Introduction
Both genetic and epigenetic mechanisms are obligatory for physiologically responsive activation and suppression of genes that govern cell growth, proliferation, phenotype and metabolic homeostasis for development, differentiation and tissue remodelling (reviewed in (Lee and Workman 2007;Ng and Gurdon 2008;Fazzari and Greally 2004;Goldberg et al. 2007;Stein et al. 2009;Stein et al. 2006;Kim et al. 2009). Transformation, tumorigenesis, tumor progression and metastasis are dependent on signalling cascades that transduce and integrate regulatory cues to determine competency for transcription and the extent to which genes are expressed. We will focus on emerging evidence for prominent contributions from regulatory factor-mediated epigenetic parameters of transcriptional and posttranscriptional mechanisms that are operative in biology and pathology. We will explore epigenetic mechanisms that include transcription factor control of chromatin organization, cell fate and lineage commitment and microRNA-dependent gene expression for biological control and cancer.
Transcription factors strategically locate regulatory machinery at promoter sites
It is well established that histone acetylation, methylation and phosphorylation epigenetically influence the accessibility of promoter elements to transcription factors by modifying histone-DNA and histone-histone interactions with accompanying changes in chromatin structure and nucleosome organization. These posttranslational alterations of histones are reversible to accommodate cellular requirements for gene expression (Liu et al. 2005;Westendorf et al. 2002;Yang et al. 2007;Sun et al. 2007;Delcuve et al. 2009). A histone code, based on histone modifications and histone subtypes, extend the informational content of the genome by contributing to options for sequence recognition by regulatory proteins (Berger 2007;Richards and Elgin 2002;Jenuwein and Allis 2001). DNA methylation has similarly been shown to determine access of transcription factors to gene regulatory sequences (Jones and Baylin 2007;GOLD et al. 1963;Tucker et al. 1996). Therefore, fundamental questions are the mechanisms that mediate specificity of histone and DNA modifications that are responsive to biological cues.
Emerging evidence indicates that transcription factors, through protein-protein interactions, strategically localize co-regulatory proteins to modify histones (acetylases, deacetylases, kinases, phosphotases, methylases and demethylases) at sites of target gene promoters, where they modulate chromatin structure and competency for transcription to accommodate cellular requirements (Gutierrez et al. 2007;Shen et al. 2003;Cruzat et al. 2009;Montecino et al. 2007). Biologically responsive control at specific sites of target gene promoters is further facilitated by interactions of transcription factors with cohorts of co-regulatory proteins that include but are not confined to co-activators and co-repressors, endpoints of signalling pathways and steroid hormone receptors (reviewed in (Zaidi et al. 2007;Zaidi et al. 2005;Zaidi et al. 2006)).
Transcription factors control chromatin organization for biological regulation
The scaffolding of co-regulatory proteins by transcription factors at regulatory domains of target genes is illustrated by the Runx/AML transcription factors that control hematopoiesis (Runx1, AML1), osteogenesis (Runx2, AML3) and the gastrointestinal/neurological phenotypes (Runx3, AML2) (Westendorf and Hiebert 1999;Ito et al. 2005;Ben-Ami et al. 2009;Zeng et al. 1997;Speck and Gilliland 2002;Blyth et al. 2005;Blyth et al. 2005). As shown in Figure 1, Runx proteins function as scaffolds for the organization and assembly of regulatory machinery that supports tissue-specific gene expression at sites of promoter regulatory elements in a manner that is consistent with biological control. Runx/AML factors also participate in cell cycle control (Durst and Hiebert 2004;Galindo et al. 2005) (Figure 1). To accommodate responsiveness to diverse and changing cues that are both systemic and confined to tissue microenvironments, the composition and organization of transcription factor-associated co-regulatory protein complexes at target gene promoter elements are not static. Dynamic modifications in the representation of factors reflect context-dependent changes in biological requirements. The promoter localization of factors by Runx proteins that mediate histone modifications and chromatin remodelling (Gutierrez et al. 2004;Shen et al. 2003;Montecino et al. 2007;Vradii et al. 2006) illustrates the architectural organization of regulatory machinery for epigenetic control within the context of genomic sequences that are principal components of physiological function.
