Abstract
AeSCP-2, a sterol carrier protein, is involved in sterol trafficking in mosquitoes. The activity of the AeSCP-2 gene is important for mosquito development. An earlier study demonstrated that the transcription of this gene was up-regulated by 20-hydroxyecdysone (20E) in cultured gut tissues. To investigate 20E-regulated transcription of AeSCP-2 gene we truncated the upstream flanking region of AeSCP-2 gene and linked it to a reporter gene. The mosquito Aag-2 cell line was transfected with these promoter/reporter constructs and treated with 20E at various concentrations. Expression vectors of different transcription factors such as HR3 and βFTZ-F1 were also co-transfected with the AeSCP-2 promoter/reporter constructs. The observed results demonstrated that varied combinations of transcription factors produce different promoter activities of the AeSCP-2 gene. This observation leads us to the conclusion that the partnership of transcription factors is crucial in regulating transcriptional activity of AeSCP-2 gene.
Keywords: cholesterol, sterol carrier protein, transcription factor, insect hormone
Introduction
Insects as all other animals require cholesterol for their cellular membranes and for steroid hormone syntheses. Cholesterol is the precursor for ecdysteroid biosyntheses (Borovsky et al., 1986; Grieneisen et al., 1991). Insects do not however have the ability to synthesize cholesterol de novo and depend on dietary intake or symbionts to provide cholesterol for their physiological needs (Clayton, 1964; Dwivedi et al., 1982; Nes et al., 1997; Noda et al., 1979; Ritter et al., 1981).
Sterol carrier protein-2 is a small intracellular protein that facilitates the uptake of lipids in vivo (Dyer et al., 2008; Gallegos et al., 2001). The first insect SCP-2 was identified in the yellow fever mosquito, Aedes aegypt (Krebs et al., 2003). Aedes aegypti SCP-2 (AeSCP-2) binds to cholesterol (Krebs and Lan, 2003) and fatty acids (Dyer et al., 2003). AeSCP-2 is localized mostly in cytosol with some presence in mitochondria and nucleus, but not in membranous vesicles (Lan et al., 2004). The vertebrate SCP-2/x gene transcribes SCP-2 and SCP-x mRNAs via two independent promoters of the gene (Gallegos et al., 2001). Previous studies in SCP-2 transcriptional regulation were conducted in vertebrates (McLean et al., 1989; Rennert et al., 1991; Trzeciak et al., 1987) and these studies have demonstrated that SCP-2 gene expression appears to be under the control of factors such as adrenocorticotropic hormone and gonadotropins via cyclic adenosine monophosphate (cAMP) activation (Lopez et al., 2007), which in turn stimulates steroidogenesis (Lopez et al., 2001). It has been demonstrated that both the vertebrate SCP-2 and SCP-x are up-regulated in the presence of cAMP and SF-1, a steroidogenic factor, homologous to βFTZ-F1 in insects (Lopez et al., 2007; Lopez et al., 2001). In insects, experiments with AeSCP-2 gene expression regulation demonstrated that addition of 20-hydroxyecdysone (20E) to cultured larval gut tissues increased mRNA levels by 2 fold after a 12 hour exposure (Krebs and Lan, 2003). However, the molecular mechanism of 20E-induced AeSCP-2 expression is unknown.
According to the Ashburner model of 20-hydroxyecdysone (20E) action, 20E rapidly and directly induces a small set of early regulatory genes (Ashburner, 1973). The protein products of these early genes exhibit two opposing regulatory functions: first to repress their own expression, and second to activate large sets of secondary-response genes to provide the competence for subsequent regulatory responses (Karim et al., 1992). The ecdysteroid acts through a heterodimeric receptor complex consisting of the ecdysone receptor (EcR) and ultraspiracle (USP) to initiate the expression cascade of transcription factors to inactivate and/or activate tissue specific genes, as well as to activate directly several structural genes (Lan et al., 1999). While ecdysone pulses directly induce a small set of early genes, induction of other genes is delayed. One of these delayed early genes is hormone receptor 3 (HR3), a homolog of the vertebrate retinoid orphan receptor (ROR), that has additional requirements for early ecdysone-induced protein synthesis (Thummel, 1995). The βFTZ-F1 gene is a late gene in the 20E-induced gene expression cascade, and is induced in mid-prepupae in Drosophila as HR3 is repressed (Woodard et al., 1994). βFTZ-F1 is repressed both by ecdysone and its own expression, defining a narrow window of activity during the period of lowered hormone titer in mid-prepupae. A recent study in the levels of different transcription factors in Ae. aegypti 4th instar revealed that HR3 expression starts increasing in the period between ten to twenty four hours after the molt, while βFTZ-F1 expression stays low and is increased only much later in the development (Nishiura et al., 2007).
Based on these previous findings in SCP-2 transcriptional regulation in vertebrates and our knowledge of insect hormonal regulation of transcription, it was of interest to determine whether 20E has an effect on AeSCP-2 promoter activity in cultured cells and what transcription factors may be involved in the regulation of AeSCP-2 gene expression. To answer this question, we cloned and sequenced the flanking region of AeSCP-2 gene and studied the regulatory function of the 5’ flanking sequences upstream of the translation start site by using transient transfection assays in Ae. aegypti Aag-2 cells.
Results
Organization of the AeSCP-2 gene and promoter region
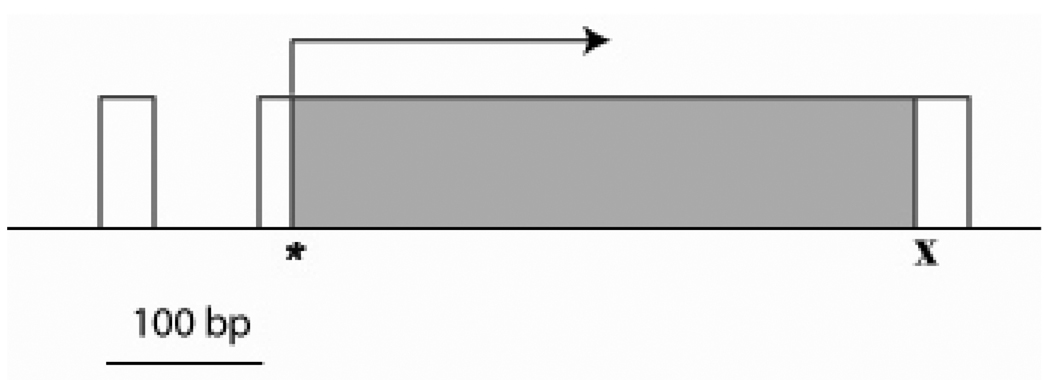
The 6.5 kb subclone of the genomic DNA containing AeSCP-2 gene was sequenced from both directions (FJ554568). There are over 30 independent ESTs from various sources that match to the AeSCP-2 gene sequence (http://compbio.dfci.harvard.edu/tgi/temp/blastn-a_aegypti-16513-1200078472.html), including the 5’UTR and 3’ UTR (Krebs and Lan, 2003). Therefore, the existence of AeSCP-2 gene in the Ae. aegypti genome is in no doubt. Interestingly, AeSCP-2 genomic DNA sequence could not be found in the newly annotated Aedes aegypti genome due to the lack of full coverage of the annotated genome. The 6.5 kb genomic sequence was analyzed for gene structure and putative regulatory elements. The AeSCP-2 gene is composed of two exons, the first exon is 36 bp and includes 5’ UTR and is followed by an intron which is 75 bp in length (Fig. 1). The second exon is 568 bp in length and contains both start and stop codons (Fig. 1). AeSCP-2 promoter lacks the TATA box-like element, but contains TCAGT sequence that matches to the arthropod capsite consensus sequence (Cherbas et al., 1993) near the transcription start site (Fig. 1).
Figure 1.
