Introduction
Sphingosine 1-phosphate (S1P) is a polar sphingolipid metabolite that regulates cell migration, differentiation, survival and complex physiological processes (Bartke et al. 2009). S1P functions primarily by activating a subgroup of the endothelial differentiation gene (EDG) family of G-protein coupled cell surface receptors now referred to as S1P1–5 (Lee et al. 1998; Rosen et al. 2009; Van Brocklyn et al. 1998). The biological effects of S1P receptor signaling involve several pathways including mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase (JNK), extracellular-signal-regulated kinase (ERK), phosphoinositide 3-kinase (PI3K), phospholipase C, phospholipase D and other downstream mediators (Cuvillier 2002; Pyne et al. 2009; Pyne et al. 2000; Sanchez et al. 2004; Yu et al. 2009). S1P signaling plays diverse roles in physiology, generally enhancing cell survival in response to stressful conditions, promoting vascular maturation and vascular permeability by fostering homotypic and heterotypic cell-cell interactions, and regulating lymphocyte egress from the thymus and peripheral lymphoid organs. These and other functions of S1P have been reviewed extensively elsewhere and, thus, will be mentioned herein only as they pertain to the specific functions of S1P lyase (SPL) (Bandhuvula et al. 2007b; Fyrst et al. 2008a; Kumar et al. 2009; Oskouian et al. 2007).
Pools of S1P available for autocrine, paracrine and potentially more distant signaling events are regulated through the actions of three enzyme activities: sphingosine kinase (SK) (E.C. 2.7.1.91) which synthesizes S1P by phosphorylating the long chain base sphingosine, phosphohydrolases which reverse the actions of SK, and S1P lyase (SPL) (E.C.4.1.2.27), a membrane-bound enzyme which irreversibly degrades S1P by cleaving the acyl chain between C2–3. The reaction catalyzed by SPL results in formation of hexadecenal and ethanolamine phosphate and depletion of intracellular S1P.
Many of the observed effects of SPL modulation can be attributed to alteration of intracellular pools of S1P, and as such this review will focus primarily on the regulation of SPL and its contribution to S1P-mediated biology. However, we emphasize that the products of the SPL reaction and more global effects of SPL on sphingolipid metabolism should be considered when investigating the potential mechanism of SPL’s influence in different experimental systems.
SPL in physiology and disease
SPL is the final enzyme in the sphingolipid degradative pathway and an important regulator of S1P as well as the levels of other sphingolipid intermediates which influence various aspects of cell growth, proliferation and death (Saddoughi et al. 2008). Ceramide is the backbone of all higher order sphingolipids and the molecule from which S1P is generated. Ceramide functions as a signaling molecule but, unlike S1P, ceramide generally mediates anti-proliferative responses. Ceramide generation is considered to be a key mechanism by which chemotherapeutic agents induce apoptosis in cancer cells. Conversely, S1P promotes cell growth, migration, tumor angiogenesis, invasion and metastasis (Figure 1). A dynamic balance between ceramide and S1P is maintained inside of cells, influencing cellular outcome in response to stress. SPL has the ability to shift the balance towards cell death by attenuating the proliferative S1P signal. Thus, SPL is necessary for maintaining lipid homeostasis and appropriate cell fate responses, whereas its dysregulation could potentially contribute to the pathophysiology of diseases, especially those in which the balance of cell growth and death is abnormal, such as cancer and degenerative diseases (Merrill 2002). SPL, as well as sphingosine 1-phosphate phosphatase (EC 3.1.3.-), have been shown to be downregulated in human colorectal carcinomas (Oskouian, 2006), and SPL expression and activity are significantly reduced in adenomas that develop in the Min mouse model of colon cancer in comparison to uninvolved surrounding intestinal tissue (Oskouian, 2006). These findings suggest that, whereas sphingosine kinase functions as an oncogene, SPL may serve an anti-oncogenic role in the gut.
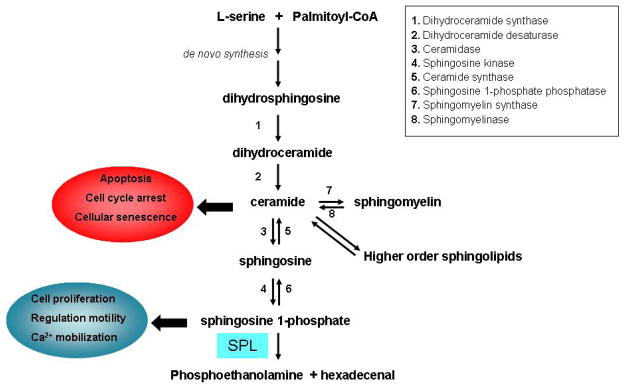
Figure 1.
Scheme of sphingolipid metabolism. Long-chain bases are formed by de novo synthesis or by degradation of higher order sphingolipids. S1P and ceramide have opposite effects in the regulation of cell metabolism.
The role of SPL in cell fate, physiology and development has been studied in several model systems. For example, an insertional mutagenesis study revealed that mutations in the Dictyostelium sglA gene, which encodes SPL, confer resistance to the anticancer drug cysplatin (Li et al. 2000). Moreover, overexpression of SPL enhanced the sensitivity of Dictyostelium cells to the drug (Min et al. 2004). These studies have translational relevance, as SPL has also been shown to sensitize mammalian cells to chemotherapeutic agents and DNA damage (Min et al. 2005; Reiss et al. 2004).
The lack of SPL expression in Drosophila Sply mutants results in dysregulation of sphingolipid metabolism. Sply mutants exhibited decreased fecundity and increased apoptosis in the reproductive organs (Herr et al. 2003; Phan et al. 2007). The sphingolipid metabolite responsible for this effect is unknown, although these tissues show a profound accumulation of long-chain bases including Δ4,6-sphingadienes, which promote apoptosis in Drosophila cell lines (Fyrst & Saba 2008a; Fyrst et al. 2008b). A similar accumulation of long chain bases (LCBs) was also found in the D. melanogaster cell line Schneider-2 (S2) following RNA interference against Sply (Fyrst et al. 2008b). Sply flies were also flightless, and histological analysis revealed a severe myopathy affecting the thoracic flight muscles (Herr et al. 2003). There is growing evidence that S1P regulates skeletal muscle differentiation and regeneration (Bruni et al. 2008; Donati et al. 2005). Thus, the Sply myopathy may foretell a role for SPL in maintenance of muscle homeostasis. Additional studies of the Sply model to explore the molecular mechanisms by which sphingolipids influences muscle biology are ongoing.
In the nematode C. elegans, spl-1 expression is critical for the maintenance of intestinal integrity and, as in the fruitfly model, normal reproduction. Without spl-1 expression, animals become bloated, pale, congested with eggs and hatched larvae, and demonstrate smaller gut and gonadal structures (Mendel et al. 2003).
In the protozoan parasite L. major, SPL mutants were found to be defective in stationary phase differentiation and virulence, raising the possibility that SPL may serve as a useful target for antibiotic therapy for leishmanial diseases (Zhang et al. 2007). Interestingly, it is the lack of ethanolamine phosphate rather than accumulation of S1P that is responsible for the organism’s lack of infectivity in the absence of SPL function. This case highlights the importance of considering SPL’s products as well as S1P in mediating its various effects in vivo.
