In addition, we also transfected cells with either empty vector or Pim-2 (WT or KD) only. As shown in Figure 8A, cells transfected with WT Pim-2 have a lower proliferation capacity compared to empty vector control. However, KD Pim-2 does not have any significant effect on the proliferation of HCT116 cells. When the cells were co-transfected with p21 and Pim-2 (either WT or KD), only WT Pim-2, not KD Pim-2, is able to inhibit the proliferation of HCT116 cells. To determine whether Pim-2 inhibition of cell proliferation is mediated by phosphorylation of p21, we also compared the effect of Pim-2 on cell proliferation together with p21 WT or Thr145 mutants (T/D and T/A). As shown in Figure 8B, cells transfected with p21 T/D only or together with Pim-2 have significant slower proliferation compared with cells transfected with p21 T/A or p21 WT only. This result indicates that Pim-2 inhibits cell proliferation through phosphorylating p21 on Thr145. To verify the importance of Thr145 phosphorylation of p21, we compared effects of p21 WT and its different phosphomimic mutants on cell proliferation. As shown in Figure 8C, HCT116 cells were transfected with equal amount of various p21 plasmids, and as expected, different proliferation rates were detected. Compared to p21 WT, cells transfected with the p21 T/D or DD mutant have a marked decrease in cell proliferation, whereas those transfected with p21 T/A or AA mutant have no obvious effect. Taken together, these results indicate that Pim-2 kinase inhibit HCT116 cell proliferation by phosphorylation of p21 on Thr145.
Figure 8. Pim-2 inhibits cell proliferation via phosphorylation of p21.
A) Pim-2 inhibits cell proliferation. HCT116 cells were transfected with Pim-2 (WT or KD) only or co-transfected with wild-type p21 and Pim-2 (WT or KD). Forty hours post transfection, cells were trypsinized and used for the proliferation assay as described in the Materials and Methods. Relative proliferation rates were calculated as percentage of the sample with the highest luminescence reading. B) Pim-2 inhibits cell proliferation via phosphorylation of p21 on Thr145. HCT116 cells were co-transfected with Pim-2 and p21 (either T/D or T/A). Forty hours post transfection, the cell proliferation rate was determined as above. C) Phosphorylation of p21 on Thr145 is required for the inhibition of cell proliferation. HCT116 cells were transfected with different forms of p21 (WT, T/D, T/A, DD, or AA). Forty hours post transfection, cell proliferation rate were determined as above. Data represent means ± SD for four independent experiments, *=P< 0.01 as determined by student’s t-test.
Discussion
It is known that both Pim-1 and Pim-2 kinase share a high degree of sequence homology, consensus sequence for substrate phosphorylation, and functional activities in cell survival and proliferation. Therefore, it is likely that they act through similar biochemical pathways and can phosphorylate the same substrates. Consistent with this notion, it has been shown that both Pim-1 and Pim-2 can phosphorylate the SOCS-1 protein (Chen et al., 2002) and pro-apoptotic protein BAD (Yan et al., 2003;Macdonald et al., 2006). In this report, we show that p21 is another common substrate for Pim family members. Like Pim-1, Pim-2 kinase is able to phosphorylate p21 on Thr145 and promote its stability. More importantly, with regard to phosphorylation of p21, we found that Pim-1 and Pim-2 have very similar functions when examined under identical genetic backgrounds which is further evidence that the Pim kinase family members have redundant functions.