Genomic DNA organization of the AeSCP-2 gene. “*”- indicates a start codon, “x” – indicates a stop codon. Exons are shown as boxes, coding sequences are shown in gray, arrow indicate the transcription direction
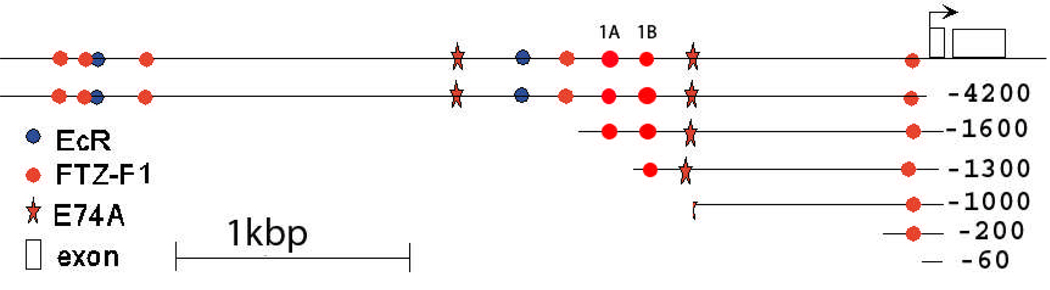
In order to identify possible regulatory elements for the AeSCP-2 gene in the 5’ upstream region, a search for known regulatory motifs and transcription factors was conducted on 4200 bp of upstream sequence from the transcription start site using both MatInspector (Cartharius et al., 2005) and AliBaba 2.1 (Grabe, 2002), the web-based promoter analysis software programs. Several putative regulatory elements with high similarity scores were identified and include seven βFTZ-F1 responsive elements (4162 bp, 3830 bp, 3490 bp, 1720 bp, 1345 bp, 1265 bp and 210 bp upstream of transcription start site), two E74A responsive elements (2531 bp and 1109 bp) and two EcR responsive elements (3808 bp and 2088 bp) (Fig. 2).
Figure 2.
The map of putative responsive elements of transcription factors identified by computational search in upstream, flanking sequence of the AeSCP-2 gene and constructs of truncated AeSCP-2 promoter.
Effects of 20E treatment on AeSCP-2 mRNA levels in cultured tissues
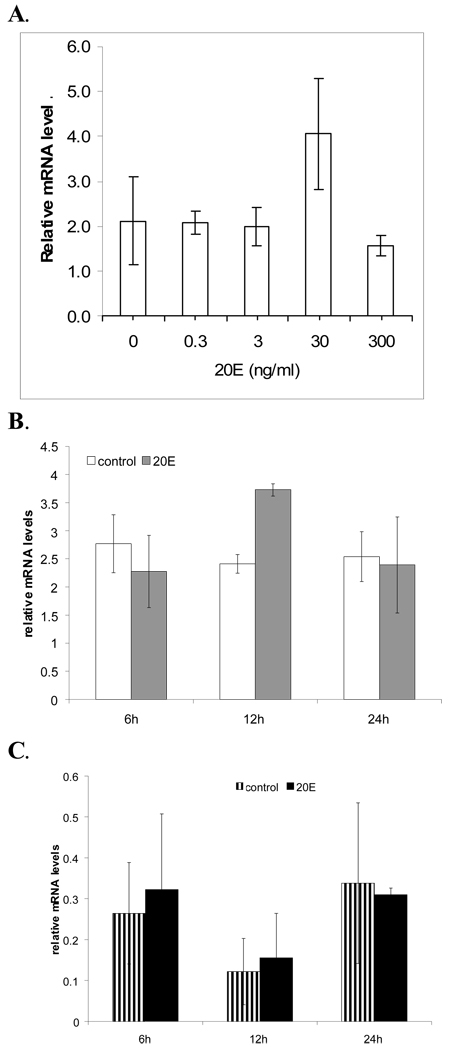
It was previously demonstrated that AeSCP-2 mRNA levels increase in cultured larval gut tissue after being exposed to 20E (60 ng/ml or 0.13 µM) for 12 hours (Krebs and Lan, 2003). The 20E-induced AeSCP-2 expression is detected in a dosage-dependent fashion. At concentrations between 3 to 30 ng/ml, 20E-treatment significantly increased AeSCP-2 mRNA levels (Fig. 3A and B; p<0.02). However, at a much higher concentration (300 ng/ml), 20E did not affect AeSCP-2 expression in the gut tissue cultures (p=0.3). To investigate the time course of ecdysteroid-induced AeSCP-2 expression, we treated gut tissue from 24 hour old Ae. aegypti 4th instar in in vitro cultures with 20E (60 ng/ml) for six, twelve and twenty four hours. Relative mRNA levels of AeSCP-2 did not change in the case of six and twenty four hour treatments, but mRNA levels significantly increased at 12 hours as expected (Fig. 3B, p<0.01), indicating that 20E-induced expression of AeSCP-2 gene was transient. We repeated the experiment with carcasses of the same 24 hour old Ae. aegypti from which the gut tissue were obtained. The results from this experiment demonstrate that relative mRNA levels of AeSCP-2 were not affected by 20E in the carcasses (Fig 3C, p=0.4). These results indicate that effect of 20-induced expression of AeSCP-2 was not only transient but also tissue specific.
Figure 3.
The induction of AeSCP-2 expression by 20E in tissue cultures. A. Dosage-response of AeSCP-2 gene transcription in larval gut tissue cultures after 12 hours incubation. B. Time course-response of AeSCP-2 gene transcription in larval gut tissue cultures. C. Time course-response of AeSCP-2 gene transcription in larval carcasses tissue cultures. Bar = mean values ± S.D. (N= 3–6). Relative mRNA level = normalized to the level detected in newly molted 4th instars which is arbitrarily set at 1.
Effects of HR3 and βFTZ-F1 on 20E induction of AeSCP-2 promoter
The sequential expression patterns of 20E-regulated transcription factors have been well established in insect model systems (Riddiford et al., 2003). It is known that expression of 20E-induced early genes such as EcR, USP, and E75 precede the early-late genes such as HR3 and HR39 (Horner et al., 1995). It was previously reported that the ecdysone titer peaks during the last larval stage in insects coinciding with increased expression of the HR3 transcription factor, which precedes the peak in βFTZ-F1 expression (Sun et al., 1994; Weller et al., 2001). AeSCP-2 expression peaks along with the small increase of 20E titer at 24 hours in the last larval stage (Lan et al., 2004), coinciding with the small increase of HR3 (Nishiura et al., 2007). The fact that 20E-induced AeSCP-2 expression in vitro gut tissue cultures took longer than 6 hours suggests that AeSCP-2 gene is neither early nor early-late 20E-responsive genes. To determine whether AeSCP-2 promoter activity is affected by early-late genes such as transcription factors βFTZ-1 and HR-3, we co-transfected Aag-2 cells with AeSCP-2 promoter/reporter constructs and pIE1hr-expression vectors of several 20E-responsive transcription factors (Hiruma et al., 2001). These transfected cells were treated with 5 µM 20E and the relative CAT levels were measured.