Homozygous Sgpl1 knockout mice do not survive beyond 3–4 weeks after birth and demonstrate significant growth failure and anemia. Several congenital defects were reported, including vascular abnormalities, skeletal defects, thoracic malformations of sternum, ribs and vertebrae, and renal abnormalities. Embryonic fibroblasts from Sgpl1 knockout mice demonstrated defects of migration in vitro (Schmahl et al. 2007). This “wound healing” assay result is consistent with a previous report indicating that S1P can enhance wound healing in diabetic mice (Kawanabe et al. 2007). These findings suggest that SPL could be a useful therapeutic target for promoting traumatic or surgical wound healing (Francis-Goforth et al. 2009). SPL-deficient mice also showed lesions in lungs and alveoli that contained smooth homogeneous material accompanied by increased numbers of alveolar macrophages (Vogel et al. 2009). Whether these deposits represent inflammatory material or some other substance has not been determined, and thus the role of SPL in lung function remains uncertain. SPL null mice also developed cardiac lesions characterized by expansion of the interstitium by vacuoles and vacuolated mesenchymal cells separated by cardiomyocytes (Vogel et al. 2009). The physiological import of these changes has not been determined yet, but these findings suggest that SPL expression is important in the heart, even though baseline expression is very low in cardiac tissue (our unpublished observations).
S1P signaling plays an important role in the immune system by affecting cell survival, migration, cytokine secretion and immune cell differentiation (Rosen et al. 2007). It was recently demonstrated that partial loss of SPL activity affects immune function (Vogel et al. 2009). The immunomodulatory drug FTY720 functions as a sphingosine analog (Mandala et al. 2002). FTY720 treatment deprives thymocytes and lymphocytes of an S1P signal that stimulates their egress from thymus and secondary lymphoid tissues (Matloubian et al. 2004) and has also been shown to inhibit SPL activity (Bandhuvula et al. 2005). Inhibition of SPL by the food colorant 2-acetyl-4-(tetrahydroxybutyl)imidazole (THI) or reducing SPL expression through genetic approaches prevents lymphocyte egress from the thymus and secondary lymphoid organs (Schwab et al. 2005; Vogel et al. 2009). Replacement of the human SPL gene in the Sgpl1 null background resulted in SPL expression at 10–20% of normal mouse SPL levels, yet failed to restore normal T-cell development and trafficking (Vogel et al. 2009). Together, these data indicate that, like the S1P receptor analogs, SPL inhibitors may define a new class of immunosuppressant drugs. Toward that end, Lexicon Pharmaceuticals, Inc. has developed an orally delivered small molecule inhibitor of SPL, LX2931 (Lexicon Pharmaceuticals). Conversely, there is evidence that SPL may play a role in mediating inflammatory responses. Atopic dermatitis is a chronic relapsing inflammatory skin disease often associated with autoimmune diseases. Increased SPL expression has been reported in the skin of patients affected by atopic dermatitis (Seo et al. 2006; Wood et al. 2009). SPL upregulation might be related to inflammatory changes in the skin associated with the disease (Holleran et al. 2006). However, it is not clear whether SPL is mediating the disease or is instead a secondary response to inflammatory changes in the skin, which is replete with unique forms of ceramide that may require enhanced degradation under these conditions.
Recently, it has been reported that sphingolipid metabolism is altered in Alzheimer’s disease patients (He et al. 2008; Katsel et al. 2007). Accumulation of ceramide and reduction of S1P levels in the brains of Alzheimer’s patients shift sphingolipid signaling in the direction of cell death. The mRNA levels of SPL and other S1P-degrading enzymes are upregulated in Alzheimer’s disease (Katsel et al. 2007). Upregulation of SPL expression also correlated with clinical dementia progression (Katsel et al. 2007). Polymorphisms in Sglp1 gene have been described to confer susceptibility to late-onset Alzheimer’s disease (Morgan et al. 2007). Together, these findings suggest that SPL activity and function may contribute to neurodegenerative disease.
In summary, metazoan models illustrate that SPL is an enzyme that is essential for normal lipid homeostasis, and its absence leads to significant developmental and functional defects. Similarly, murine SPL knockout models exhibit early death and developmental abnormalities indicative of a role for SPL in specific organ functions. Inhibition of SPL appears to confer immunomodulatory effects, whereas upregulation of SPL and SPL polymorphisms are associated with human pathologic conditions including atopic dermatitis and Alzheimer’s disease. Whether SPL mutations are compatible with life and/or are responsible for human disease remains to be determined. Nonetheless, SPL appears to be a regulator of critical physiological processes and a target for pharmacological modulation of human disease.
Genes encoding SPL enzymes
Dihydrosphingosine phosphate lyase (Dpl1), the first SPL gene to be identified, was cloned through yeast genetics in which a screen for survival in the presence of D-erythro-sphingosine was employed (Saba et al. 1997), The identification of Dpl1 led to nucleotide homology-based searches resulting in the cloning and characterization of SPL enzymes from other organisms including C. elegans (Mendel et al. 2003), Mus musculus (Zhou et al. 1998), Dictyostelium discoideum (Li et al. 2001), Leishmania major (Zhang et al. 2007), Drosophila melanogaster (Herr et al. 2003) and Arabidopsis thaliana (Tsegaye et al. 2007)(Figure 2).
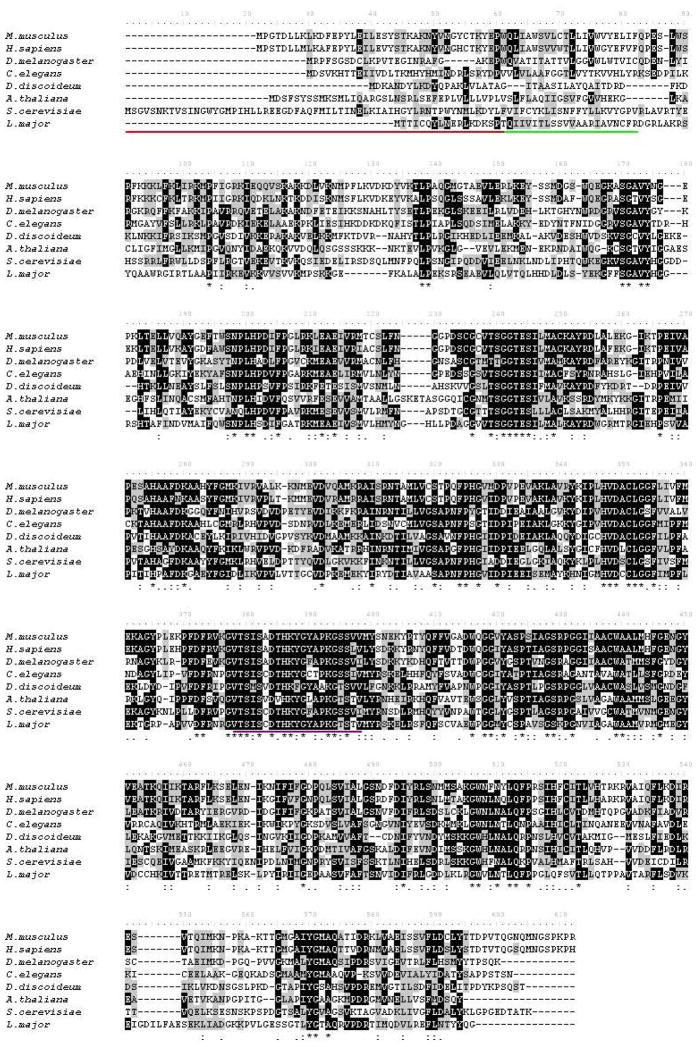
Figure 2.
Sequence alignment of SPL homologs. ClustalW alignment of SPL amino acid sequences of Homo sapiens (NP 003892), Mus musculus (NP 033189), Drosophila melanogaster (NP 652032), Caenorhabditis elegans (NP 499913), Saccharomyces cerevisiae (XP 751779), Dictyostelium discoideum (XP 639378), Leishmania major (CAJ06500) and Arabidopsis thaliana (AAG51494). Identical residues are indicated in black, and similar residues are indicated in gray. The underlined regions indicate the ER luminal domain (red), the transmembrane spanning domain (green) and the predicted pyridoxal phosphate binding (purple).