Although few studies have examined the functional or biochemical redundancy within the Pim family, the expression of pim-1 and pim-2 genes appear to respond similarly to growth factors and various cellular stresses (Wang et al., 2001). Their complementary effect observed between pim-1 and pim-2 knock-out mice (Allen and Berns, 1996;Mikkers et al., 2004) leads to the idea that Pim-1 and Pim-2 should have similar functions. Both Pim-1 and Pim-2 are able to phosphorylate p21 in vitro and in vivo and enhance p21 stability in vivo when examined in two different cell lines, the lung cancer cell line, HCT116 (p53+/+) and the colon cancer cell line, H1299 (p53−/−). However, several differences do exist in the phosphorylation pattern of p21 by Pim-1 and Pim-2 in these two cell lines. First, Pim-1 mediates an indirect phosphorylation of p21 on Ser146 in H1299 cells, but phosphorylation of p21 on Ser146 by Pim-2 is undetectable in HCT116 cells. Secondly, Pim-1 phosphorylation of p21 on Ser146 in H1299 cells leads to the cytoplasmic localization of p21, but Pim-2 phosphorylation of p21 in HCT116 cells has no detectable effect on its nuclear localization. Finally, Pim-1 phosphorylation of p21 in H1299 cells promotes cell proliferation, however, Pim-2 phosphorylation of p21 inhibits cell proliferation in HCT116 cells. Based on the similarity of substrate identification, induced expression pattern, and functional activities reported so far between Pim-1 and Pim-2, these differences in the phosphorylation of p21 were not expected. We suspected that the differences in the Pim kinase phosphorylation pattern of p21 might actually involve differences in the genetic background of the two cell lines. When we compared phosphorylation of p21 by Pim-1 and Pim-2 side by side in HCT116 cells, there was no distinction in terms of either subcellular localization or influence on cell proliferation (data not shown). Therefore, we conclude that Pim-2 and Pim-1 kinases do indeed function similarly in HCT116 cells, which indicates that the Pim family members are as redundant as previously suspected.
The cellular localization of p21 has been proposed to be critical for the regulation of p21 function (Coqueret, 2003). Multifaceted functions of p21 are achieved through p21’s direct interaction with numerous binding partners. Furthermore, the subcellular localization of p21 also plays an important part in dictating the binding partners to which p21 is exposed (Child and Mann, 2006). The Thr145 and Ser146 sites lie adjacent to the experimentally defined p21 nuclear localization signal. Phosphorylation of p21 on either Thr145 or Ser146 has been reported to result in the translocation of p21 from the nucleus to the cytoplasm in NIH3T3 cells, H1299 cells and MDA-MB453 cells, a breast cancer cell line (Zhou et al., 2001;Xia et al., 2004;Zhang et al., 2007). Nevertheless, the subcellular localization of p21 appears to depend on the cellular background. Rossig et al showed that p21 remains in nucleus after phosphorylation by Akt in human umbilical vein endothelial cells (Rossig et al., 2001) and in a keratinocyte transformation model, elevated Akt activity did not correlate with an enrichment of p21 to the cytosol (Segrelles et al., 2006). Therefore, although it is unclear at present how the cellular localization of p21 is controlled, the genetic background (i.e. differential expression of key components in the export/import pathways and of p21 binding partners) among different cell types appears to have dominant effect on the subcellular localization of p21.
The different outcomes on cell proliferation that are observed with p21 phosphorylation are most likely due to the different subcellular localizations of p21 that occur between H1299 and HCT116 cells. Cytoplasmic localization of p21 has been reported to promote cell proliferation and cell survival (Zhou et al., 2001;Xia et al., 2004;Perez-Tenorio et al., 2006;Zhang et al., 2007). On the other hand, nuclear p21 would be expected to function as cell cycle inhibitor to slow down cell proliferation. Given its important role in cell proliferation, differentiation and survival, p21 is very likely regulated through multiple signaling pathways via various protein kinases. To date, p21 has been reported to be regulated by several different kinases including Akt/PKB, GSK3, MAPK p38, PKC, PKA, and Pim-1 (Li et al., 2002;Rossig et al., 2001;Kim et al., 2002;Kashiwagi et al., 2000;Suzuki et al., 2000;Zhang et al., 2007;Rossig et al., 2001). Pim-2 now joins this group as a known kinase of p21. It is not clear how the ambient levels of those kinases take part in phosphorylation of endogenous p21, but it is well known that other p21 kinases need to be activated to be functional while the Pim kinase family members are constitutively active and ready to phosphorylate p21. Therefore, the Pim kinases appear to provide an extra layer of concise regulation of p21 stability and function, which can have a significant influence on cell proliferation and/or survival depending on expression levels of these kinases.