Figure 1. Runx proteins are context dependent regulators of gene expression.
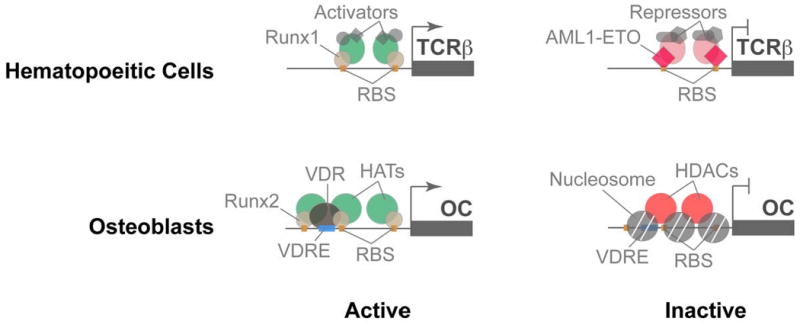
Runx transcription factors share a highly conserved DNA binding domain that recognizes Runx binding sites (shown in this figure as orange boxes) present in promoters of genes involved in cell proliferation, growth and differentiation. Runx proteins are also targeted to specific nuclear microenvironment where they occupy target gene promoters and function as scaffolding proteins that recruit co-regulators to gene promoters. In hematopoietic cells (top panel), Runx1, a master regulator of definitive hematopoiesis (shown here as light orange oval), interacts with and recruits co-activators (green and grey ovals) to the promoters of several genes, that include TCR beta, required for differentiation of hematopoietic stem cells into various lineages. In acute myelogenous leukemias, wild type Runx1 protein is replaced by the chimeric AML1-ETO protein (red diamond), which binds to the same promoter sites, but recruits repressors of gene transcription (light red and grey ovals). In osteoblasts, Runx2, a master regulator of bone cell lineage, binds and recruits co-activators to promoters of osteoblast-specific genes that include osteocalcin (OC). When Runx binding sites are mutated in the OC gene promoter, the promoter acquires a close chromatin conformation (grey circles; nucleosomes), thus inhibiting gene transcription.
Transcription factors control chromatin structure for tumorigenesis
In addition to serving as a paradigm for the architectural organization of regulatory machinery for biological control of tissue-specific genes, the Runx proteins illustrate that changes in the cohort of co-regulatory proteins binding to Runx/AML transcription factors can contribute to transformation and tumorigenesis. As strikingly demonstrated by the Runx1/AML1-ETO translocation fusion that is associated with approximately 15% of acute myelogenous leukemia, loss of the C terminal domain and acquisition of a chromosome 8-encoded C terminal sequence changes co-regulatory factor interactions (McNeil et al. 1999;Barseguian et al. 2002;Bakshi et al. 2008). Because the N terminal Runx1/AML sequence that contains the DNA binding domain is retained in the AML/ETO translocation fusion protein, a different complement of factors interact with cognate sequences of target genes to alter chromatin structure, nucleosome organization, response to regulatory signals and transcription. The consequence is a preleukemia/leukemia phenotype (Vradii et al. 2005) (Figure 1).
The involvement of transcription factor-mediated epigenetic control in leukemogenesis is associated with well documented differences in the chromatin organization of the AML locus in AML leukemia patients (Erickson et al. 1992). The chromatin confirmation of the AML locus supports the recombination events required for the reciprocal chromosome 8;21 translocation. Recent findings suggest that interactions of AML transcription factors at sites within the AML locus “scaffolds” the enzymology for histone modifications to selectively render the sites permissive to recombination (Stuardo et al. 2009).