Van Veldhoven and collaborators were the first to identify the human SPL gene, which encodes a predicted protein of 568 amino acids with a molecular mass of 63.5 kDa (Van Veldhoven et al. 2000). The amino acid sequence of the murine SPL homolog displays 84% identity and 92% similarity to human SPL. Similarity in primary sequence has also been found between SPL homologs from S. cerevisiae, D. melanogaster, L. major, C. elegans and D. discoideum (Fyrst & Saba 2008a).
Tissue distribution of SPL
SPL is expressed in many mammalian tissues. In rodents, SPL activity and expression appear to be highest in the small intestine, colon, thymus, spleen and harderian gland, whereas moderate expression is observed in liver, kidney, lung, stomach and testis (our unpublished data). The lowest expression levels are found in heart, skeletal muscle and brain, with the exception of the olfactory mucosal epithelium, where the enzyme is highly enriched (Genter et al. 2003). The olfactory mucosa is a neuronal tissue unique in its ability to sustain continuous neurogenesis (Genter 2006). This tissue exemplifies the fact that SPL expression appears to be highest in tissues marked by rapid cell turnover, consistent with its ability to promote apoptosis. The major example of this is the pronounced expression and activity of SPL in intestinal epithelial cells, which are renewed every 12 h. SPL in intestinal epithelial cells also likely plays an important role in catabolizing dietary sphingolipids (Vesper et al. 1999). SPL might also be required to maintain low S1P levels in the cells at the villus tips, facilitating cell death and tissue turnover in response to oxidative stress and as a mechanism of gut immunity that helps to eliminate parasitic infestation (Cliffe et al. 2005; Radtke et al. 2005).
The high expression of SPL in thymus maintains low S1P levels in thymic tissues compared with the surrounding plasma and lymph, thereby generating S1P gradients that facilitate T cell egress into the circulation (Schwab et al. 2005). SPL expression is low in lymphocytes and absent in platelets and erythrocytes, which are the major sources of S1P in plasma (Ito et al. 2007; Yatomi et al. 1997a; Yatomi et al. 1997b). SPL is expressed in some inflammatory cells, but its activity in macrophages, monocytes, dendritic cells and neutrophils has not been characterized to date.
Subcellular localization of SPL
The first reports describing SPL activity by Stoffel found the majority of cellular SPL activity to be associated with microsomal fractions, with a lesser amount enriched in the inner mitochondrial membrane (Stoffel et al. 1969). Subsequent immunofluorescence studies established that mammalian SPL localizes primarily to the endoplasmic reticulum (ER). However, LegS2 (the Legionella homologue of eukaryotic SPL) was recently shown to be localized to the host cell mitochondria, rather than the ER (Degtyar et al. 2009). This raises the intriguing possibility that a pool of mammalian SPL may be located in mitochondria, where its product ethanolamine phosphate could contribute to formation of PE, which is enriched in the mitochondrial inner membrane. Interestingly, SPL was also found by proteomics approaches to be present in the mitochondrial associated membrane (MAM), a unique membrane structure involved in cell signalling and which is associated with both the mitochondria and the endoplasmic reticulum (Hayashi et al. 2009) (Anamaris Colberg-Poley, personal communication). These recent observations raise interesting questions regarding the potential participation of SPL in functions unique to the mitochondria and MAM.
Biochemical characterization of SPL
SPL belongs to the class of pyridoxal 5′-phosphate-dependent carbon-carbon lyases subclass of aldehyde lyases. The enzyme acts on the derivatives of sphingoid bases that contain a phosphate group in position 1. The cleavage products are an aliphatic fatty aldehyde and ethanolamine phosphate (Van Veldhoven 2000). SPL only recognizes the D-(+) erythro isomer of its substrates, which is the configuration of the natural occurring sphingoid bases (Van Veldhoven et al. 1991).
SPL activity has been measured using both radioactive and fluorescent substrates. Given the membrane-associated nature of SPL, accurate enzyme kinetic parameters are difficult to obtain and depend on the conditions used for the analysis. Using the radioactive LCBP substrates, the Km values for DHS1P determined for rat liver microsomes were from 9 to 16μM (Van Veldhoven & Mannaerts 1991), and the Km value found for DHS1P in HEK293 overexpressing human SPL was 20μM (Bandhuvula et al. 2007a). Using a fluorescent NBD-S1P substrate, the calculated Km was 15μM (Bandhuvula et al. 2007a), and for the BODIPY-C14S1P the value of Km was 35 μM (Bandhuvula et al. 2009)*
SPL topology
The topology of the enzyme in the endoplasmic reticulum (ER) membrane has been predicted by biochemical characterization and structural modelling. SPL belongs to the family of single-pass type III membrane proteins also referred to as type I without a cleavable N-terminal signal sequence (Fyrst & Saba 2008a; Ikeda et al. 2004; Van Veldhoven & Mannaerts 1991). There is only one transmembrane segment close to the N-terminus, which faces the ER lumen. The catalytic domain faces the cytosol facilitating the availability of the substrate to the active site (Van Veldhoven & Mannaerts 1991). In human and murine SPL, a stretch of amino acids that spans amino acids 344–364 showed 76% similarity to the pyridoxal 5′-phosphate binding site of class V aminotransferase (Fyrst & Saba 2008a; Van Veldhoven et al. 2000). This region contains two lysine residues which are highly conserved in all SPL homologs (human SPL positions 353 and 359) predicted to be involved in the aldimide link with pyridoxal 5′-phosphate. Mutagenesis studies of human SPL defined two cysteine residues C218 and C317 that are required for enzymatic activity (Van Veldhoven et al. 2000). C317 is conserved throughout all of the SPL sequences identified to date. In contrast, C218 is not found in D. melanogaster, D. discoideum, L. major and plant SPL genes. Interestingly, recombinant SPL lacking the transmembrane domain and the ER luminal sequence was found to be active in vitro when expressed in bacteria (Van Veldhoven et al. 2000).
The binding of the pyridoxal 5′-phosphate cofactor requires a Lys residue at the C-terminal domain that forms an internal Schiff base with the cofactor, which is sandwiched between Cys and His residues (Ikeda et al. 2004). The pyridinium nitrogen of the pyridoxal 5′-phoshate ring interacts electrostatically with the side chain carboxylate of an Asp residue (Mukhopadhyay et al. 2008).
An extensive study carried out by Capitani and co-workers evaluated the key residues required for activity of the S. cerevisiae SPL gene (Dpl1p) (Mukhopadhyay et al. 2008). According to this study, Dpl1p must oligomerize as a tetramer or hexamer to become active. The luminal domain was found to be important for protein stability. Dpl1p was also found to undergo N-linked glycosylation, which affected SPL stability, conformation and targeting (Mukhopadhyay et al. 2008).
Regulation of SPL
SPL activity is regulated at multiple levels, including epigenetic, transcriptional and post-translational. In simple metazoan organisms including Drosophila and C. elegans, SPL expression is largely restricted to the developing and adult gut (Herr et al. 2003; Mendel et al. 2003; Renault et al. 2002). During murine development, SPL mRNA expression is temporally regulated and is involved in early endodermal differentiation (Ikeda et al. 2004; Kihara et al. 2003). A conserved mechanism of transcriptional regulation of SPL expression by GATA transcription factors was identified and appears to be operational in nematodes and humans (Oskouian et al. 2005). SPL was also identified as an immediate early gene and transcriptional target of platelet-derived growth factor (PDGF) (Chen et al. 2004). Analysis of the 5′-flanking region of the SPL gene suggests that other mechanisms of transcriptional control are involved in the regulation of SPL expression. An investigation of the promoter region of murine Sgpl1 identified cis-elements, early growth response factor, zinc-binding protein factor and GC-box factor motifs (Hutton et al. 2004). Sgpl1 was also found along with other pro-apoptotic genes to be a downstream target of mSin3A, a member of a co-repressor complex involved in embryonic development and in regulating cellular functions such as cell cycle progression, proliferation, DNA repair, apoptosis and mitochondrial metabolism (Dannenberg et al. 2005).