In term of pathological mechanisms, the CDK inhibitor p21 is often responsible for stress-induced p53-dependent and p53-independent cell cycle arrest (Gartel et al., 1996). Cell cycle arrest permits cells to pause and to repair DNA damage, therefore, protecting cells from stress-induced apoptosis (Gartel and Tyner, 2002). The clinical importance of p21 is demonstrated by accumulating data that p21 can function as an anti-apoptotic factor and is involved in the responsiveness to chemotherapy and radiotherapy (Abbas and Dutta, 2009;Abukhdeir and Park, 2008;Gartel, 2009). Deregulated expression of p21 has been reported in a variety of human cancers. Therefore, regulation of p21 protein levels in tumor cells will apparently an important consideration in the clinical treatment of cancers. In this study, we found that p21 levels could be increased by Pim-2 phosphorylation in HCT116 cells and that knockdown of Pim-2 results in decreased levels of p21. Recently, it was reported that the inhibitor of Pim kinase, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells (Chen et al., 2009), and resensitizes chemoresistant prostate cancer cells to taxanes (Mumenthaler et al., 2009). Interestingly, SGI-1776 inhibits not only Pim-2 kinase activity, but also p21 phosphorylation (Mumenthaler et al., 2009), which could result in a reduced levels of p21 protein in tumor cells. The application of newly identified Pim-2 inhibitors may contribute greatly to cancer therapy in the near future (Amaravadi and Thompson, 2005;Tao et al., 2009;Li et al., 2009;Akue-Gedu et al., 2009;Qian et al., 2009;Lin et al., 2010).
Supplementary Material
Acknowledgments
This study was supported by NIH grant R01-CA104470. We thank Dr. Michael Lilly (University of California at Irvine) for providing us mouse Pim-2 cDNA, Dr. Bert Vogelstein (John Hopkins University) for human p21 cDNA and Angela K. Hinz (Washington State University) for flow cytometry analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akue-Gedu R, Rossignol E, Azzaro S, Knapp S, Filippakopoulos P, Bullock AN, Bain J, Cohen P, Prudhomme M, Anizon F, Moreau P. Synthesis, kinase inhibitory potencies, and in vitro antiproliferative evaluation of new Pim kinase inhibitors. J Med Chem. 2009;52:6369–6381. doi: 10.1021/jm901018f. [DOI] [PubMed] [Google Scholar]
- Allen JD, Berns A. Complementation tagging of cooperating oncogenes in knockout mice. Semin Cancer Biol. 1996;7:299–306. doi: 10.1006/scbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- Allen JD, Verhoeven E, Domen Jd, van V, Berns A. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene. 1997;15:1133–1141. doi: 10.1038/sj.onc.1201288. [DOI] [PubMed] [Google Scholar]
- Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amson R, Sigaux F, Przedborski S, Flandrin G, Givol D, Telerman A. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc Natl Acad Sci U S A. 1989;86:8857–8861. doi: 10.1073/pnas.86.22.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohensky J, Shapiro IM, Leshinsky S, Watanabe H, Srinivas V. PIM-2 is an independent regulator of chondrocyte survival and autophagy in the epiphyseal growth plate. J Cell Physiol. 2007;213:246–251. doi: 10.1002/jcp.21117. [DOI] [PubMed] [Google Scholar]
- Breuer ML, Cuypers HT, Berns A. Evidence for the involvement of pim-2, a new common proviral insertion site, in progression of lymphomas. EMBO J. 1989;8:743–748. doi: 10.1002/j.1460-2075.1989.tb03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010 doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Chen JL, Limnander A, Rothman PB. Pim-1 and Pim-2 kinases are required for efficient pre-B-cell transformation by v-Abl oncogene. Blood. 2008;111:1677–1685. doi: 10.1182/blood-2007-04-083808. [DOI] [PubMed] [Google Scholar]
- Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009;114:4150–4157. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Losman JA, Cowan S, Donahue E, Fay S, Vuong BQ, Nawijn MC, Capece D, Cohan VL, Rothman P. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc Natl Acad Sci USA. 2002;99:2175–2180. doi: 10.1073/pnas.042035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child ES, Mann DJ. The intricacies of p21 phosphorylation: protein/protein interactions, subcellular localization and stability. Cell Cycle. 2006;5:1313–1319. doi: 10.4161/cc.5.12.2863. [DOI] [PubMed] [Google Scholar]