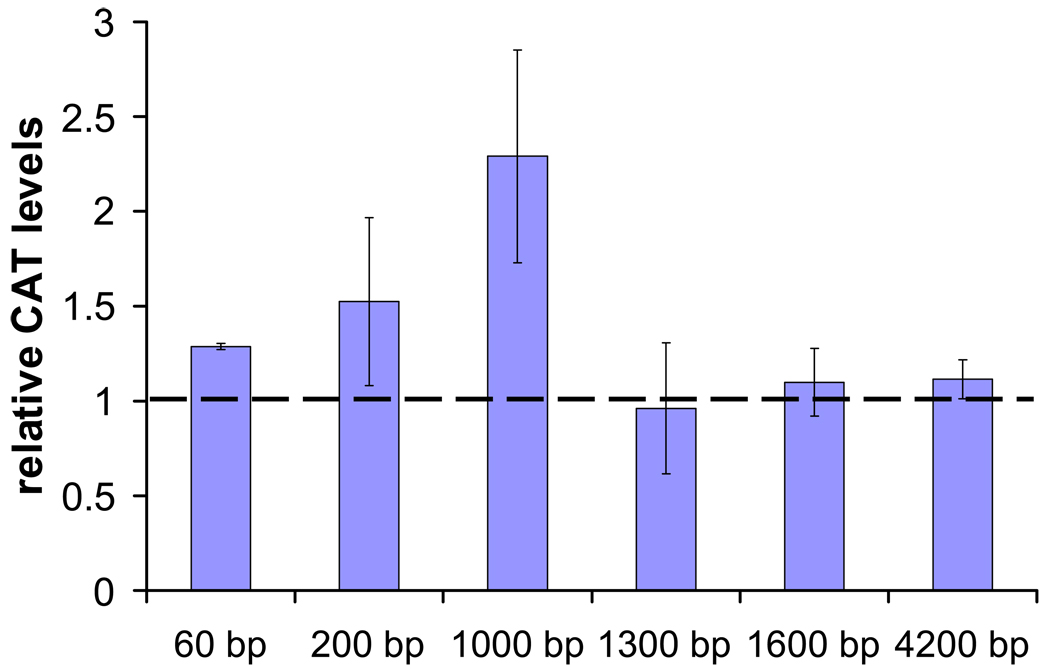
AeSCP-2 promoter activity was examined in Aag-2 cells transfected with AeSCP-2 promoter/reporter gene. The −4.2 kb construct contains two putative EcREs (Fig. 2), addition of 5 µM 20E did not changed the promoter activities (Table 2, −.4.2 kb). However, when −4.2 kb construct was co-transfected with pIE1hr-EcRb/ pIE1hr-USPa, there were significant 20E-induced promoter activities (Table 2, −4.2 kb/EcRb/USPa). Whereas, there was no change in −4.2 kp construct promoter activity when co-transfected with pIE1hr-EcRa/ pIE1hr-USPb (data not shown). Truncation of the putative EcREs from the upstream region abolished the EcRb/USPa mediated 20E-induced promoter activity (Table 2, −1.6 kb/EcRa/USPb). The results indicate that EcRb/USPa may regulate AeSCP-2 gene expression via one or both putative EcREs up stream of the promoter. For the −1.6 kb construct, addition of 20E caused a significant decrease of promoter activity by 34% (Table 2, −1.6 kb). On the other hand, 20E induced a significant increase of promoter activity with −1.3 kb upstream promoter/report gene construct (Table2, −1.3 kb). The results suggest that sequences within −1.6 and −1.3 kb were responsive to 20E, even though classic EcR responsive element was not found via computation (Fig. 2).
Table 2.
The effects of 20E on the promoter activity of AeSCP-2 gene constructs
| constructs | control | 5 µM 20E | t test (p values) | replicates |
|---|---|---|---|---|
| −4.2 kb | 1.00 | 0.88 | 0.352 | 18 |
| −4.2 kb/EcRb/USPa | 1.00 | 1.53 | 0.0004 | 6 |
| −1.6 kb/EcRb/USPa | 1.00 | 0.91 | 0.331 | 6 |
| −1.6 kb | 1.00 | 0.66 | 0.0001 | 31 |
| −1.3 kb | 1.00 | 1.29 | 0.028 | 6 |
| −60 bp | 1.00 | 0.94 | 0.665 | 12 |
When pIE1hr-HR3 was co-transfected with −4.2 kb, −1.6 kb or −1.3 kb AeSCP-2 promoter constructs, the addition of 5 µM 20E did not cause any significant change in relative CAT level after 12 hour exposure (Fig. 4; p≤0.2). The same treatment of 5 µM 20E caused a 2 fold increase in relative CAT level in cells co-transfected with pIE1hr-HR3 with the −1.0 kb AeSCP-2 promoter construct (Fig. 4; p≤0.001). The same effect was observed when pIE1hr-HR3 was co-transfected with shorter AeSCP-2 promoter constructs of −200 bp and −60 bp (Fig. 4; p≤0.001 and 0.003, respectively). The results indicated that HR3 may be partially responsible for the 20E-induced AeSCP-2 expression and the HR3 responsive element(s) is located within 1 kb upstream of the transcription start site. However, within the 1 kb 5’ flanking region of AeSCP-2 gene, computational programs did not detect any consensus responsive element for HR3.
Figure 4.
Transient transfection assay of AeSCP-2 promoter activity as determined by the increase in CAT protein after co-transfection with pIE1hr-HR3 and treated with 5 µM of 20E for 12 hours. Bar = mean values ± S.D. (N= 6–12). Relative CAT levels = normalized to the level detected in untreated cells which is arbitrarily set at 1.
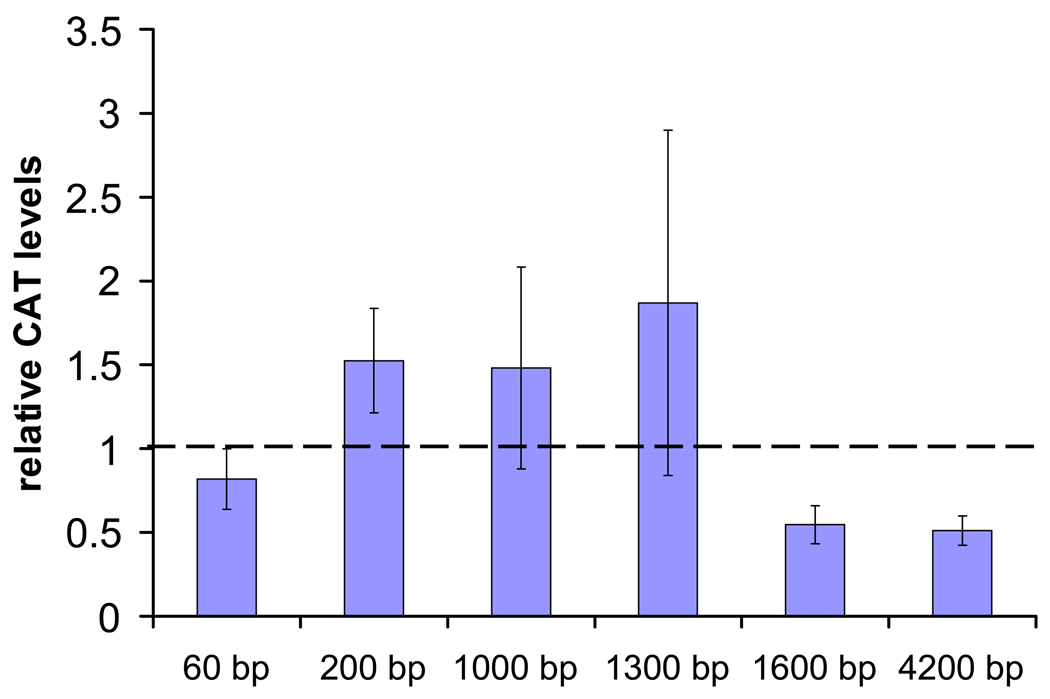
When pIE1hr-βFTZ-F1 was co-transfected with −4.2 kb and −1.6 kb AeSCP-2 promoter constructs the addition of 5 µM 20E caused significant decrease of 2 fold in relative CAT level after 12 hour exposure (Fig. 5; p≤0.04 and 0.02, respectively). The same treatment of 5 µM 20E did not cause significant changes in relative CAT level in cells co-transfected with pIE1hr-βFTZ-F1 and −1.3 kb, −1.0 kb, −200 bp or −60 bp AeSCP-2 promoter construct (Fig. 5; p≥0.1). The results suggest that βFTZ-F1, a 20E-responsive late gene, may be involved in the down regulation of AeSCP-2 gene and the βFTZ-F1 responsive element(s) is located between 1.3 to 1.6 kb upstream of the transcription start site (Fig. 2).
Figure 5.
Transient transfection assay of AeSCP-2 promoter activity as determined by the increase in CAT protein after co-transfection with pIE1hr-βFTZ-F1 and treated with 5 µM of 20E for 12 hours. Bar = mean values ± S.D. (N= 6–12). Relative CAT levels = normalized to the level detected in untreated cells which is arbitrarily set at 1.
Binding of βFTZ-F1 to the putative regulatory elements
There are two potential βFTZ-F1 responsive elements within 1.6 kb in the 5’ upstream region of AeSCP-2 gene. βFTZ-F1-1A and -1B elements are located at 1345 and 1265 bp upstream of the transcription start site, respectively (Fig. 2). Truncation of βFTZ-F1-1A from the promoter region resulted in 20E-induced up-regulation of AeSCP-2 transcription activities (Table 2, −1.3 kb), whereas the inclusion of βFTZ-F1-1A in the promoter region led to the 20E-induced suppression of transcription activities (Table 2, −1.6 kb). Moreover, βFTZ-F1 over-expression suppressed promoter activities of the −1.6 kb construct when exposed to 20E, whereas under the similar conditions, there was no significant change in promoter activities of the −1.3 kb construct (Fig. 5). To examine whether the putative βFTZ-F1 regulatory elements in the −1.6 kb upstream region of the AeSCP-2 gene bind to βFTZ-1, we used oligonucleotides corresponding to the sequences of βFTZ-F1-1A and -1B for electrophoretic mobility shifting assays (EMSAs) to detect DNA/protein interactions.