The identification of human SPL in a screen for nitrosylated proteins suggests that the enzyme is regulated by post-translational modifications, specifically by nitrosylation on tyrosine residues Y356 and Y366 (Zhan et al. 2006). The alteration of the charge of those tyrosine residues, which reside inside the active site, could interfere with enzyme-substrate binding. Multiple protein kinase specific phosphorylation sites (7 Ser, 3 Thr and 7 Tyr residues) are also predicted by protein sequence analysis, although this fact has not been verified experimentally (Huang et al. 2005). In a recent study, Dpl1p was shown to form higher order complexes, and oligomerization of Dpl1p was found to be required for in vivo function (Mukhopadhyay et al. 2008). Although these findings have not been tested in the human SPL protein, the notion that the enzyme could function as a multimer subject to regulation by control over subunit assembly is intriguing.
Sphingosine 1-phosphate lyase activity assays
Radiometric Assays
The measurement of SPL activity was originally described using a radioactive assay in which conversion of a radiolabeled substrate to a long chain aldehyde that retained the label was monitored. The most accepted radiometric method for SPL activity involves the use of [4,5-3H]dihydrosphingosine phosphate (Van Veldhoven & Mannaerts 1991). SPL is solubilized in Triton X-100, which does not interfere with enzyme activity. The reaction mixture also contains pyridoxal 5′-phosphate cofactor, as well as phosphatase inhibitors. Radiolabeled products are separated by TLC and the regions of interest are scraped into scintillation vials and the radioactivity is counted. The drawback of this assay system is the necessity of using radioactive materials, the time required to develop the autoradiograms, and the lack of commercial sources of high quality substrate.
Fluorescence Assay
More recently, our laboratory has developed several fluorescence-based methods to measure SPL activity. One method uses 7-nitrobenz-2-oxa-1,3-diazole-sphingosine 1-phosphate (NBD-S1P) as a substrate, which leads to production of NBD-hexadecenal and phosphoethanolamine (Bandhuvula et al. 2007a). Another method relies on a substrate containing a boron dipyrromethene difluoride (BODIPY) group. Advantages associated with the BODIPY fluorophore include better photochemical stability, high fluorescent intensity (Pagano et al. 1991) and insensitivity to polarity and pH of the environment (Bandhuvula et al. 2009). In both cases, the fluorophore is attached to the omega carbon to avoid steric hindrance that could interfere with the SPL reaction. Another approach to measure SPL activity makes use of a fluorogenic substrate as was previously described for measuring activity of hydrolytic enzymes (Badalassi et al. 2000). An example of this technique involves the utilization of a coumarinic sphinganine 1-phosphate analog as an SPL substrate. After SPL catalyzes the cleavage if the substrate, a coumarinic aldehyde is produced which then undergoes β-elimination at neutral-alkaline pH to release the fluorescent product, umbelliferone. The umbelliferone group is not fluorescent while it is still linked to the sphinganine 1-phosphate backbone. This method allows the use of microtiter plates and could be employed for the screening of potential SPL inhibitors (Bedia et al. 2009)(Figure 3).
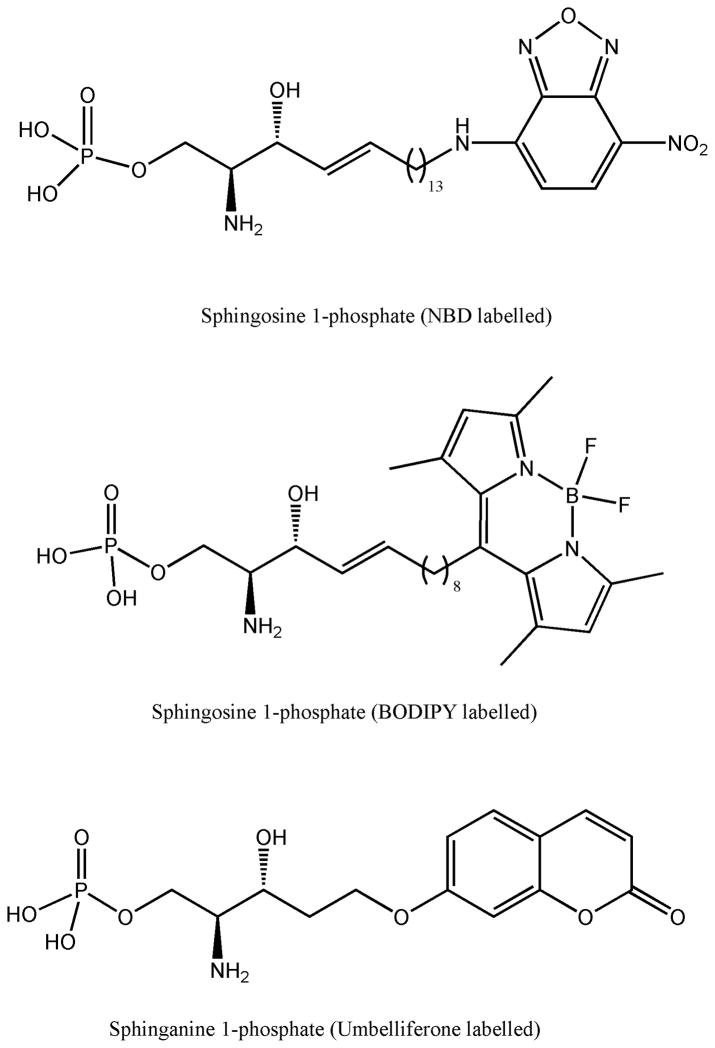
Figure 3.
Structure of fluorescent substrates for SPL activity assays. BODIPY substrates have a different emission wavelength depending on the concentration of the label. Coumarinic substrate only shows fluorescence when it is not linked to the S1P backbone.
THI and other pharmacological agents that modulate SPL activity
The S1P receptor super-agonist FTY720 produces lymphopenia by preventing T lymphocytes from recognizing the S1P gradient between tissues and circulating lymph. This gradient is required for egress of mature S1P1-expressing T lymphocytes from the thymus as well as their egress from peripheral lymphoid organs after interacting with antigen (Schwab et al. 2007). It was recently discovered that THI, a component of caramel food coloring, also produces lymphopenia through inhibition of lymphocyte trafficking, but in the latter case the effect appears to be due to inhibition of SPL, which results in attenuation of the S1P gradient (Schwab & Cyster 2007). This observation revealed the potential of SPL to serve as a therapeutic target for immune modulation (Schwab et al. 2005). Oral administration of THI results in reduced tissue SPL activity and elevated levels of tissue and circulating S1P; however, the drug does not inhibit SPL in vitro. It has been suggested that THI is neither an active-site inhibitor of SPL nor acts by inhibiting pyridoxal 5′-phosphate incorporation to generate the holoenzyme. THI may act indirectly, may require metabolism to an active state or may be dependent on the formation of a higher-order complex (Bagdanoff et al. 2009; Mukhopadhyay et al. 2008; Nussbaumer 2008).
