Introduction
The enormous progress in genomic research over the past decade has had a huge impact on experimental directions designed to understand gene function and regulation in mammalian cells. Research has rapidly shifted from analysis of individual genes to using microarray expression analysis and similar global approaches to decipher expression changes and their regulation among thousands of genes. While this progress is impressive, our understanding of how gene expression is organized and coordinated within the context of the three dimensional structure of the cell nucleus has lagged behind. With the development of more sophisticated microscopy and computer imaging approaches, however, a new view of the cell nucleus is emerging based on genomic function. This new “functional view” of the cell nucleus has already revealed discrete domains in the nucleus where genomic functions are arranged such as sites of DNA replication, transcription, splicing and DNA repair (Berezney 2002; Berezney, et al. 1996; Dundr and Misteli 2001; Meaburn and Misteli 2007; Stein, et al. 2007; Stein, et al. 2004; Stein, et al. 2003; Strouboulis and Wolffe 1996).
Correlating with these functional domains is the hierarchial organization of genomic DNA in the cell nucleus from nucleosomes to chromatin fibers to chromatin loop domains to higher order arrangements leading up to the entire chromosomes which maintain discrete territorial arrangements within the interphase nucleus (Berezney 2002; Cremer, et al. 2006; Razin, et al. 2007). Considerable progress has been made in deciphering higher order chromatin domains in the cell nucleus through investigation of functional sites of DNA replication. By combining 3-D microscopy and computer imaging approaches with appropriate pulse and pulse-chase experiments, it was demonstrated that chromatin is arranged into ~1 mbp higher order domains during DNA replication which are maintained throughout the cell cycle and in subsequent daughter cells as a fundamental feature of higher order chromatin arrangement in the interphase cell nucleus (Berezney, et al. 1995; Jackson and Pombo 1998; Ma, et al. 1998; Sparvoli, et al. 1994). Moreover these 1 mbp chromatin domain units have been readily identified in living cells using appropriate in vivo labeling or microinjection approaches which have further revealed their dynamic nature (Pliss, et al. 2009; Zink, et al. 1998).
These findings provided the basis for further experimentation and extension of these studies in several directions. For example, by simultaneously labeling DNA replication and transcription sites in mammalian cells and applying 3-D microscopy and imaging techniques, our group has determined that replication and transcription sites in the cell nucleus are sequestered into separate higher order zones each of which contains numerous chromatin domains (Berezney 2002; Wei, et al. 1998). Moreover, the individual chromatin domains and their higher order arrangement into nuclear zones is maintained following extraction of cells for nuclear matrix (Berezney and Wei 1998; Wei, et al. 1998; Wei, et al. 1999). This further indicated the importance of higher order nuclear architecture in the organization and perhaps regulation of genomic function in the cell nucleus.
This review focuses on our recent investigations of the spatio-temporal relationships of DNA replication and transcription sites and their functional components in the cell nucleus. Combining pulse-chase experiments with 3-D microscopy and newly developed computer imaging algorithms has enable us to elucidate functional neighborhoods of DNA replication/transcription factories within the interphase cell nucleus.