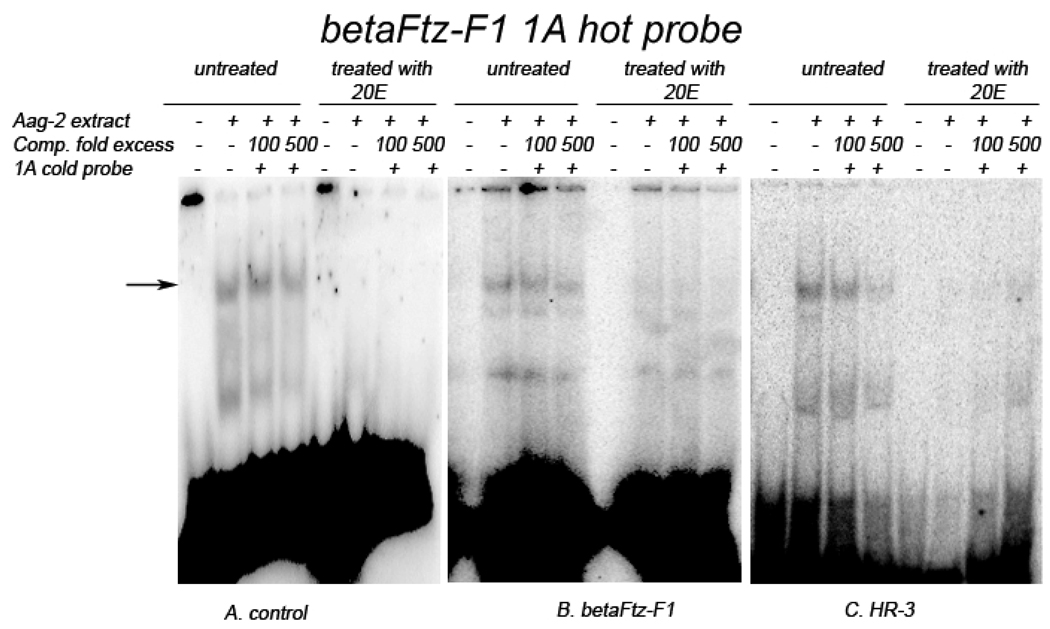
Two shifted bands appeared after βFTZ-F1-1A oligo probes were incubated with Aag-2 cellular extracts. The lower molecular weight shifted band did not seem to change its intensity under various treatment conditions, and considered to be nonspecific binding. The higher molecular weight shifted band showed that βFTZ-F1-1A specifically binds to a protein in the Aag-2 cell extracts, which was weakly competed off by 500-fold cold probes (Fig. 6, the arrowed). However, 20E-treatment abolished the protein binding to βFTZ-F1-1A probes (Fig. 6A, treated). When βFTZ-F1-1A was incubated with extracts from cells over-expressing βFTZ-F1, the DNA/protein binding was much weaker (Fig. 6B, untreated), and was almost undetectable after 20E treatment (Fig. 6B, treated). The same effect was observed with protein-binding of βFTZ-F1-1A to extracts from cells over-expressing HR-3, the addition of 20E abolished protein-binding to βFTZ-F1-1A (Fig. 6C, treated). However, with the cellular extract from the HR3 over-expressing cells that were not treated with 20E, protein-binding to βFTZ-F1-1A was not reduced as was seen with extract from βFTZ-F1 over-expressing cells (Fig. 6B and C, untreated). The results suggest that βFTZ-F1-1A element interacts with in an endogenous factor and that a 20E-induced factor or 20E/FTZ-F1 significantly weaken this interaction. This finding is consistent with the observation that 20E/βFTZ-F1 suppressed the promoter activity of the −1.6 kb AeSCP-2 construct in Aag-2 cell culture (Fig. 5). The results suggest that βFTZ-F1 may be responsible for the weakening of βFTZ-F1-1A/protein-binding under the influence of 20E.
Figure 6.
EMSAs for protein-binding of βFTZ-F1-1A. 32P-labaled double-stranded βFTZ-F1-1A oligonucleotide probes (20,000 cmp) were incubated with 5 µg of Aag-2 cell extract in the presence or absence of unlabeled probe-specific competitors. A. Cell extracts with no co-transfection, B. Cell extracts cotransfected with pIE1hr-βFTZ-F1, and C. Cell extracts cotransfected with pIE1hr-HR3
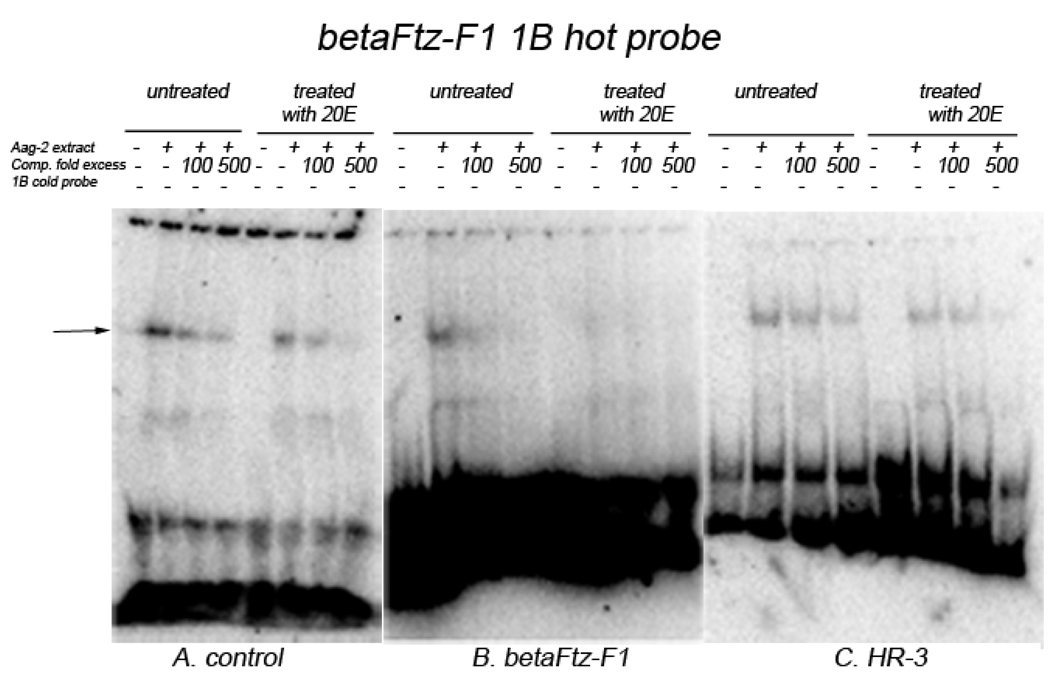
The βFTZ-F1-1B probe bound specifically to a protein in Aag-2 cell extract, which was strongly competed off by 100-fold excess of cold probes (Fig. 7A, the arrowed). Moreover, βFTZ-F1-1B/protein interaction was more intense in cell extracts from the untreated experiments than that of extracts from cells treated with 20E (Fig 7A). Under the similar conditions, βFTZ-F1-1B/protein interaction was noticeably stronger when compared with βFTZ-F1-1A protein-binding in protein extracts from both 20Etreated and untreated cells (Fig. 6A and 7A). When the βFTZ-F1-1B probe was incubated with extracts from cells over-expressing βFTZ-F1, the probe/protein band was weaker (Fig. 7B, untreated) and was almost undetectable after 20E treatment (Fig. 7B, treated). The binding of βFTZ-F1-1B probes to the protein in extracts from cells over-expressing HR3 did not change noticeably in protein extracts from either untreated or treated with 20E cells (Fig. 7C), even though overall protein-binding intensity was slightly weaker than that to the cell extracts without HR3 over-expression (Fig. 7A, untreated).