THI derivatives designed based on the structure of S1P and pyridoxal 5′-phosphate have been synthesized and screened for their ability to induce lymphopenia in mice (Bagdanoff et al. 2009). Several modifications were made to THI in the core heterocycle, in the side chain and in the position 2 of the imidazole ring. In this latter one, the α,α-difluoro analogue has remarkable ability to deplete lymphocyte levels but generated high toxicity due to HF poisoning. Adding another heterocycle in this position showed depletion of lymphocyte levels (Bagdanoff et al. 2009). Those compounds, however, have not been tested in vitro and their mechanism of action remains unknown. As described above, LX2931 Phase I clinical trials are now complete and Phase II trials for the treatment of rheumatoid arthritis have been initiated (Lexicon Pharmaceuticals). LX2931 has been described to reduce inflammatory responses in challenge models of inflammation, autoimmunity and transplantation (Oravecz et al. 2008). For example, preclinical studies showed a consistent reduction in circulating lymphocyte counts in multiple species and reduced joint inflammation and prevented arthritic destruction of joints in mouse and rat models of arthritis (Brown et al. 2008; Frazier et al. 2009; Oravecz et al. 2008; Oravecz et al. 2007). The pyridoxal 5′phosphate analog 4′-deoxypyridoxine (DOP) inhibits the activity of SPL and other enzymes dependent on this cofactor. Interestingly, DOP is also known to induce lymphopenia, presumably by reducing the S1P gradient. SPL activity is profoundly inhibited in the thymus of DOP-treated mice, an effect which can be overcome by treatment with excess cofactor coincident with restoration of SPL activity (Schwab et al. 2005). Since DOP inhibition is not specific for SPL and affects the activity of many other enzymes including those involved in DNA synthesis, long-term exposure is associated with considerable toxicity (Billich et al. 2008)(Figure 4).
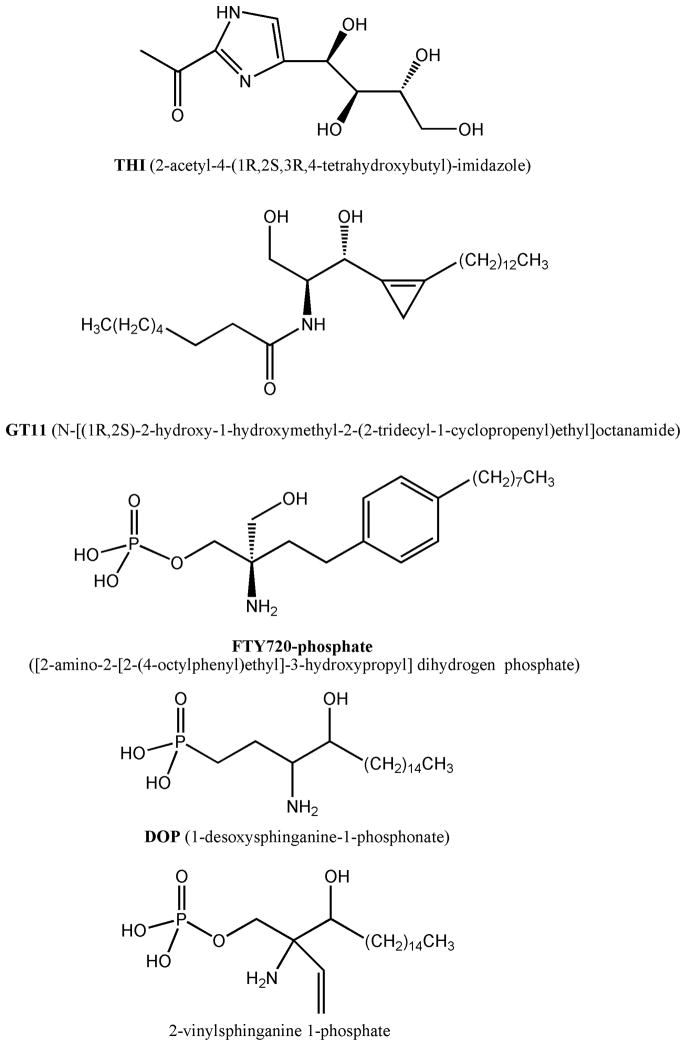
Figure 4.
Chemical structure of known SPL inhibitors. Other than THI, all the inhibitors shown have a chemical structure mimicking S1P.
The active, phosphorylated form of FTY720 remains stable after 24 h in the presence of SPL in vitro indicating that the drug is not degraded by the actions of this enzyme. Interestingly, unphosphorylated FTY720 serves as a weak inhibitor of SPL activity both in vitro and in vivo (Bandhuvula et al. 2005). SPL inhibition by FTY720 occurs in murine tissues with the same kinetics as drug-induced lymphopenia through a mechanism that does not appear to involve changes in SPL gene transcription or protein expression.
A number of other compounds have been shown to inhibit SPL activity, but generally they are not appropriate for in vivo use due to lack of specificity and high toxicity. For example, one of the first reported SPL inhibitors was the S1P analogue 1-desoxysphinganine 1-phosphonate which acts as a competitive inhibitor with a Ki of 5 μM (Stoffel et al. 1974). Similarly, the S1P analog 2-vinyl dihydrosphingosine was synthesized and exhibited an IC50 of 2.4 μM against SPL (Boumendjel et al. 1994). Another compound that may function as an SPL inhibitor is the ceramide analog N-[(1R,2S)-2-hydroxy-1-hydroxymethyl-2-(2-tridecyl-1-cyclopentenyl)ethyl]octanamide (GT11). This compound was found to be a dihydroceramide desaturase inhibitor but was additionally observed to induce accumulation of S1P in vivo when plasma concentrations above 5 μM were achieved. Like THI, GT11 fails to inhibit SPL in vitro (Triola et al. 2004). Thus, metabolism of GT11 to an active intermediate or indirect effect may be involved in its mechanism of action upon SPL.
None of the SPL inhibitors thus far identified demonstrates high specificity, and the complete toxicity profiles have not been evaluated. Considering that spl knockout murine models have demonstrated a number of developmental and postnatal abnormalities, it will be important to fully examine the effects of prolonged complete or partial inhibition of SPL as inhibitors are deployed in the clinic(Hagen et al. 2009; Kihara et al. 2003; Vogel et al. 2009).
Future perspectives
S1P is a potent signaling molecule involved in cell stress responses, cancer, angiogenesis and lymphocyte trafficking. Tight control of S1P levels in tissues and the circulation is required to prevent untoward cell growth, to maintain immune function and to control vascular physiology. SPL may serve as a useful target for modulating intracellular S1P pools and may provide a sink for extracellular S1P as well. SPL inhibition has already been shown to modulate lymphocyte trafficking. Its potential to modulate other physiological endpoints such as inflammatory responses, malignant growth and tissue homeostasis remain to be tested. To date, no specific small molecule inhibitors of SPL have been described. Generating specific inhibitors is an important goal that would facilitate the study of SPL activity and function and potentially offer a therapeutic strategy to treat certain pathological conditions by increasing circulating and/or tissue S1P levels. The development of high-throughput screening methods to measure SPL activity would aid in the discovery of new specific inhibitors.
Summary
SPL is a highly conserved enzyme responsible for the irreversible degradation of the bioactive lipid, S1P. SPL regulates intracellular pools of S1P available for ligation of its five receptors and as such has a strong influence upon autocrine, paracrine and distant S1P signaling events. SPL is differentially expressed in mammalian tissues, is itself highly regulated at transcriptional and posttranslational levels, and requires oligomerization for normal function. Inhibitors are being developed to SPL, which may be useful therapeutic agents for a variety of diseases in which S1P has been implicated, including cancer, autoimmune disorders and inflammation.