Spatio-temporal arrangement of DNA replication and transcription sites
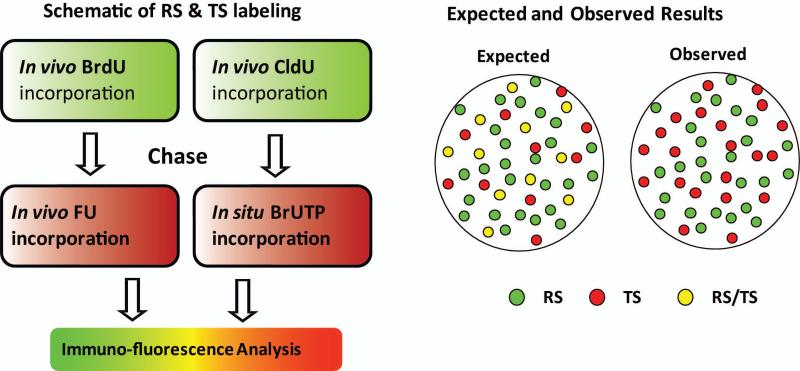
Our earlier studies of nuclear zones of replication and transcription in the cell nucleus focused on early S phase (Wei, et al. 1998) when most of the actively transcribed genes in the cell are replicated (Goldman, et al. 1984; Hatton, et al. 1988; MacAlpine, et al. 2004; Schubeler, et al. 2002). This immediately raised a fundamental issue: “If chromatin domains are involved with DNA replication in early S phase, when and where will gene transcription occur within these newly replicated chromatin domain and does this involve a highly coordinated spatio-temporal programming of DNA replication and transcription?” As a first step in addressing this question, we performed pulse-chase-pulse dual labeling experiments of replication and transcription sites. Replication sites (RS) in exponentially growing HeLa cells were labeled with BrdU or CldU, chased between 0 and 24 hours followed by labeling of transcription sites (TS) either in vivo with FU or in permeabilized cells (in situ labeling) with BrUTP (Fig. 1). The rationale behind these experiments is that at different times following DNA replication (green labeling), transcription (red labeling) will occur within subpopulations of the replicated chromatin domains. We would then be able to visualize those chromatin domains that were actively transcribing their genes by the merged red and green or yellow color. Much to our surprise, extensive experimentation did not reveal significant levels of yellow color throughout the extensive chase period. Instead, separate sites of green replication and red transcription were always observed (Malyavantham, et al. 2008a).
Fig. 1. The labeling scheme of early S phase replication sites (RS) and transcription sites (TS).
Nucleotide analogs are incorporated in living cells (referred to as in vivo labeling) or permeabilized cells (referred to as in situ labeling). The predicted and observed distributions of RS and TS for both in vivo and in situ approaches are represented as a cartoon. The legend describes the colors assigned to the RS, TS and the expected merged sites.
The inability to observe conversion of RS to TS by a color change to yellow sites led us to develop a computer imaging approach to directly measure the spatial position of labeled TS in relation to the RS-labeled chromatin domains (Malyavantham, et al. 2008a). We term this “nearest neighbor proximity analysis”. Following application of a high resolution segmentation program for the individual sites (Bhattacharya, et al. 2005), our proximity program determines the distance between each individual site (center-to-center distance between centroids) within one population of sites (e.g, the TS) to its nearest neighbor in another population of sites (e.g., the RS). The percent of total TS that are within a certain distance or proximity of their nearest neighbor early RS chromatin domain is then determined over a range of pixel values (Malyavantham, et al. 2008a; Malyavantham, et al. 2008b).Our analysis demonstrated that over 70% of the TS centroids are within 7 pixels (0.5 μ) of the nearest neighbor RS centroids and over 80% are within 10 pixels (0.7 μ). Since RS and TS sites average ~0.5 μ in diameter (Ma, et al. 1998; Wei, et al. 1998; Wei, et al. 1999), these results show a strikingly high degree of proximity for the great majority of the TS to RS chromatin domains (Malyavantham, et al. 2008a). For example, a 0.5 μ distance between sites indicates an average edge to edge distance between the two sites of zero pixels. In other words, the two sites are in complete juxtaposition. Virtually identical proximity associations were measured independent of the chase period (0-24 hours) between RS and TS labeling (Malyavantham, et al. 2008a). These findings were directly supported by visualization of the RS and TS centroids following segmentation. The TS centroids were preferentially associated along the borders of the early RS chromatin domains but never directly co-localized with the RS centroids and virtually identical results were observed independent of the chase time (Malyavantham, et al. 2008a).
The proximity of RS and TS to RNA polymerase II was then investigated using our imaging and computer program approaches. Much to our initial surprise we found that the proximity distributions were nearly identical with no preference of the overall population of RNA polymerase II sites to either TS or RS chromatin domain sites (Malyavantham, et al. 2008a). We conclude that the overall populations of RNA pol II sites are positioned approximately midway between the centers of the early RS chromatin domains and the TS. It is intriguing to consider that pol II sites may represent linchpins for the close spatial arrangement and coordinate expression of replicating chromatin domains and transcription sites in the cell nucleus. This possibility is further suggested in the last section of this review on functional neighborhoods.