Figure 7.
EMSAs for protein-binding of βFtz-F1-1B. 32P-labaled double-stranded βFTZF1-1B oligonucleotide probes (20,000 cmp/lane) were incubated with 5 µg of Aag-2 cell extract in the presence or absence of unlabeled probe-specific competitors. A. Cell extracts with no co-transfection, B. Cell extracts co-transfected with pIE1hr-βFTZ-F1 and C. Cell extracts co-transfected with pIE1hr-HR3
Discussion
When AeSCP-2 genomic organization was compared to Anopheles gambiae SCP-2 gene we found that An. gambiae SCP-2 (AGAP003091) has a similar organization with two exons (40 bp and 659 bp) and one intron (186 bp) in the 5’ UTR. On the other hand, Drosophila melanogaster SCP-2 (CG12269), mosquito SCP-2like genes (AAEL012703, AAEL015661, AGAP004094, AGAP004093) have two exons with the starting codon in ExonI (Vyazunova et al., 2007). These differences in this gene organization might indicate repeated independent evolutionary events.
Previous studies in vertebrates (humans, mice and rats) demonstrated that SCP-2/SCP-x played a role in steroidogenesis in adrenocortical cells, and suggested that SCP2 may not be the putative high turnover "labile protein#x00022; involved in acute steroidogenesis (Chanderbhan et al., 1986). It was also demonstrated that steroid hormones have a direct effect on SCP-2 expression in vertebrates. For example injection of estradiol (steroid hormone) into roosters causes a 2-fold increase of SCP-2 mRNA (Pfeifer et al., 1993). In rats treatments with gonadotropic hormones, which stimulate luteinization of the cells, also increase SCP-2 gene expression several fold (Rennert et al., 1991). In this study we also found that the addition of 20E to gut tissue culture (Fig.3A) has an effect on AeSCP-2 expression after 12 hour incubation period, corresponding to the second ecdysone peak in 4th instar Ae. aegypti at 44–66 hours that induces the larval-pupal molt (Lan et al., 2004; Margam et al., 2006). This physiological ecdysone peak might be one of the explanations why 20E applications did not increase AeSCP-2 expression after 6 and 24 hours treatments in gut tissue cultures (Fig. 3B). However, we also report here that 20E treatment did not change expression of AeSCP-2 in carcasses tissues (Fig. 3C). Both these findings support the hypothesis that AeSCP-2 is regulated by 20E but has a narrow window during the development of 4th instar when this regulation might occur. It is also evident that 20E regulation of AeSCP-2 requires tissue specific factors that are present in gut tissues but are absent in carcasses.
In mosquito, isoforms of EcR and USP have different expression profiles in larvae and in blood-fed adult females. It seems that expression of Aedes EcRb and USPa peak at the same time, whereas Aedes EcRa and USPb have more coordinated expression patterns (Margam et al., 2006; Wang et al., 2002). AeSCP-2 −4.2 kb upstream sequence contains two putative EcREs (Fig. 2) that only responded to the over-expression of AeEcRb/USPa under the influence of 20E (Table 2, −4.2 kb EcRb/USPa). The result is consistent with the hypothesis that Aedes EcRb and USPa heterodimer complex may be the functional partnership in vivo (Parthasarathy et al., 2007).
It was observed that 20E-responsiveness in AeSCP-2 upstream regulatory region was not restricted to the putative EcREs since −1.6 and −1.3 kb constructs showed altered promoter activities when 20E was present (Table 2, −1.6 kb and −1.3 kb). It is speculated that other 20E-induced transcription factor(s) may be involved in the AeSCP-2 gene transcription regulation. In vertebrates it was demonstrated that SCP-2/SCP-x gene expression is up-regulated by SF-1 transcription factor and cAMP (Lopez et al., 2007; Lopez et al., 2001). Regulation by both these factors is similar to the regulatory mechanisms observed for other genes involved in steroid hormone production (Menon et al., 1974). In insects the homolog of SF-1 transcription factor is βFTZ-F1 and it has been studied in detail in several model organisms. However, studies of mosquito βFTZ-F1’s function have been mostly focused on its role in the reproduction cycle of Ae. aegypti (Zhu et al., 2003; Zhu et al., 2006). In the fat body of Ae. aegypti females the protein-protein interaction between βFtz-F1 and a p160/SRC coactivator of the ecdysone receptor forms a multiple protein complex with EcR/USP to activate vitellogenin promoter, which is shown to be critical in the 20E-mediated stage-specific expression of genes during vitellogenesis. Drosophila βFTZ-F1 is also an essential competence factor for stage-specific responses to ecdysone signals (Broadus et al., 1999).
Our findings demonstrate that βFTZ-F1 does not induce expression of AeSCP-2 in the presence of 20E, but suppresses it (Fig. 5). The fact that βFTZ-F1-mediated suppression is observed only in the case of longer promoter/reporter constructs of −1600 bp and 4200 bp suggests that putative transcription factor βFTZ-F1-1A located at −1345 bp may be involved in this regulation. Interestingly, βFTZ-F1 did not seem to bind directly to βFTZ-F1-1A (Fig. 6B). EMSA experiments showed that the addition of 20E reduced protein-binding to βFTZ-F1-1A (Fig. 6A) and over-expression of βFTZ-F1 abolished protein/βFTZ-F1-1A interaction (Fig. 6B). The results imply that βFTZ-F1 blocked protein-binding to βFTZ-F1-1A, which in turn may affect the AeSCP-2 promoter activities. Although βFTZ-F1-1B (included in the −1.3 kb construct, Fig. 2) and protein-binding was also blocked by over-expression of Ftz-F1 (Fig. 7B), the promoter activities of the −1.3 kb construct were not affected by over-expression of βFTZ-F1 in the presence of 20E (Fig. 5). The results indicate that βFTZ-F1 may interact with a 20E-responsive co-activator at the βFTZ-F1-1A site (Fig. 6B), and the over-expression of βFTZ-F1 led to the reduced promoter activities (Fig. 5). The results indicate that SCP-2 transcriptional regulation in vertebrates and in insects has different mechanisms. This might be attributed to the fact that insects lost their cholesterol synthesis pathway and developed unique cholesterol/sterol hormone pathways to regulate SCP-2 expression.
Another transcription factor that has been demonstrated to be affected by 20E and play an important role in larval-pupal molt is HR-3. This transcription factor is known to be up-regulated by ecdysone during both ecdysone peaks in last larval stadium (Riddiford et al., 2003). Highest levels of HR-3 transcript coincide with ecdysone peaks (Palli et al., 1992) and precede the expression of βFTZ-F1 that shows an expression peak later in the development after 20E titers have declined (Lam et al., 1997). Computational search for transcription factors binding sites did not locate any putative HR-3 recognizing sequences (data not shown). However, experiments with 20E treated Aag-2 cells demonstrated that HR-3 up-regulates AeSCP-2 expression of shorter promoter/reporter constructs of <−1000 bp (Fig. 4), and does not have any effect on expression levels of longer promoter/reporter constructs. This finding might be an indication that there is a repressor binding site located upstream in 5’UTR region between −1000 bp and −4200 bp. The repressor might also explain why we saw only a transient tissue specific up-regulation of AeSCP-2 in gut tissue cultures, since factors releasing the repressor might be present only in gut tissues but not in other tissues.
In summary we report here the first study that demonstrates the genomic structure of AeSCP-2, the first described sterol carrier protein in insects (Krebs and Lan, 2003). We demonstrated that AeSCP-2 is up-regulated by 20E and this up-regulation requires tissue specific factors. We also report that AeSCP-2 transcription is not up-regulated by βFTZ-F1 unlike the vertebrate SCP-2 promoter (Lopez et al., 2001), but is up-regulated by HR-3, an ecdysone-inducible transcription factor. This study is the first step in understanding the intricate hormonal regulation of AeSCP-2 gene that is important in mosquito reproduction and development (Blitzer et al., 2005).
Experimental Procedures
Chemicals
Chemicals and reagents were purchased from Sigma (St Louis, MO, USA), Fisher Scientific (Pittsburgh, PA, USA) and ICN (Costa Mesa, CA, USA) unless otherwise mentioned.