Mitotic retention of transcription factors epigenetically conveys competency for gene expression
Biological control
Cell fate and lineage commitment requires the sustained expression of genes that are associated with tissue specificity. Competency for phenotype-restricted gene expression must be retained during mitosis for parental and progeny cells to remain structurally and functionally equivalent. Recent findings suggest transcription factors that control tissue specificity remain complex with target gene loci of mitotic chromosomes to support post-mitotic gene expression for the hematopoietic, bone, muscle and adipocyte phenotypes (Zaidi et al. 2003;Young et al. 2007a;Young et al. 2007b;Ali et al. 2008;Bakshi et al. 2008) (Figure 2). While all regulatory proteins do not remain complex with cognate sequences during cell division (He and Davie 2006), mitotic retention of regulatory proteins is a component of epigenetic control. The retention of regulatory factors with gene loci is not confined to DNA polymerase II-mediated control. Several lines of biochemical and in situ evidence indicate transcription factors that regulate DNA polymerase I-mediated expression of ribosomal cells are retained during mitosis to epigenetically support competency for post-mitotic protein synthesis (Young et al. 2007a;Young et al. 2007b;Ali et al. 2008). The control of cell fate, cell cycle and cell growth by Runx transcription factors is consistent with mechanistic linkage of cell fate, proliferation and protein synthesis in a manner that invokes epigenetic regulation.
Figure 2. Mitotic retention of sequence-specific transcription factors as an epigenetic mechanism for lineage maintenance in normal cells and for the sustained tumor phenotype in cancer cells.
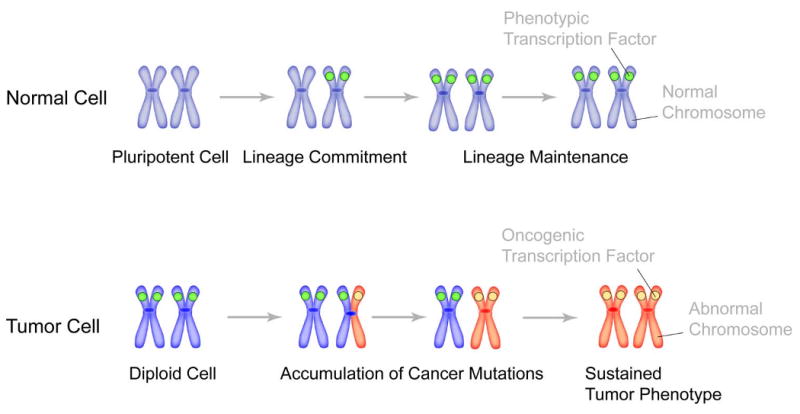
Gene loci on metaphase chromosomes (blue color) are occupied by phenotypic transcription factors (green circle) for lineage maintenance through successive cell divisions in normal cells. In tumors, as cells undergo genomic instability and accumulate cancer mutations, it is possible that one allele (shown here as red chromosome arm) is occupied by oncogenic transcription factor (which can be a phenotypic protein, ectopically expressed in cancer cells). Successive cell divisions and acquisition of additional cancer mutations then lead to the replacement of phenotypic regulatory proteins with oncogenic transcription factors. Occupancy of target gene loci with oncogenic proteins results in sustained tumor phenotype.
Cancer
Transcription factor-mediated epigenetic control that is obligatory for initiating and sustaining transformation and tumorigenesis is reflected by retention of the AML/ETO translocation fusion protein with target gene loci of pre-leukemic and leukemia cells (Bakshi et al. 2008). Consistent with observations in normal diploid cells, the AML/ETO factor epigenetically regulates genes transcribed by both RNA polymerase I and RNA polymerase II to coordinate control of proliferation and protein synthesis (Young et al. 2007a;Young et al. 2007b).
Epigenetic control plays a key role in transformation and tumorigenesis and is increasingly becoming a therapeutic target. Both histone and DNA modifications accompany and have been functionally associated with leukemias and solid tumors. Alterations in the activities of tumor suppressor genes have been implicated. The effectiveness of epigenetic control as a platform for therapy has been established as a “proof of principle” and “emerging standards of care” include modulation of DNA methylation (Yoo and Jones 2006) and histone acetylation (Marks et al. 2004).