Acknowledgments
Dr. Serra is awarded with a Beatriu de Pinós postdoctoral fellowship from Generalitat de Catalunya (2007 BP-A 00101). Amb el support del Comissionat per a Universitats i Recerca del Departament d’Innovació, Universitats i Empresa de la Generalitat de Catalunya. This work was supported by Public Health Service grants CA77538, GM066954 and CA129438 (JDS).
Footnotes
The difference in value is probably due to the difference in chain length of the two substrates.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badalassi F, Wahler D, Klein G, Crotti P, Reymond J. A Versatile Periodate-Coupled Fluorogenic Assay for Hydrolytic Enzymes. Angewandte Chemie. 2000;39 (22):4067–70. doi: 10.1002/1521-3773(20001117)39:22<4067::aid-anie4067>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Bagdanoff JT, Donoviel MS, Nouraldeen A, Tarver J, Fu Q, Carlsen M, Jessop TC, Zhang H, Hazelwood J, Nguyen H, Baugh SD, Gardyan M, Terranova KM, Barbosa J, Yan J, Bednarz M, Layek S, Courtney LF, Taylor J, Digeorge-Foushee AM, Gopinathan S, Bruce D, Smith T, Moran L, O’Neill E, Kramer J, Lai Z, Kimball SD, Liu Q, Sun W, Yu S, Swaffield J, Wilson A, Main A, Carson KG, Oravecz T, Augeri DJ. Inhibition of sphingosine-1-phosphate lyase for the treatment of autoimmune disorders. J Med Chem. 2009;52 (13):3941–53. doi: 10.1021/jm900278w. [DOI] [PubMed] [Google Scholar]
- Bandhuvula P, Fyrst H, Saba JD. A rapid fluorescence assay for sphingosine-1-phosphate lyase enzyme activity. J Lipid Res. 2007a;48 (12):2769–78. doi: 10.1194/jlr.D700010-JLR200. [DOI] [PubMed] [Google Scholar]
- Bandhuvula P, Li Z, Bittman R, Saba JD. Sphingosine 1-phosphate lyase enzyme assay using a BODIPY-labeled substrate. Biochem Biophys Res Commun. 2009;380 (2):366–70. doi: 10.1016/j.bbrc.2009.01.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandhuvula P, Saba JD. Sphingosine-1-phosphate lyase in immunity and cancer: silencing the siren. Trends Mol Med. 2007b;13 (5):210–7. doi: 10.1016/j.molmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Bandhuvula P, Tam YY, Oskouian B, Saba JD. The immune modulator FTY720 inhibits sphingosine-1-phosphate lyase activity. J Biol Chem. 2005;280 (40):33697–700. doi: 10.1074/jbc.C500294200. [DOI] [PubMed] [Google Scholar]
- Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res. 2009;50 (Suppl):S91–6. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedia C, Camacho L, Casas J, Abad JL, Delgado A, Van Veldhoven PP, Fabrias G. Synthesis of a fluorogenic analogue of sphingosine-1-phosphate and its use to determine sphingosine-1-phosphate lyase activity. Chembiochem. 2009;10 (5):820–2. doi: 10.1002/cbic.200800809. [DOI] [PubMed] [Google Scholar]
- Billich A, Baumruker T. Sphingolipid metabolizing enzymes as novel therapeutic targets. Subcell Biochem. 2008;49:487–522. doi: 10.1007/978-1-4020-8831-5_19. [DOI] [PubMed] [Google Scholar]
- Boumendjel A, Miller SPF. Synthesis of an inhibitor of sphingosine-1-phosphate lyase. Tetrahedron Lett. 1994;35 (6):819–22. [Google Scholar]
- Brown P, Frazier K, Augeri D, Walke DW, Pappas C, Brooks B, Donoviel M, Oravecz T. LX2931: A potential small molecule treatment for autoimmune disorders. 2008 Annual Meeting of the American College of Rheumatology; 2008. [Google Scholar]
- Bruni P, Donati C. Pleiotropic effects of sphingolipids in skeletal muscle. Cell Mol Life Sci. 2008;65 (23):3725–36. doi: 10.1007/s00018-008-8236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat Genet. 2004;36 (3):304–12. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308 (5727):1463–5. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- Cuvillier O. Sphingosine in apoptosis signaling. Biochim Biophys Acta. 2002;1585 (2–3):153–62. doi: 10.1016/s1388-1981(02)00336-0. [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19 (13):1581–95. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyar E, Zusman T, Ehrlich M, Segal G. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- Donati C, Meacci E, Nuti F, Becciolini L, Farnararo M, Bruni P. Sphingosine 1-phosphate regulates myogenic differentiation: a major role for S1P2 receptor. Faseb J. 2005;19 (3):449–51. doi: 10.1096/fj.04-1780fje. [DOI] [PubMed] [Google Scholar]
- Francis-Goforth KN, Harken AH, Saba JD. Normalization of diabetic wound healing. Surgery. 2009 doi: 10.1016/j.surg.2009.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier K, Brooks B, Freiman J, Oravecz T, Augeri D, Brown P. LX2931: Potential Small Molecule Treatment for Rheumatoid Arthritis (RA) and Other Inflammatory Disorders. 2009 Annual Meeting of the European League Against Rheumatism (EULAR); 2009. [Google Scholar]
- Fyrst H, Saba JD. Sphingosine-1-phosphate lyase in development and disease: sphingolipid metabolism takes flight. Biochim Biophys Acta. 2008a;1781 (9):448–58. doi: 10.1016/j.bbalip.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrst H, Zhang X, Herr DR, Byun HS, Bittman R, Phan VH, Harris GL, Saba JD. Identification and characterization by electrospray mass spectrometry of endogenous Drosophila sphingadienes. J Lipid Res. 2008b;49 (3):597–606. doi: 10.1194/jlr.M700414-JLR200. [DOI] [PubMed] [Google Scholar]
- Genter MB. Molecular biology of the nasal airways: how do we assess cellular and molecular responses in the nose? Toxicol Pathol. 2006;34 (3):274–80. doi: 10.1080/01926230600713491. [DOI] [PubMed] [Google Scholar]
- Genter MB, Van Veldhoven PP, Jegga AG, Sakthivel B, Kong S, Stanley K, Witte DP, Ebert CL, Aronow BJ. Microarray-based discovery of highly expressed olfactory mucosal genes: potential roles in the various functions of the olfactory system. Physiol Genomics. 2003;16 (1):67–81. doi: 10.1152/physiolgenomics.00117.2003. [DOI] [PubMed] [Google Scholar]
- Hagen N, Van Veldhoven PP, Proia RL, Park H, Merrill AH, Jr, van Echten-Deckert G. Subcellular origin of sphingosine 1-phosphate is essential for its toxic effect in lyase-deficient neurons. J Biol Chem. 2009;284 (17):11346–53. doi: 10.1074/jbc.M807336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19 (2):81–8. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Huang Y, Li B, Gong CX, Schuchman EH. Deregulation of sphingolipid metabolism in Alzheimer’s disease. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr DR, Fyrst H, Phan V, Heinecke K, Georges R, Harris GL, Saba JD. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development. 2003;130 (11):2443–53. doi: 10.1242/dev.00456. [DOI] [PubMed] [Google Scholar]