Mosquitoes
The mosquitoes used in these experiments, Ae. aegypti, were taken from an inbred laboratory strain (Rockefeller). Larvae were reared at 26°C in 70–80% humidity at a light:dark cycle of 14:10 with fish food (TetraMin, Tetra Holding, Inc., Blacksburg, VA, USA). Larvae hatching during a 15-min period were collected and used in experiments. Under these conditions, development took 64 (mostly male mosquitoes) to 72 h (mostly female mosquitoes) to complete the fourth stadium. Adults were maintained at 26°C.
Pharate fourth instars were staged by physical appearance using the visible dark black hairs of fourth stadium larva that are wrapped around the body under the thorax and abdominal cuticle of the third instar (Christophers, 1960). Larvae selected by this criteria ecdysed during a 1-hour period. Pharate fourth instars were selected and transferred into a new container and newly molted fourth instar larvae were collected as a synchronized cohort for experimentation.
Cell cultures
Ae. aegypti Aag-2 cell line was maintained in Eagle's medium (Invitrogen Corporation, Carlsbad, CA) supplemented with 5% fetal bovine serum (Lan et al., 1993) at 28°C under a 5% CO2 atmosphere. Passages of cells were conducted every 7 days with a 1:5 dilution of cells.
Ae. aegypti fosmid DNA library construction and isolation of AeSCP-2 clone
Genomic DNA was isolated from forth instar Ae.aegypti larvae using the Masterpure complete DNA and RNA purification kit (Epicentre, Madison, WI). The CopyControl fosmid library production kit (Epicenter, Madison, WI) was then used to construct a genomic library. The isolated DNA was primarily 40–50 kb as compared to 40kb T7 size marker and was repaired and cloned directly into the fosmid vector without further size selection. The library was calculated to contain 2.4 × 106 individual fosmid clones.
Using labeled AeSCP-2 cDNA as the probe, five clones were isolated from Ae.aegypti fosmid library after three consecutive rounds of screening. A 6.4 kb segment containing the entire AeSCP-2 gene and it’s 5’ and 3’ flanking regions was subcloned into pBS (SK+) vector, sequenced, and used for deletion construction.
Plasmid construction and deletion analysis
The 4.2-kb 5’ upstream flanking region (Fig. 2) was ligated to the bacterial chloramphenicol acetyltransferase gene (CAT) that had been excised from pCAT basic (Promega, Madison, Wis.). Sequence truncation was conducted from the 5' end of the upstream region toward the 3' end by using the ExoIII deletion kit (Stratagene, La Jolla, Calif.). Series deletion clones of 4200 bp, 1600 bp, 1300 bp, 1000 bp, 200 bp and 60 bp of upstream promoter sequence, including IntronI, were then selected and sequenced to ensure that the sequence matched the genomic DNA.
Expression vector of βFZT-F1 was constructed by cloning the 2.5 kb fragment of Drosophila βFZT-F1 cDNA containing the entire ORF (Lavorgna et al., 1993) into the pBS-hsp70 vector (Lan et al., 1997). The hsp70 promoter has constitutive activity in insect cells (Lan and Riddiford, 1997). The pIE1hr expression vectors of MHR-3 (Manduca sexta) are previously described (Hiruma et al., 2004). The pIE1hr expression vectors for EcRa (Ae.aegypti), EcRb (Ae.aegypti), USPa (Ae.aegypti), USPb (Ae.aegypti) were constructed using cDNAs cloned via PCR (Table 1) from an Ae. aegypti cDNA pool of 4th instars. PCR products of AeEcRa, EcRb, USPa, and USPb coding regions were cloned into the pBluntZero vector (Invitrogen, Carlsbad, CA, USA) and sequenced to ensure the orientation and the sequence correctness of the cDNAs. The entire open reading frame of cloned cDNA was excised with HindIII/XhoI (each is on one side of the MCS) from the pBluntZero vector (Invitrogen) and cloned into the pIE1hr expression vector at HindIII/SalI, which put the expression of the cDNA under IE1hr control.
Table 1.
DNA sequences of primers used for cloning of AeEcRa, AeEcRb, AeUSPa and EaUSPb.
| Prime name | Primer sequense |
|---|---|
| AeEcRA F | 5′ tcgaaaggtcatgtaccga 3′ |
| AeEcRA R | 5′ ctatacctggtcgagcatgc 3′ |
| AeEcRB F | 5′ gctagagcaaaatgatgaaaagaag 3′ |
| AeEcRB R | 5′ ctagctatacctggtcgagcat 3′ |
| AeUSPa F | 5′ caccgcgaagaatgctga 3′ |
| AeUSPa R | 5′ ttaaatcggcatgtcgagca 3′ |
| AeUSPb F | 5′ tcagcagaggatggatccc 3′ |
| AeUSPb R | 5′ ctaggatactttaaatcggcatgtc 3′ |
Transient transfection assays
Pure 20E (Sigma) was dissolved in 95% ethanol, and the concentration was determined by UV spectrum absorption analysis at 243 nm wavelength (20E mg/ml = (OD243 × 480)/ε and the ε = 12,670). Four microliters of 30 ng/µl of 20E in 95% ethanol were added in the medium for hormone-treated cell cultures to make the final concentration of 1, 5, and 10 µM/ml 20E.
All DNA transfection experiments were conducted as described previously (Lan and Riddiford, 1997) with 4 µg of total plasmid DNA per ml and 8 µl of Lipofectin reagent (Invitrogen) in 1 ml of transfection medium (culture medium without FBS and antibiotics). An hsp70-β-galactosidase plasmid, pXH70ZT, was co-transfected in all experiments for normalization of transfection efficiency. Equal amounts of each vector were used for co-transfections. Cells were treated for 12 hours with 1, 5 and 10 µM 20E at 45 hours after the initial transfection. After 9 hours of incubation at 28°C cells were transferred into 37°C for 3 hours for heat shock. Both β-galactosidase activity and CAT protein assays were performed as previously described (Lan and Riddiford, 1997). Cellular proteins were extracted in a lysis buffer of 250 mM Tris-HCl (pH 8.0), 0.3% Triton X-100, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonylfluoride (PMSF) by three cycles of freeze-thaw followed by centrifugation at 4°C (12,000 × g, 5 min). The supernatant was collected and the protein concentration was determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). Aag-2 cell protein extracts were stored at −80°C.
Levels of CAT protein in each sample were determined using a CAT assay kit (Boehringer Mannheim, Mannheim, Germany). The β-gal activity was determined via colorimetric assay as described (Lan and Riddiford, 1997). The relative CAT protein level was calculated by normalizing CAT protein level with the β-galactosidase activity from the same sample. The fold increased CAT was determined by comparing the CAT level in a 20E-treated cell extract with that of untreated cells.
Electrophoretic mobility shift assay (EMSA)
Expression vectors were transfected into Aag-2 cells in different combinations, and the whole-cell extract of the cellular proteins (about 107 cells) was prepared 45 h after transfection according to the method described (Lan and Riddiford, 1997). Cell pellets were lysed in 200 µl of lysis buffer (60 mM KCl, 25 mM HEPES [pH 7.5], 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 1 µg of antipain per ml), sonicated at 4°C for 30 s, and centrifuged at 12,000 × g at 4°C for 10 min. The supernatant was collected as 40-µl aliquots and stored at −80°C. As a control, an extract of Aag-2 cells that was transfected with the empty plasmid pIE1hr/PA was used. All of the extracts were frozen only once at −80°C until use.
The oligonucleotides used as probes for EMSA were synthesized by ITD (Coralville, IA): βFTZ-F1-1A, 5'- ATTGGGAATTTCAATTACTTTCAG -3'; βFTZ-F1-1B, 5'- TTATGAAAACAATTACCCCTTTAT -3'. The probes were 5’ end-labeled as described (Hiruma et al., 1995). Each oligonucleotide was 32P labeled at the 5' end with T4 polynucleotide kinase and [γ-32P]ATP, 10-fold molar excess of the cold complementary strand was added, and the mixture was heated at 95°C for 2 min, followed by a slow cooling to the room temperature. The double-stranded labeled probes were purified with Sephadex G-50 spin columns. The cold oligonucleotides used for specific competitors were prepared as follows: equal molar amounts of both strands of oligonucleotides were heated at 95°C for 2 min and then annealed by cooling slowly to the room temperature in TE buffer 10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]).