MicroRNA-mediated epigenetic activation and suppression of regulatory networks
There is growing evidence that microRNAs are another dimension to epigenetic control of hematopoiesis and leukemogenesis (Garzon and Croce 2008;Ambros 2001;Murchison and Hannon 2004;Bartel 2004). Having established a Runx/ETO intranuclear trafficking defect in AML patients and a requirement for fidelity of Runx localization within the nucleus for myeloid differentiation, we investigated the relationship between subnuclear organization of transcription factors and microRNA-related activation and suppression of regulatory networks (Zaidi et al. 2009). Genome-wide microRNA expression profiling identified Runx/AML subnuclear location-dependent microRNAs in AML patients. We demonstrated that the modified Runx1 transcriptional program and altered nuclear localization is linked to the activity of specific microRNAs that perturb MAP kinase signaling (Figure 3). As observed with AML1/ETO or a modified Runx1 that exhibits altered subnuclear targeting of AML/ETO (Vradii et al. 2005), expression of mir24 renders myeloid progenitor cells hyperproliferative and growth factor independent. A block in granulocyte differentiation occurs. These findings provide the first experimental evidence for a regulatory network in which Runx-responsive microRNAs control myeloid cell proliferation and differentiation by targeting a negative regulator of the MAP-kinase signaling cascade. On the basis of these findings, we are exploring mechanistic linkage between downregulation of mir24 in leukemic blast cells and reversal of the disease phenotype. Further studies are required to confirm an obligatory relationship between upregulation of mir24 and the onset and progression of leukemia in vivo. However, our results are consistent with mir24 as a novel and selective epigenetic therapeutic target for the treatment of acute myelogenous leukemia and illustrate the complexity of transcription-factor mediated epigenetic control of biological processes and tumorigenesis.
Figure 3. Runx1 responsive miR 24 regulates myeloid cell proliferation and differentiation by inhibiting an antagonist of the MAPK signaling pathway.
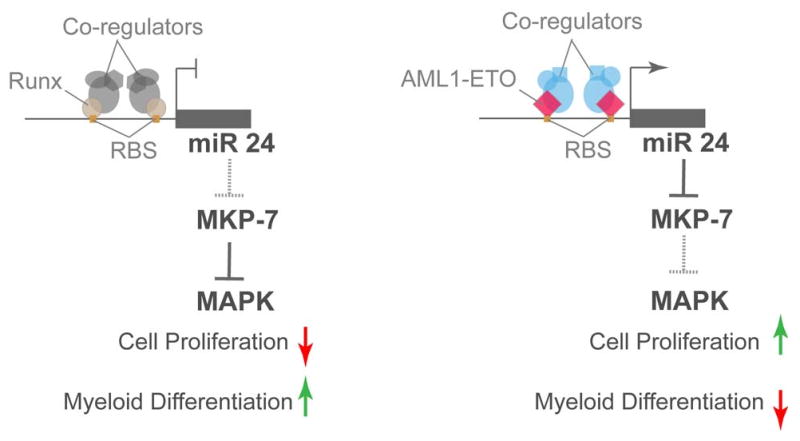
In normal myeloid progenitors (left side schematic), miR-24 is repressed by Runx1. This activates MAP kinase phosphatase 7 (MKP-7), which in turn blocks the activity of MAP kinase, thus down-regulating myeloid cell proliferation, concomitant with an increase in granulocytic differentiation. In AML patients, the leukemic AML1-ETO occupies the same site in the miR-24 promoter, but activates it. Activation of miR-24 down-regulates MKP-7 and up-regulates down-stream MAPK signaling. Activation of MAPK then leads to activated cell proliferation and inhibited myeloid cell differentiation, hallmarks of leukemia blast cells.