Models of transcriptional regulation at RS chromatin domains
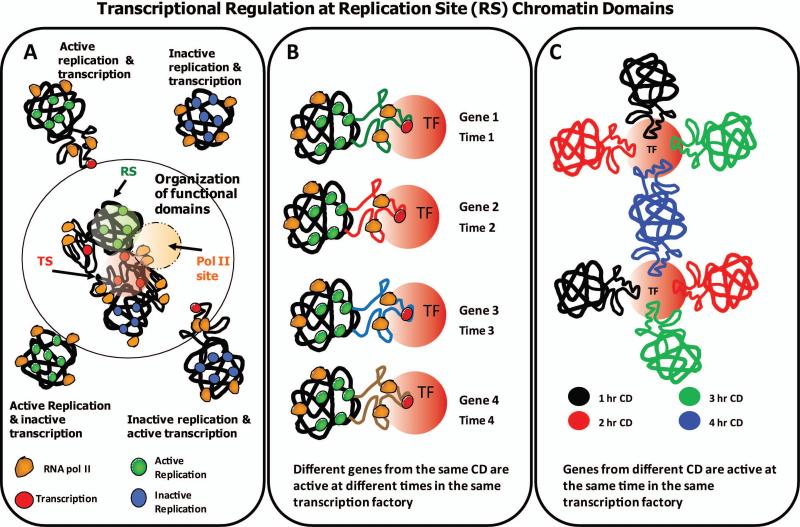
Based on our proximity data and visualization of centroids following segmentation of RS chromatin domains and TS, we propose that chromatin activated for transcription, dynamically unfolds or “loops out” of densely packed discrete chromatin domains (Fig. 2 A & B) where it engages in active transcription within “transcription factories” (Jackson, et al. 2000; Pombo, et al. 2000). It is possible that the local configuration changes from more condensed (discrete) regions of individual chromatin domains, into more diffuse (decondensed) regions occurs during activation of gene transcription. Several studies have indicated the unfolding/folding of chromatin domains during the activation or inactivation of both artificial and endogenous gene loci (Bickmore, et al. 2004; Chambeyron and Bickmore 2004; Chambeyron and Bickmore 2004; Mahy, et al. 2002; Mahy, et al. 2002; Tumbar and Belmont 2001; Volpi, et al. 2000). Our findings extend this analysis to the global level of the genome and suggest that chromatin unfolding may be a fundamental event required for transcriptional activation.
Fig. 2. Transcriptional regulation at early S replication site (RS) chromatin domains.
A hypothetical model extends our understanding of the 1 Mbp chromatin domains and the coordination of replication and transcription. (A) Chromatin domains are either active or inactive for replication and transcription giving rise to various possible configurations as depicted. Active and inactive replication is represented over chromatin domains with green and blue colored circles, respectively. Transcriptional activity and pol II are visualized as red and orange colored circles, respectively. Despite the absence of colocalization (yellow color), proximity analysis revealed a high degree of spatial association between RS and TS. These results led us to propose that genes encompassing the chromatin domains either active or inactive for replication (green or blue), loop out or decondense, when activated for transcription (red). Based on proximity results, some pol II (orange) are placed at ~ equal distance from both chromatin domains and transcriptional activity. (B) Proximity analysis of pulse-chase experiments also suggests that different genes from the same chromatin domain are active at different times in the same transcription factory. (C) Our analysis suggests that genes from different temporally labeled subsets of chromatin domains are active at the same time in the same transcription factory. The organization of functional domains is also illustrated in (A) where transcription factories are visualized as the product of the genes encompassing different chromatin domains.
It has been suggested that gene transcription occurs stochastically in bursts of activity throughout the cell cycle (Chubb, et al. 2006; Levsky, et al. 2002; Levsky and Singer 2003; Raj, et al. 2006; Ross, et al. 1994). Using multi-FISH approaches, this process has been directly visualized using a series of gene probes (Levsky, et al. 2007; Levsky, et al. 2002). Thus, the constant high level of transcription associated with the early RS chromatin domains may be a manifestation of burst transcription in which different subsets of genes within each chromatin domain are unfolded and activated for transcription at different times over the cell cycle (Fig. 2 B). The final result of this stochastic process at the level of individual chromatin domains is that all the active genes within the domain are transcribed at different times and at the required levels for the given cell.