Five micrograms of cell extract in 19 µl of buffer containing 1 µg of poly(dI-dC)(dI-dC), 12 mM HEPES (pH 7.9), 60 mM KCl, 65 mM NaCl, 7.5 mM MgCl2, 6.6 mM EDTA, 1.2 mM DTT, and 12% glycerol was pre-incubated on ice for 15 to 20 min, and then 50 fmol of labeled probe and competitor DNA were added. The reaction mixture was incubated for 20 min on ice. The mixture was then run on a 4% polyacrylamide gel (16×18 cm) containing 2.5% glycerol in 1× TBE (45 mM Tris-borate, 1 mM EDTA), which had been pre-run for 40–60 min in 1× TBE. The gels were dried and were exposed to a Phosphor image screen for at least 1 h and scanned with a Storm820 (Molecular Dynamics, Piscataway, NJ). Data was analyzed using Storm800 software.
In vitro tissue culture
24 hour old staged fourth instars were dissected in insect saline (Riddiford et al., 1979) to obtain gut and carcass for in vitro tissue culture study. Dissected tissues were first washed in Grace's basal medium (Invitrogen) and cultured at 26°C for 6 h in 60 mm glass dishes in 2 ml of Grace's basal medium containing 50 µg/ml penicillin, 50 µg/ml streptomycin and 100 µg/ml neomycin. A stock solution of 20E in 100% ethanol was diluted with 95% ethanol to desired concentrations and was added in the medium for hormone-treated tissue cultures. The experiment was repeated twice each with replicate samples. Total RNA was extracted from cultured tissues and analyzed by dot blotting analysis.
RNA extraction
Ten staged animals were washed with ddH2O, rinsed once with DEPC-H2O, and excess water was blotted off using clean Kimwipes. Cleaned animals were put into a 1.5 ml Eppendorf test tube and homogenized with a micropestle in 1 ml of Trizol reagent according to the manufacturer's recommendations (Invitrogen). The quantity of the total RNAs was determined by UV absorption at 260 nm.
Dot blot analysis
For dot blot analysis, 5 µg of total RNA from three parallel samples (10 to 15 larval guts per sample) of each treatment were blotted on to positively charged Nylon membrane as described (Millipore). The membranes were UV cross-linked (Stratagene Cloning system, La Jolla, CA) and baked at 80 °C under vacuum for 1 h.
AeSCP-2 probes were generated using 50 µCi [α-32P] dCTP (3000 Ci/mm) the PCR reaction with DNA polymerase Tfl (Epicentre, Madison, WI) using AeSCP-2 cDNA (Krebs and Lan, 2003) as the template and two primers matching to the 5' and 3' of ORF 5'-atgtctctgaagtccg-3' and 5'-ttacttcagcgagg-3'). The PCR reaction was performed under 1 cycle of 94°C, 3 min to denature the template, then 3 cycles of 94°C for 2 min, 50 C for 3 min and 72 C for 5 min. The probes were cleaned using QiaSpin column (QIAGEN, Valencia, CA).
Membranes were prehybridized at 42°C for at least 2 h in a hybridization oven (Robbins Scientific, Sunnyvale, CA) in pre/hybridization solution (4 × SSC, 10 × Denhardt's solution, 50% formamide, 0.1% SDS, 50 mm NaPO4 (pH 7.2), 1 mM EDTA and 100 µg/ml sheared herring sperm DNA). Denatured labelled cDNA probes were added in the prehybridization solution at more than 2–3 × 106 cpm/ml and incubated at 42°C overnight in the hybridization oven. The membranes were washed once each at 65°C for 15 min in 2 × SSC/0.1% SDS, 1 × SSC/0.1% SDS, 0.5 × SSC/0.1% SDS and 0.2 × SSC/0.1% SDS. The membranes were exposed to a Phosphor Image Screen for at least 1 h and scanned with a Storm820 (Molecular Dynamics). The data were analyzed using the Storm800 software.
Acknowledgements
This work was supported by the Graduate School Research Grant from the University of Wisconsin-Madison, by the National Institute of Health research grant #5R01AI067422 to Q.L. and by the Alex and Lillian Feir Graduate Fellowship to I.V.
References
- Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster : I. Dependence upon ecdysone concentration. Dev Biol. 1973;35:47–61. doi: 10.1016/0012-1606(73)90006-7. [DOI] [PubMed] [Google Scholar]
- Blitzer EJ, Vyazunova I, Lan Q. Functional analysis of AeSCP-2 using gene expression knockdown in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2005;14:301–307. doi: 10.1111/j.1365-2583.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- Borovsky D, Whisenton LR, Thomas BR, Fuchs MS. Biosynthesis and distribution of ecdysone and 20-hydroxyecdysone in Aedes aegypti. Archives of Insect Biochemistry and Physiology. 1986;3:19–30. [Google Scholar]
- Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila βFTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Molecular Cell. 1999;3:143–149. doi: 10.1016/s1097-2765(00)80305-6. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chanderbhan RF, Kharroubi AT, Noland BJ, Scallen TJ, Vahouny GV. Sterol carrier protein2: further evidence for its role in adrenal steroidogenesis. Endocr Res. 1986;12:351–370. doi: 10.3109/07435808609035445. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Cherbas P. The arthropod initiator: The capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- Christophers SR. Aedes aegypti (L.) Cambridge: Cambridge University Press; 1960. The Yellow Fever Mosquito. Its Life History, Bionomics and Structure. [Google Scholar]
- Clayton RB. The utilization of sterols by insects. J Lipid Res. 1964;5:3–19. [PubMed] [Google Scholar]
- Dwivedi A, Shukla S. Utilization of cholesterol by the adults of house fly, Musca domestica. Entomology. 1982;7:411–422. [Google Scholar]
- Dyer DH, Lovell S, Thoden JB, Holden HM, Rayment I, Lan Q. The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35-A resolution. J Biol Chem. 2003;278:39085–39091. doi: 10.1074/jbc.M306214200. [DOI] [PubMed] [Google Scholar]
- Dyer DH, Wessely V, Forest KT, Lan Q. Three-dimensional structure/function analysis of SCP-2-like2 reveals differences among SCP-2 family members. J Lipid Res. 2008;49:644–653. doi: 10.1194/jlr.M700460-JLR200. [DOI] [PubMed] [Google Scholar]
- Gallegos AM, Atshaves BP, Storey SM, Starodub O, Petrescu AD, Huang H, McIntosh AL, Martin GG, Chao H, Kier AB, Schroeder F. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In silico biology. 2002;2:S1–S15. [PubMed] [Google Scholar]
- Grieneisen ML, Warren JT, Sakurai S, Gilbert LI. A putative route to ecdysteroids: metabolism of cholesterol in vitro by mildly disrupted prothoracic glands of Manduca sexta. Insect Biochem. 1991;21:41–51. [Google Scholar]
- Hiruma K, Riddiford LM. Regulation of transcription factors MHR4 and betaFTZ-F1 by 20-hydroxyecdysone during a larval molt in the tobacco hornworm, Manduca sexta. Dev Biol. 2001;232:265–274. doi: 10.1006/dbio.2001.0165. [DOI] [PubMed] [Google Scholar]
- Hiruma K, Riddiford LM. Differential control of MHR3 promoter activity by isoforms of the ecdysone receptor and inhibitory effects of E75A and MHR3. Developmental Biology. 2004;272:510–521. doi: 10.1016/j.ydbio.2004.04.028. [DOI] [PubMed] [Google Scholar]
- Hiruma KM, Carter S, Riddiford LM. Characterization of the dopa decarboxylase gene of Manduca sexta and its suppression by 20-hydroxyecdysone. Dev Biol. 1995;169:195–209. doi: 10.1006/dbio.1995.1137. [DOI] [PubMed] [Google Scholar]
- Horner MA, Chen T, Thummel CS. Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev Biol. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- http://compbio.dfci.harvard.edu/tgi/temp/blastn-a_aegypti-16513-1200078472.html.