Transcription factor-mediated epigenetic control: A perspective
Transcription factors contribute to epigenetic control of gene expression in several contexts. Localization of the regulatory machinery for histone and DNA modifications through protein-protein interactions with transcription factors that occupy strategic genomic sequences is becoming increasingly evident. Mitotic retention of regulatory factor cohorts with target genes that are controlled RNA polymerase I and RNA polymerase II implicates transcription factors in epigenetic control of cell fate and lineage commitment. And, regulation of microRNA expression by transcription factors points to epigenetic control that is primarily operative at the level of mRNA stability and/or translatability. Taken together, there is emerging evidence that transcription factor control of gene expression extends beyond influencing the processive assembly of transcripts. Evidence is accruing for a central role of transcription factors in epigenetically regulating processes that include controlling the organization and placement of proteins that determine accessibility and transcriptional competency of genomic sequences for expression and supporting proliferation, growth, phenotype, and homeostatic regulation at both transcriptional and posttranscriptional levels (Figure 4). Epigenetic regulation by transcription factors provides a strategy for targeting gene expression in transformed and tumor cells with minimal offtarget consequences.
Figure 4. Epigenetic mechanisms operative in mammalian cells.
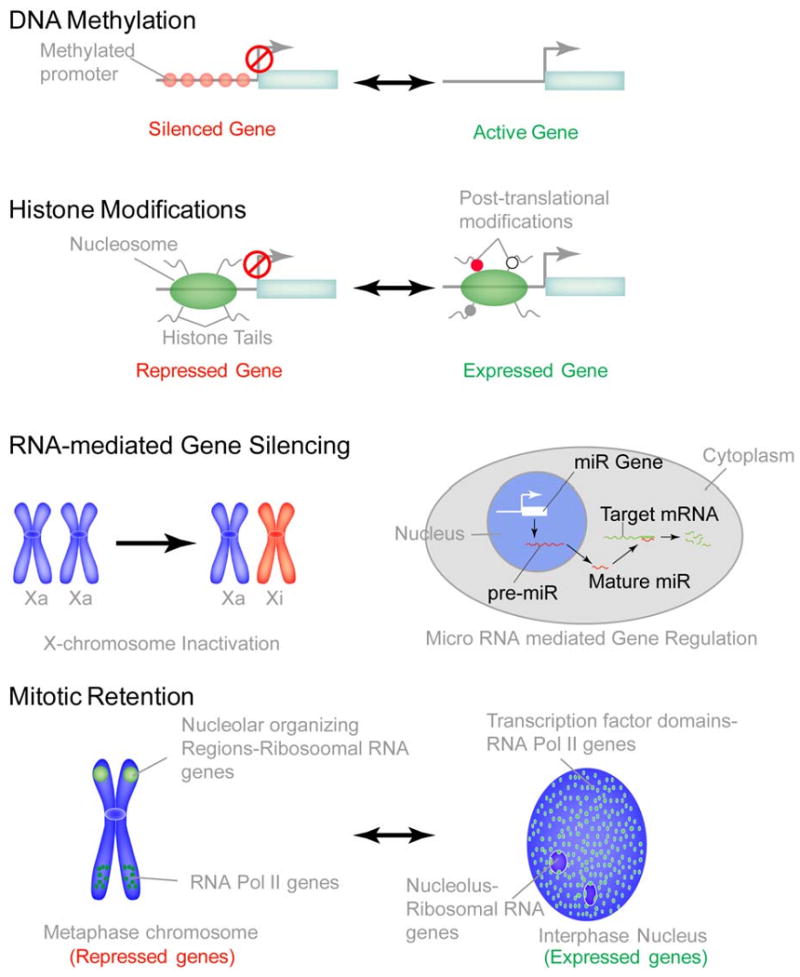
In addition to genetically inheritable mechanisms that coordinate cell proliferation, growth and differentiation, several epigenetic mechanisms are operative in mammalian cells to ensure the fidelity of gene expression through generations. DNA methylation is a well established mechanism that epigenetically controls the silencing of many developmental genes, often irreversibly, to ensure lineage commitment. DNA methylation is also an important mechanism for silencing tumor suppressor genes during tumor progression. Histone modifications are another established parameter of epigenetic control. Over 60 histone modifications regulate and fine-tune gene regulation under various biological conditions. Recently, RNA mediated gene silencing is gaining attention as another epigenetic mechanism. The most notable example is the inactivation of one copy of X chromosome in females. One of the two copies of active X-chromosomes (labeled here as Xa) is inactivated (labeled here as Xi) by Xist RNA. Recently, micro-RNA mediated down-regulation of gene expression is being extensively investigated. miRs are typically transcribed by RNA ploymerase II as precursor-miRs, that are processed by enzymes Drosha and Dicer to generate 22 nt long mature miRs. Mature miRs recognize the seed sequence in target mRNA and lead to their degradation, thus negatively regulating gene expression. A novel epigenetic mechanism that is emerging is the mitotic retention of phenotypic proteins on target gene loci to ensure lineage commitment and maintenance in progeny cells. Depicted here is the mitotic localization of Runx2 transcription factor to nucleolar organizing regions (NORs) were the ribosomal RNA genes reside, as well as to the RNA Pol II regulated genes that are involved in cell cycle control and lineage commitment.