- Claudio JO, Masih-Khan E, Tang H, Goncalves J, Voralia M, Li ZH, Nadeem V, Cukerman E, Francisco-Pabalan O, Liew CC, Woodgett JR, Stewart AK. A molecular compendium of genes expressed in multiple myeloma. Blood. 2002;100:2175–2186. doi: 10.1182/blood-2002-01-0008. [DOI] [PubMed] [Google Scholar]
- Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Dai H, Li R, Wheeler Td, Diaz V, Frolov A, Tahir S, Agoulnik I, Thompson T, Rowley D, Ayala G. Pim-2 upregulation: biological implications associated with disease progression and perinueral invasion in prostate cancer. Prostate. 2005;65:276–286. doi: 10.1002/pros.20294. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Dotto GP. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta. 2000;1471:M43–M56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- Eichmann A, Yuan L, Breant C, Alitalo K, Koskinen PJ. Developmental expression of pim kinases suggests functions also outside of the hematopoietic system. Oncogene. 2000;19:1215–1224. doi: 10.1038/sj.onc.1203355. [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. The Pim kinases control rapamycin-resistant T cell survival and activation. J Exp Med. 2005;201:259–266. doi: 10.1084/jem.20042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL. The conflicting roles of the cdk inhibitor p21(CIP1/WAF1) in apoptosis. Leuk Res. 2005;29:1237–1238. doi: 10.1016/j.leukres.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Gartel AL. p21(WAF1/CIP1) and cancer: a shifting paradigm? Biofactors. 2009;35:161–164. doi: 10.1002/biof.26. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Serfas MS, Tyner AL. p21--negative regulator of the cell cycle. Proc Soc Exp Biol Med. 1996;213:138–149. doi: 10.3181/00379727-213-44046. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- Hammerman PS, Fox CJ, Cinalli RM, Xu A, Wagner JD, Lindsten T, Thompson CB. Lymphocyte transformation by Pim-2 is dependent on nuclear factor-kappaB activation. Cancer Res. 2004;64:8341–8348. doi: 10.1158/0008-5472.CAN-04-2284. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi M, Ohba M, Watanabe H, Ishino K, Kasahara K, Sanai Y, Taya Y, Kuroki T. PKCeta associates with cyclin E/cdk2/p21 complex, phosphorylates p21 and inhibits cdk2 kinase in keratinocytes. Oncogene. 2000;19:6334–6341. doi: 10.1038/sj.onc.1204028. [DOI] [PubMed] [Google Scholar]
- Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem. 2002;277:29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Laird PW, van der Lugt NM, Clarke A, Domen J, Linders K, McWhir J, Berns A, Hooper M. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 1993;21:4750–4755. doi: 10.1093/nar/21.20.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Helfman DM. Cytoplasmic p21Cip1 is involved in Ras-induced inhibition of the ROCK/LIMK/cofilin pathway. J Biol Chem. 2004;279:1885–1891. doi: 10.1074/jbc.M306968200. [DOI] [PubMed] [Google Scholar]
- Li Y, Dowbenko D, Lasky LA. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J Biol Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- Li YY, Wang YY, Taniguchi T, Kawakami T, Baba T, Ishibashi H, Mukaida N. Identification of stemonamide synthetic intermediates as a novel potent anti-cancer drug with an apoptosis-inducing bility. Int J Cancer. 2009 doi: 10.1002/ijc.25048. [DOI] [PubMed] [Google Scholar]