Using FISH studies of individual genes, several investigations have demonstrated that distally located genes along a chromosome or between chromosomes cluster together at transcription factories (Fraser 2006; Ling, et al. 2006; Mitchell and Fraser 2008; Nunez, et al. 2008; Osborne, et al. 2004). Finding high levels of proximity of transcription sites to the early S chromatin domains is consistent with these results and suggests that assembly/activation of dynamic transcription factories containing distally located genes may be a fundamental event involved in the global regulation of genomic transcription.
To further explore this possibility we performed proximity measurements of TS compared with two temporal (two hour chase between pulses) and spatially distinct populations of early S labeled RS (Malyavantham, et al. 2008a). Since this by definition must represent two different subsets of actively transcribed genes, our finding of identical proximity of transcription sites to each spatio-temporal population of labeled chromatin domains, is likely reflecting the participation of genes from different chromatin domains (i.e., distal genes) at common transcription factories as illustrated in Figure 2 C.
Our model (Fig. 2 A) also predicts that transcription can occur simultaneously with replication at chromatin domains but without colocalization at the discrete RS due to the unfolding of the transcriptionally active regions of the chromatin domain. The chromatin domains are either active or inactive for replication and transcription giving rise to several possible states as depicted in the schematic. For simplicity, we depicted the position of pol II sites as equidistant from both replicational and transcriptional activities as indicated by the proximity values (Fig. 2 A). To produce the large number of transcripts of abundantly transcribed genes, it is conceivable that these genes are maintained in an unfolded (diffuse) state for prolonged periods of time within a transcription factory. In contrast, genes that are transcribed only a very limited number of times per cell cycle (e.g. 1-10 copies), may need to be in a more diffuse chromatin state for transcription for only very limited number of times in each cell cycle. This less abundantly transcribed class of genes, which composes the vast majority of transcribed genes in a typical mammalian cell (Chubb, et al. 2006; Shav-Tal, et al. 2006), may, therefore, reversibly extend into the transcription factories surrounding the early RS replicated chromatin domains for active transcription and fold back into the discrete chromatin domains following termination of transcription (Fig. 2 A). Several previous studies employing FISH have demonstrated massive chromatin unfolding of individual genes following transcriptional activation (Chambeyron and Bickmore 2004; Mahy, et al. 2002; Ragoczy, et al. 2003; Tumbar and Belmont 2001; Volpi, et al. 2000).
Functional neighborhoods of replication/transcription
The close proximity of early S replicated chromatin domains to transcription sites and of RNA polymerase II to both populations of these sites led us to consider whether defined neighborhoods of replication/transcription activity are present in the cell nucleus. This is consistent with previous findings of higher order zoning of replication and transcription in the cell nucleus (Berezney 2002; Berezney, et al. 2005; Berezney and Wei 1998; Cook 1998; Wei, et al. 1998). In addition, our studies of TS and chromatin domain centroids following segmentation, suggested that RNA polymerase II mediated transcription in the cell nucleus is concentrated in the regions along the periphery of the chromatin domains where multiple chromatin domains could participate in providing genes for coordinate expression in so-called transcription factories or regions of enriched gene transcription (Fig. 2 C). We, therefore, extended our studies of nearest neighbor proximity to fundamental protein components that are directly involved in either DNA replication (PCNA), transcription (RNA polymerase II) as well as HP1γ which is enriched in the euchromatic active gene regions of chromatin (de Wit, et al. 2007; Lomberk, et al. 2006; Malyavantham, et al. 2008b; Minc, et al. 2000) and might, therefore, be a suitable marker for defining functional neighborhoods in the cell nucleus.
The investigation demonstrated a high degree of proximity of all these factors to each other. This supports our view of defined functional neighborhoods for DNA replication/transcription function (Malyavantham, et al. 2008b). Moreover, detailed analysis revealed a defined order in the degree of proximities for the different components that was highly reproducible (Malyavantham, et al. 2008b). For example, PCNA, which decorates RS, was always the component with the least degree of proximity when determining the nearest neighbors to TS, RNA polymerase II or HP1γ while pol II had the highest relative proximity in nearest neighbor analysis to TS, HP1γ and PCNA (Malyavantham, et al. 2008b).