Summary
There is a requirement to retain regulatory information and parameters of nuclear organization during cell division when transcription is globally silenced to sustain competency for expression of genes that control proliferation, cell growth and phenotype in progeny cells. Histone modifications, DNA methylation and RNA-mediated silencing are well defined, DNA-independent epigenetic mechanisms that regulate gene expression. Recent results suggest that retention of lineage-specific transcription factors with promoter elements and sequential assembly of gene regulatory machinery is a novel parameter of epigenetic control to sustain cellular identity post-mitotically. Collectively, these epigenetic signatures “bookmark” genes for activation or suppression as cells exit mitosis and resume cell cycle progression. We propose a model for cell fate and lineage commitment, coordination and maintenance where the regulatory proteins are retained on mitotic chromosomes to convey information for biological control to progeny cells as well as for controlling expression of the transformed and tumor phenotypes. MicroRNAs are another dimension to transcription factor-mediated epigenetic regulation of gene expression in biological control and cancer. Our findings demonstrate that fidelity of transcription factor localization is required for microRNA-related activation and suppression of regulatory networks in myeloid progenitor cells providing a novel and selective epigenetic target for treatment of acute myelogenous leukemia.
Acknowledgments
Studies reported in this chapter were supported by the National Institutes of Health (AR048818 and CA82834). The authors thank Ms. Patricia Jamieson for editorial assistance with preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, Stein GS, Stein JL. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci U S A. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Bakshi R, Zaidi SK, Pande S, Hassan MQ, Young DW, Montecino M, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The leukemogenic t(8;21) fusion protein AML1-ETO controls ribosomal RNA genes and associates with nucleaolar organizing regions at mitotic chromosomes. J Cell Sci. 2008;21:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barseguian K, Lutterbach B, Hiebert SW, Nickerson J, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc Natl Acad Sci U S A. 2002;99:15434–15439. doi: 10.1073/pnas.242588499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The runx genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Cruzat F, Henriquez B, Villagra A, Hepp M, Lian JB, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS, Montecino MA. SWI/SNF-independent nuclease hypersensitivity and increased histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry. 2009 doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J Cell Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- Gold M, Hurwitz J, Anders M. The enzymatic methylation of RNA and DNA. Biochem Biophys Res Commun. 1963;11:107–114. doi: 10.1016/0006-291x(63)90075-5. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gutierrez J, Paredes R, Cruzat F, Hill DA, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Imbalzano AN, Montecino M. Chromatin remodeling by SWI/SNF results in nucleosome mobilization to preferential positions in the rat osteocalcin gene promoter. J Biol Chem. 2007;282:9445–9457. doi: 10.1074/jbc.M609847200. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Liu J, Javed A, Montecino M, Stein GS, Lian JB, Stein JL. The vitamin D response element in the distal osteocalcin promoter contributes to chromatin organization of the proximal regulatory domain. J Biol Chem. 2004;279:43581–43588. doi: 10.1074/jbc.M408335200. [DOI] [PubMed] [Google Scholar]