- Lin YW, Beharry ZM, Hill EG, Song JH, Wang W, Xia Z, Zhang Z, Aplan PD, Aster JC, Smith CD, Kraft AS. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood. 2010;115:824–833. doi: 10.1182/blood-2009-07-233445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JS. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7:1. doi: 10.1186/1471-2121-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiussi S, Turrini P, Testolin L, Martelli F, Zaccagnini G, Mangoni A, Barlucchi LM, Antonini A, Illi B, Cirielli C, Padron J, Nicolo C, Testi R, Osculati F, Biglioli P, Capogrossi MC, Gaetano C. p21(Waf1/Cip1/Sdi1) mediates shear stress-dependent antiapoptotic function. Cardiovasc Res. 2004;61:693–704. doi: 10.1016/j.cardiores.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumenthaler SM, Ng PY, Hodge A, Bearss D, Berk G, Kanekal S, Redkar S, Taverna P, Agus DB, Jain A. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Mol Cancer Ther. 2009;8:2882–2893. doi: 10.1158/1535-7163.MCT-09-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Knebel A, Morrice NA, Li X, Barringer K, Li J, Jakes S, Werneburg B, Wang L. Pim kinase substrate identification and specificity. J Biochem. 2007;141:353–362. doi: 10.1093/jb/mvm040. [DOI] [PubMed] [Google Scholar]
- Perez-Tenorio G, Berglund F, Esguerra MA, Nordenskjold B, Rutqvist LE, Skoog L, Stal O. Cytoplasmic p21WAF1/CIP1 correlates with Akt activation and poor response to tamoxifen in breast cancer. Int J Oncol. 2006;28:1031–1042. [PubMed] [Google Scholar]
- Qian K, Wang L, Cywin CL, Farmer BT, Hickey E, Homon C, Jakes S, Kashem MA, Lee G, Leonard S, Li J, Magboo R, Mao W, Pack E, Peng C, Prokopowicz A, III, Welzel M, Wolak J, Morwick T. Hit to lead account of the discovery of a new class of inhibitors of Pim kinases and crystallographic studies revealing an unusual kinase binding mode. J Med Chem. 2009;52:1814–1827. doi: 10.1021/jm801242y. [DOI] [PubMed] [Google Scholar]
- Rossig L, Jadidi AS, Urbich C, Badorff C, Zeiher AM, Dimmeler S. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol Cell Biol. 2001;21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrelles C, Moral M, Lara MF, Ruiz S, Santos M, Leis H, Garcia-Escudero R, Martinez-Cruz AB, Martinez-Palacio J, Hernandez P, Ballestin C, Paramio JM. Molecular determinants of Akt-induced keratinocyte transformation. Oncogene. 2006;25:1174–1185. doi: 10.1038/sj.onc.1209155. [DOI] [PubMed] [Google Scholar]
- Sheaff RJ, Singer JD, Swanger J, Smitherman M, Roberts JM, Clurman BE. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K. Procaspase 3/p21 complex formation to resist fas-mediated cell death is initiated as a result of the phosphorylation of p21 by protein kinase A. Cell Death Differ. 2000;7:721–728. doi: 10.1038/sj.cdd.4400706. [DOI] [PubMed] [Google Scholar]
- Tao ZF, Hasvold LA, Leverson JD, Han EK, Guan R, Johnson E, Stoll VS, Stewart KD, Stamper G, Soni N, Bouska JJ, Luo Y, Sowin TJ, Lin NH, Giranda VS, Rosenberg SH, Penning TD. Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as potent, highly selective, and orally bioavailable inhibitors of the human protooncogene proviral insertion site in moloney murine leukemia virus (PIM) kinases. J Med Chem. 2009;52:6621–6636. doi: 10.1021/jm900943h. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Verhoeven E, Linders K, van der GH, Allen J, Berns A. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J. 1995;14:2536–2544. doi: 10.1002/j.1460-2075.1995.tb07251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, Berns A. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Wang Z, Bhattacharya N, Weaver M, Petersen K, Meyer M, Gapter L, Magnuson NS. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J Vet Sci. 2001;2:167–179. [PubMed] [Google Scholar]
- Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y, Zhou BP, Hung MC. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res. 2004;10:3815–3824. doi: 10.1158/1078-0432.CCR-03-0527. [DOI] [PubMed] [Google Scholar]
- Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, Lilly M. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003;278:45358–45367. doi: 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kaneita Y, Aoki Y, Seto M, Mori S, Moriyama M. Identification of heterologous translocation partner genes fused to the BCL6 gene in diffuse large B-cell lymphomas: 5′-RACE and LA - PCR analyses of biopsy samples. Oncogene. 1999;18:7994–7999. doi: 10.1038/sj.onc.1203293. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res. 2007;5:909–922. doi: 10.1158/1541-7786.MCR-06-0388. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.