- Holleran WM, Takagi Y, Uchida Y. Epidermal sphingolipids: metabolism, function, and roles in skin disorders. FEBS Lett. 2006;580 (23):5456–66. doi: 10.1016/j.febslet.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Huang HD, Lee TY, Tzeng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33 (Web Server issue):W226–9. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton JJ, Jegga AG, Kong S, Gupta A, Ebert C, Williams S, Katz JD, Aronow BJ. Microarray and comparative genomics-based identification of genes and gene regulatory regions of the mouse immune system. BMC Genomics. 2004;5 (1):82. doi: 10.1186/1471-2164-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Kihara A, Igarashi Y. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5′-phosphate binding domain exposed to the cytosol. Biochem Biophys Res Commun. 2004;325 (1):338–43. doi: 10.1016/j.bbrc.2004.10.036. [DOI] [PubMed] [Google Scholar]
- Ito K, Anada Y, Tani M, Ikeda M, Sano T, Kihara A, Igarashi Y. Lack of sphingosine 1-phosphate-degrading enzymes in erythrocytes. Biochem Biophys Res Commun. 2007;357 (1):212–7. doi: 10.1016/j.bbrc.2007.03.123. [DOI] [PubMed] [Google Scholar]
- Katsel P, Li C, Haroutunian V. Gene expression alterations in the sphingolipid metabolism pathways during progression of dementia and Alzheimer’s disease: a shift toward ceramide accumulation at the earliest recognizable stages of Alzheimer’s disease? Neurochem Res. 2007;32 (4–5):845–56. doi: 10.1007/s11064-007-9297-x. [DOI] [PubMed] [Google Scholar]
- Kawanabe T, Kawakami T, Yatomi Y, Shimada S, Soma Y. Sphingosine 1-phosphate accelerates wound healing in diabetic mice. J Dermatol Sci. 2007;48 (1):53–60. doi: 10.1016/j.jdermsci.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Kihara A, Ikeda M, Kariya Y, Lee EY, Lee YM, Igarashi Y. Sphingosine-1-phosphate lyase is involved in the differentiation of F9 embryonal carcinoma cells to primitive endoderm. J Biol Chem. 2003;278 (16):14578–85. doi: 10.1074/jbc.M211416200. [DOI] [PubMed] [Google Scholar]
- Kumar A, Saba JD. Lyase to live by: Sphingosine phosphate lyase as a therapeutic target. Expert Opin Ther Targets. 2009 doi: 10.1517/14728220903039722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Van Brocklyn JR, Thangada S, Liu CH, Hand AR, Menzeleev R, Spiegel S, Hla T. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279 (5356):1552–5. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- Lexicon Pharmaceuticals I. http://www.lexicon-genetics.com/pipeline/clinical-trials/lx2931-lx2932.html.
- Li G, Alexander H, Schneider N, Alexander S. Molecular basis for resistance to the anticancer drug cisplatin in Dictyostelium. Microbiology. 2000;146 ( Pt 9):2219–27. doi: 10.1099/00221287-146-9-2219. [DOI] [PubMed] [Google Scholar]
- Li G, Foote C, Alexander S, Alexander H. Sphingosine-1-phosphate lyase has a central role in the development of Dictyostelium discoideum. Development. 2001;128 (18):3473–83. doi: 10.1242/dev.128.18.3473. [DOI] [PubMed] [Google Scholar]
- Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, Thornton R, Shei GJ, Card D, Keohane C, Rosenbach M, Hale J, Lynch CL, Rupprecht K, Parsons W, Rosen H. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296 (5566):346–9. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427 (6972):355–60. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- Mendel J, Heinecke K, Fyrst H, Saba JD. Sphingosine phosphate lyase expression is essential for normal development in Caenorhabditis elegans. J Biol Chem. 2003;278 (25):22341–9. doi: 10.1074/jbc.M302857200. [DOI] [PubMed] [Google Scholar]
- Merrill AH., Jr De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J Biol Chem. 2002;277 (29):25843–6. doi: 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- Min J, Stegner AL, Alexander H, Alexander S. Overexpression of sphingosine-1-phosphate lyase or inhibition of sphingosine kinase in Dictyostelium discoideum results in a selective increase in sensitivity to platinum-based chemotherapy drugs. Eukaryot Cell. 2004;3 (3):795–805. doi: 10.1128/EC.3.3.795-805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Van Veldhoven PP, Zhang L, Hanigan MH, Alexander H, Alexander S. Sphingosine-1-phosphate lyase regulates sensitivity of human cells to select chemotherapy drugs in a p38-dependent manner. Mol Cancer Res. 2005;3 (5):287–96. doi: 10.1158/1541-7786.MCR-04-0197. [DOI] [PubMed] [Google Scholar]
- Morgan AR, Turic D, Jehu L, Hamilton G, Hollingworth P, Moskvina V, Jones L, Lovestone S, Brayne C, Rubinsztein DC, Lawlor B, Gill M, O’Donovan MC, Owen MJ, Williams J. Association studies of 23 positional/functional candidate genes on chromosome 10 in late-onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2007;144B (6):762–70. doi: 10.1002/ajmg.b.30509. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Howell KS, Riezman H, Capitani G. Identifying key residues of sphinganine-1-phosphate lyase for function in vivo and in vitro. J Biol Chem. 2008;283 (29):20159–69. doi: 10.1074/jbc.M709753200. [DOI] [PubMed] [Google Scholar]
- Nussbaumer P. Medicinal chemistry aspects of drug targets in sphingolipid metabolism. ChemMedChem. 2008;3 (4):543–51. doi: 10.1002/cmdc.200700252. [DOI] [PubMed] [Google Scholar]
- Oravecz T, Conoviel MS, Kramer JA, Moran LB, Sun W, Swaffield J, Augeri DJ. Sphingosine 1-phosphate lyase is a potential therapeutic target in autoimmune diseases including rheumatoid arthritis. 2008 ACR Meeting; 2008. [Google Scholar]
- Oravecz T, Donoviel MS, Anderson SJ, Carson K, Sun W, Swaffield J, Liu Q, Kimball SD, Piggot JR, Main AJ, Zambrowicz BP, Sands AS, Turner CA, Augeri DJ. Genetic and chemical inhibition of sphingosine-1-phosphate lyase results in peripheral lymphopenia and alleviates disease development in animal models of inflammation and autoimmunity. 2007 Annual Meeting of the American Society of Hematology; 2007. [Google Scholar]
- Oskouian B, Mendel J, Shocron E, Lee MA, Jr, Fyrst H, Saba JD. Regulation of sphingosine-1-phosphate lyase gene expression by members of the GATA family of transcription factors. J Biol Chem. 2005;280 (18):18403–10. doi: 10.1074/jbc.M410928200. [DOI] [PubMed] [Google Scholar]
- Oskouian B, Saba J. Sphingosine-1-phosphate metabolism and intestinal tumorigenesis: lipid signaling strikes again. Cell Cycle. 2007;6 (5):522–7. doi: 10.4161/cc.6.5.3903. [DOI] [PubMed] [Google Scholar]
- Pagano RE, Martin OC, Kang HC, Haugland RP. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol. 1991;113 (6):1267–79. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan VH, Herr DR, Panton D, Fyrst H, Saba JD, Harris GL. Disruption of sphingolipid metabolism elicits apoptosis-associated reproductive defects in Drosophila. Dev Biol. 2007;309 (2):329–41. doi: 10.1016/j.ydbio.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne NJ, Long JS, Lee SC, Loveridge C, Gillies L, Pyne S. New aspects of sphingosine 1-phosphate signaling in mammalian cells. Adv Enzyme Regul. 2009;49 (1):214–21. doi: 10.1016/j.advenzreg.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Pyne S, Pyne NJ. Sphingosine 1-phosphate signalling in mammalian cells. Biochem J. 2000;349 (Pt 2):385–402. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307 (5717):1904–9. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Reiss U, Oskouian B, Zhou J, Gupta V, Sooriyakumaran P, Kelly S, Wang E, Merrill AH, Jr, Saba JD. Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J Biol Chem. 2004;279 (2):1281–90. doi: 10.1074/jbc.M309646200. [DOI] [PubMed] [Google Scholar]