- He S, Davie JR. Sp1 and Sp3 foci distribution throughout mitosis. J Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, Hiong KC, Peh BK, Han HC, Ito T, Teh M, Yeoh KG, Ito Y. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65:7743–7750. doi: 10.1158/0008-5472.CAN-05-0743. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- McNeil S, Zeng C, Harrington KS, Hiebert S, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor. Proc Natl Acad Sci U S A. 1999;96:14882–14887. doi: 10.1073/pnas.96.26.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecino M, Stein JL, Stein GS, Lian JB, van Wijnen AJ, Cruzat F, Gutierrez S, Olate J, Marcellini S, Gutierrez JL. Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence. Biochem Cell Biol. 2007;85:419–425. doi: 10.1139/O07-070. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Shen J, Hovhannisyan H, Lian JB, Montecino MA, Stein GS, Stein JL, van Wijnen AJ. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Mol Endocrinol. 2003;17:743–756. doi: 10.1210/me.2002-0122. [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Montecino M, Zaidi SK, Braastad C. An architectural perspective of cell-cycle control at the G1/S phase cell-cycle transition. J Cell Physiol. 2006;209:706–710. doi: 10.1002/jcp.20843. [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Stein JL, Lian JB, van Wijnen AJ, Montecino M, Young DW, Javed A, Pratap J, Choi JY, Ali SA, Pande S, Hassan MQ. Transcription-factor-mediated epigenetic control of cell fate and lineage commitment. Biochem Cell Biol. 2009;87:1–6. doi: 10.1139/o08-094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuardo M, Martinez M, Hidalgo K, Montecino M, Javed A, Lian JB, Stein GS, Stein JL, Gutierrez SE. Altered chromatin modifications in AML1/RUNX1 breakpoint regions involved in (8;21) translocation. J Cell Physiol. 2009;218:343–349. doi: 10.1002/jcp.21599. [DOI] [PubMed] [Google Scholar]
- Sun JM, Chen HY, Espino PS, Davie JR. Phosphorylated serine 28 of histone H3 is associated with destabilized nucleosomes in transcribed chromatin. Nucleic Acids Res. 2007;35:6640–6647. doi: 10.1093/nar/gkm737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KL, Talbot D, Lee MA, Leonhardt H, Jaenisch R. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc Natl Acad Sci U S A. 1996;93:12920–12925. doi: 10.1073/pnas.93.23.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vradii D, Doan DN, Wagner S, Nickerson JA, Lian JB, Stein JL, van Wijnen AJ, Imbalzano AN, Stein GS. Brg1, the ATPase subunit of SWI/SNF chromatin remodeling complex, is required for myeloid differentiation to granulocytes. J Cell Physiol. 2006;206:112–118. doi: 10.1002/jcp.20432. [DOI] [PubMed] [Google Scholar]
- Vradii D, Zaidi SK, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype. Proc Natl Acad Sci, USA. 2005;102:7174–7179. doi: 10.1073/pnas.0502130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf JJ, Hiebert SW. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J Cell Biochem. 1999 Suppl 32-33:51–58. doi: 10.1002/(sici)1097-4644(1999)75:32+<51::aid-jcb7>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007;26:91–101. doi: 10.1038/sj.onc.1209760. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Dowdy CR, van Wijnen AJ, Lian JB, Raza A, Stein JL, Croce CM, Stein GS. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAP kinase network. Cancer Research. 2009 doi: 10.1158/0008-5472.CAN-09-1567. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Pratap J, Schroeder TM, Westendorf J, Lian JB, van Wijnen AJ, Stein GS, Stein JL. Alterations in intranuclear localization of Runx2 affect biological activity. J Cell Physiol. 2006;209:935–942. doi: 10.1002/jcp.20791. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van WA, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SH, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue specific transcription factors in progeny cells. Proc Natl Acad Sci USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]