- Karim FD, Thummel CS. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO Journal. 1992;11:4083–4093. doi: 10.1002/j.1460-2075.1992.tb05501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs KC, Lan Q. Isolation and expression of a sterol carrier protein-2 gene from the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2003;12:51–60. doi: 10.1046/j.1365-2583.2003.00386.x. [DOI] [PubMed] [Google Scholar]
- Lam GT, Jiang C, Thummel CS. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- Lan Q, Gerenday A, Fallon AM. Cultured Aedes albopictus mosquito cells synthesize hormone-inducible proteins. In Vitro Cell Dev Biol. 1993;29:813–818. doi: 10.1007/BF02634349. [DOI] [PubMed] [Google Scholar]
- Lan Q, Grier CA. Critical period for pupal commitment in the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2004;50:667–676. doi: 10.1016/j.jinsphys.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Lan Q, Hiruma K, Hu X, Jindra M, Riddiford LM. Activation of a delayed-early gene encoding MHR3 by the ecdysone receptor heterodimer EcRB1-USP-1 but not by EcR-B1-USP-2. Mol Cell Biol. 1999;19:4897–4906. doi: 10.1128/mcb.19.7.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Massey RJ. Subcellular localization of the mosquito sterol carrier protein-2 and sterol carrier protein-x. J Lipid Res. 2004;45:1468–1474. doi: 10.1194/jlr.M400003-JLR200. [DOI] [PubMed] [Google Scholar]
- Lan Q, Riddiford LM. DNA transfection in the ecdysteroid-responsive GV1 cell line from the tobacco hornworm, Manduca sexta. In Vitro Cell Dev Biol Anim. 1997;33:615–621. doi: 10.1007/s11626-997-0111-5. [DOI] [PubMed] [Google Scholar]
- Lan Q, Wessely V. Expression of a sterol carrier protein-x gene in the Yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2004;13:519–529. doi: 10.1111/j.0962-1075.2004.00510.x. [DOI] [PubMed] [Google Scholar]
- Lavorgna G, Karim FD, Thummel CS, Wu C. Potential role for a FTZ-F1 steroid receptor superfamily member in the control of Drosophila metamorphosis. PNAS USA. 1993;90:3004–3008. doi: 10.1073/pnas.90.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Niesen M, Bedi M, Hale D, McLean MP. Activation of the SCPx promoter in mouse adrenocortical Y1 cells. Biochem Biophys Res Commun. 2007;357:549–553. doi: 10.1016/j.bbrc.2007.03.194. [DOI] [PubMed] [Google Scholar]
- Lopez D, Shea-Eaton W, McLean M. Characterization of a steroidogenic factor-1-binding site found in promoter of sterol carrier protein-2 gene. Endocrine. 2001;14:253–261. doi: 10.1385/ENDO:14:2:253. [DOI] [PubMed] [Google Scholar]
- Margam VM, Gelman DB, Palli SR. Ecdysteroid titers and developmental expression of ecdysteroid-regulated genes during metamorphosis of the yellow fever mosquito, Aedes aegypti (Diptera: Culicidae) J Insect Physiol. 2006;52:558–568. doi: 10.1016/j.jinsphys.2006.02.003. [DOI] [PubMed] [Google Scholar]
- McLean M, Puryear T, Khan I, Azhar S, Billheimer J, Orly J, G G. Estradiol regulation of sterol carrier protein-2 independent of cytochrome P450 side-chain cleavage expression in the rat corpus luteum. Endocrinology. 1989;125:1337–1344. doi: 10.1210/endo-125-3-1337. [DOI] [PubMed] [Google Scholar]
- Menon KM, Gunaga KP. Role of cyclic AMP in reproductive processes. Fertil Steril. 1974;25:732–750. doi: 10.1016/s0015-0282(16)40577-7. [DOI] [PubMed] [Google Scholar]
- Nes WD, Lopez M, Zhou W, Guo D, Dowd PF, Norton RA. Sterol utilization and metabolism by Heliothis zea. Lipids. 1997;32:1317–1323. doi: 10.1007/s11745-006-0170-5. [DOI] [PubMed] [Google Scholar]
- Nishiura JT, Burgos C, Aya S, Goryacheva Y, Lo W. Modulation of larval nutrition affects midgut neutral lipid storage and temporal pattern of transcription factor expression during mosquito metamorphosis. J Insect Physiol. 2007;53:47–58. doi: 10.1016/j.jinsphys.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Noda H, Wada K, Saito T. Sterols in Laodelphax striatellus with special reference to the intracellular yeast-like symbiotes as a sterol source. J Insect Physiol. 1979;25:443–447. [Google Scholar]
- Palli SR, Hiruma K, Riddiford LM. An ecdysteroid-inducible Manduca gene similar to the Drosophila DHR3 gene, a member of the steroid hormone receptor superfamily. Dev Biol. 1992;150:306–318. doi: 10.1016/0012-1606(92)90244-b. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Palli SR. Stage- and cell-specific expression of ecdysone receptors and ecdysone-induced transcription factors during midgut remodeling in the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2007;53:216–229. doi: 10.1016/j.jinsphys.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Pfeifer SM, Sakuragi N, Ryan A, Johnson AL, Deeley RG, Billheimer JT, Baker ME, Strauss JFI. Chicken sterol carrier protein 2-sterol carrier protein x: cDNA cloning reveals evolutionary conservation of structure and regulated expression. Arch Biochem Biophys. 1993;304:287–293. doi: 10.1006/abbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Rennert H, Amsterdam A, Billheimer JT, Strauss JF. Regulated expression of sterol carrier protein 2 in the ovary: a key role for cyclic AMP. Biochemistry. 1991;30:11280–11285. doi: 10.1021/bi00111a013. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Curtis AT, Kiguchi K. Culture of the epidermis of the tobacco hornworm, Manduca sexta. Tissue Culture Assn Man. 1979;5:975–985. [Google Scholar]
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Ritter KS, Nes WR. The effects of cholesterol on the development of Heliothis zea. J Insect Physiol. 1981;27:175–182. [Google Scholar]
- Sun G-C, Hirose S, Ueda H. Intermittent expression of BmFTZ-F1, a member of the nuclear hormone receptor superfamily during development of the silkworm Bombyx mori. Dev Biol. 1994;162:426–437. doi: 10.1006/dbio.1994.1099. [DOI] [PubMed] [Google Scholar]
- Thummel CS. From embryogenesis to metamorphosis: The regulation and function of drosophila nuclear receptor superfamily members. Cell. 1995;83:871–877. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- Trzeciak WH, Simpson ER, Scallen T, Vahouny GV, Waterman MR. Studies on the synthesis of sterol carrier protein-2 in rat adrenocortical cells in monolayer culture. Regulation by ACTH and dibutyryl cyclic 3',5'-AMP. J Biol Chem. 1987;262:3713–3717. [PubMed] [Google Scholar]
- Vyazunova I, Wessley V, Kim M, Lan Q. Identification of two sterol carrier protein-2 like genes in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2007;16:305–314. doi: 10.1111/j.1365-2583.2007.00729.x. [DOI] [PubMed] [Google Scholar]
- Wang S-F, Li C, Sun G, Zhu J, Raikhel AS. Differential expression and regulation by 20-hydroxyecdysone of mosquito ecdysteroid receptor isoforms A and B. Mol Cell Endocrinol. 2002;196:29–42. doi: 10.1016/s0303-7207(02)00225-3. [DOI] [PubMed] [Google Scholar]
- Weller J, Sun GC, Zhou B, Lan Q, Hiruma K, Riddiford LM. Isolation and developmental expression of two nuclear receptors, MHR4 and betaFTZ-F1, in the tobacco hornworm, Manduca sexta. Insect Biochemistry and Molecular Biology. 2001;31:827–837. doi: 10.1016/s0965-1748(00)00188-0. [DOI] [PubMed] [Google Scholar]
- Woodard CT, Baehrecke EH, Thummel CS. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell. 1994;79:607–615. doi: 10.1016/0092-8674(94)90546-0. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen L, Raikhel AS. Posttranscriptional control of the competence factor βFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100:13338–13343. doi: 10.1073/pnas.2234416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen L, Sun G, Raikhel AS. The competence factor βFtz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol. 2006;26:9402–9412. doi: 10.1128/MCB.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]