- Renault AD, Starz-Gaiano M, Lehmann R. Metabolism of sphingosine 1-phosphate and lysophosphatidic acid: a genome wide analysis of gene expression in Drosophila. Mech Dev. 2002;119 (Suppl 1):S293–301. doi: 10.1016/s0925-4773(03)00131-x. [DOI] [PubMed] [Google Scholar]
- Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S. Sphingosine 1-phosphate receptor signaling. Annu Rev Biochem. 2009;78:743–68. doi: 10.1146/annurev.biochem.78.072407.103733. [DOI] [PubMed] [Google Scholar]
- Rosen H, Sanna MG, Cahalan SM, Gonzalez-Cabrera PJ. Tipping the gatekeeper: S1P regulation of endothelial barrier function. Trends Immunol. 2007;28 (3):102–7. doi: 10.1016/j.it.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272 (42):26087–90. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem. 2008;49:413–40. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez T, Hla T. Structural and functional characteristics of S1P receptors. J Cell Biochem. 2004;92 (5):913–22. doi: 10.1002/jcb.20127. [DOI] [PubMed] [Google Scholar]
- Schmahl J, Raymond CS, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat Genet. 2007;39 (1):52–60. doi: 10.1038/ng1922. [DOI] [PubMed] [Google Scholar]
- Schwab SR, Cyster JG. Finding a way out: lymphocyte egress from lymphoid organs. Nat Immunol. 2007;8 (12):1295–301. doi: 10.1038/ni1545. [DOI] [PubMed] [Google Scholar]
- Schwab SR, Pereira JP, Matloubian M, Xu Y, Huang Y, Cyster JG. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309 (5741):1735–9. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- Seo EY, Park GT, Lee KM, Kim JA, Lee JH, Yang JM. Identification of the target genes of atopic dermatitis by real-time PCR. J Invest Dermatol. 2006;126 (5):1187–9. doi: 10.1038/sj.jid.5700234. [DOI] [PubMed] [Google Scholar]
- Stoffel W, Grol M. Chemistry and biochemistry of 1-desoxysphinganine 1-phosphonate (dihydrosphingosine-1-phosphonate) Chem Phys Lipids. 1974;13 (4):372–88. doi: 10.1016/0009-3084(74)90011-5. [DOI] [PubMed] [Google Scholar]
- Stoffel W, LeKim D, Sticht G. Distribution and properties of dihydrosphingosine-1-phosphate aldolase (sphinganine-1-phosphate alkanal-lyase) Hoppe Seylers Z Physiol Chem. 1969;350 (10):1233–41. doi: 10.1515/bchm2.1969.350.2.1233. [DOI] [PubMed] [Google Scholar]
- Triola G, Fabrias G, Dragusin M, Niederhausen L, Broere R, Llebaria A, van Echten-Deckert G. Specificity of the dihydroceramide desaturase inhibitor N-[(1R,2S)-2-hydroxy-1-hydroxymethyl-2-(2-tridecyl-1-cyclopropenyl)ethyl]o ctanamide (GT11) in primary cultured cerebellar neurons. Mol Pharmacol. 2004;66 (6):1671–8. doi: 10.1124/mol.104.003681. [DOI] [PubMed] [Google Scholar]
- Tsegaye Y, Richardson CG, Bravo JE, Mulcahy BJ, Lynch DV, Markham JE, Jaworski JG, Chen M, Cahoon EB, Dunn TM. Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J Biol Chem. 2007;282 (38):28195–206. doi: 10.1074/jbc.M705074200. [DOI] [PubMed] [Google Scholar]
- Van Brocklyn JR, Lee MJ, Menzeleev R, Olivera A, Edsall L, Cuvillier O, Thomas DM, Coopman PJ, Thangada S, Liu CH, Hla T, Spiegel S. Dual actions of sphingosine-1-phosphate: extracellular through the Gi-coupled receptor Edg-1 and intracellular to regulate proliferation and survival. J Cell Biol. 1998;142 (1):229–40. doi: 10.1083/jcb.142.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven PP. Sphingosine-1-phosphate lyase. Methods Enzymol. 2000;311:244–54. doi: 10.1016/s0076-6879(00)11087-0. [DOI] [PubMed] [Google Scholar]
- Van Veldhoven PP, Gijsbers S, Mannaerts GP, Vermeesch JR, Brys V. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22(1) Biochim Biophys Acta. 2000;1487 (2–3):128–34. doi: 10.1016/s1388-1981(00)00079-2. [DOI] [PubMed] [Google Scholar]
- Van Veldhoven PP, Mannaerts GP. Subcellular localization and membrane topology of sphingosine-1-phosphate lyase in rat liver. J Biol Chem. 1991;266 (19):12502–7. [PubMed] [Google Scholar]
- Vesper H, Schmelz EM, Nikolova-Karakashian MN, Dillehay DL, Lynch DV, Merrill AH., Jr Sphingolipids in food and the emerging importance of sphingolipids to nutrition. J Nutr. 1999;129 (7):1239–50. doi: 10.1093/jn/129.7.1239. [DOI] [PubMed] [Google Scholar]
- Vogel P, Donoviel MS, Read R, Hansen GM, Hazlewood J, Anderson SJ, Sun W, Swaffield J, Oravecz T. Incomplete inhibition of sphingosine 1-phosphate lyase modulates immune system function yet prevents early lethality and non-lymphoid lesions. PLoS One. 2009;4 (1):e4112. doi: 10.1371/journal.pone.0004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SH, Clements DN, Ollier WE, Nuttall T, McEwan NA, Carter SD. Gene expression in canine atopic dermatitis and correlation with clinical severity scores. J Dermatol Sci. 2009;55 (1):27–33. doi: 10.1016/j.jdermsci.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Yatomi Y, Igarashi Y, Yang L, Hisano N, Qi R, Asazuma N, Satoh K, Ozaki Y, Kume S. Sphingosine 1-phosphate, a bioactive sphingolipid abundantly stored in platelets, is a normal constituent of human plasma and serum. J Biochem. 1997a;121 (5):969–73. doi: 10.1093/oxfordjournals.jbchem.a021681. [DOI] [PubMed] [Google Scholar]
- Yatomi Y, Yamamura S, Ruan F, Igarashi Y. Sphingosine 1-phosphate induces platelet activation through an extracellular action and shares a platelet surface receptor with lysophosphatidic acid. J Biol Chem. 1997b;272 (8):5291–7. doi: 10.1074/jbc.272.8.5291. [DOI] [PubMed] [Google Scholar]
- Yu H, Okada T, Kobayashi M, Abo-Elmatty DM, Jahangeer S, Nakamura SI. Roles of extracellular and intracellular sphingosine 1-phosphate in cell migration. Genes Cells. 2009 doi: 10.1111/j.1365-2443.2009.01295.x. [DOI] [PubMed] [Google Scholar]
- Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem. 2006;354 (2):279–89. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Zhang K, Pompey JM, Hsu FF, Key P, Bandhuvula P, Saba JD, Turk J, Beverley SM. Redirection of sphingolipid metabolism toward de novo synthesis of ethanolamine in Leishmania. Embo J. 2007;26 (4):1094–104. doi: 10.1038/sj.emboj.7601565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Saba JD. Identification of the first mammalian sphingosine phosphate lyase gene and its functional expression in yeast. Biochem Biophys Res Commun. 1998;242 (3):502–7. doi: 10.1006/bbrc.1997.7993. [DOI] [PubMed] [Google Scholar]