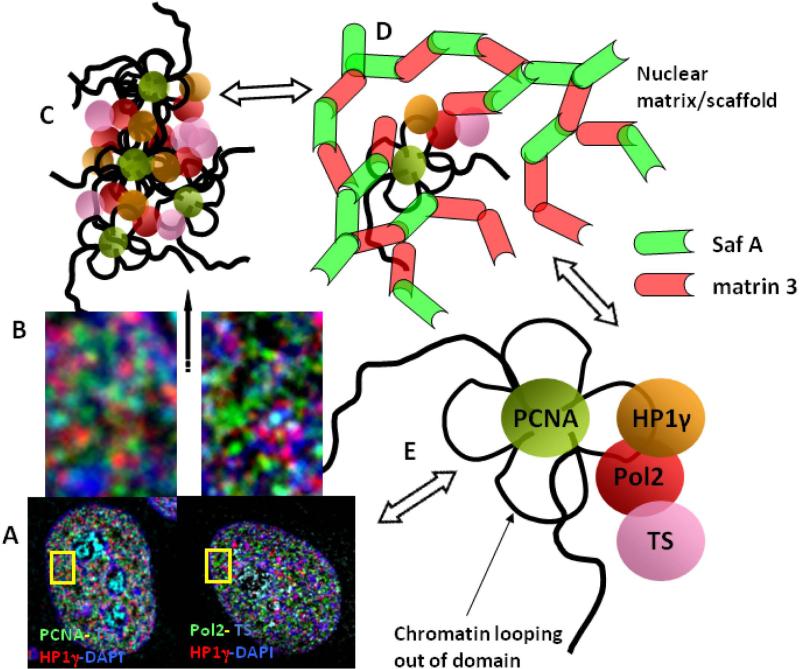
We propose that these distinctive proximity values for each pair of factors may reflect a precision in the higher order arrangement in these combinatorial arrays that constitute the functional neighborhoods. A model illustrating these relationships is shown in Figure 3. Key to the organization and functional expression of these combinatorial arrays may be the RNA pol II sites which appear to be positioned mid-way between the chromatin domains and sites of newly transcribed RNA. They may, aside from their fundamental role in transcription elongation, serve as “linchpin” for the proper assembly of the combinatorial arrays of functional factors and for the coordination of replication and transcription occurring both within individual chromatin domains and among several adjacent ones (Fig 2). This model also suggests the tethering of these combinatorial arrays to a nuclear matrix infrastructure. This is supported by findings demonstrating that these functional components and their higher order arrangements are maintained following extractions for nuclear matrix (Nakayasu and Berezney 1989; Wei, et al. 1999). In addition, proximity analysis for the nuclear matrix associated proteins matrin 3 and SAF-A (Belgrader, et al. 1991; Fackelmayer, et al. 1994; Kipp, et al. 2000; Nakayasu and Berezney 1991), demonstrates high levels of proximity to TS, PCNA, pol II and HP1γ. These levels, however, are significantly lower compared to the strikingly high proximities of matrin 3 to SAF-A sites (Malyavantham, et al. 2008b). This is consistent with a close association of the combinatorial arrays to an overall nuclear matrix structure as illustrated in Figure 3.
Fig. 3. A model for the organization of functional neighborhoods within the cell nucleus.
Based on the results of microscopy of multi-labeling experiments (A, B), proximity relationships of protein and nuclear matrix factors to functional sites, and pattern recognition analysis of matrin 3 and SAF-A using the MST algorithm for network organization, a model is proposed for the organization of these functional neighborhoods (C) that are in close association with the nuclear matrix architecture (D). Due to their propensity to form network-like structures as revealed by MST analysis and a high degree of demonstrated spatial association, the two abundant nuclear matrix proteins, matrin 3 and SAF-A have been visualized as building blocks of the nuclear architecture (D). We emphasize, however, that many other protein factors are likely components of this overall nuclear matrix/scaffold network. Proximity analysis of these nuclear matrix proteins and labeled functional and protein factor sites predict a close association between a significant portion of them as illustrated in (D). These may represent matrin 3 and SAF-A sites that are actively involved in the function and/or regulation at closely associated functional sites. Other sites of matrin 3 and SAF-A that are more distal, contribute to the overall architecture of the nuclear matrix and may correspond to future sites for functional associations. Based on the proximity distances, the functional protein factor sites are hypothesized to be arranged in a specific order within the nuclear space (E). We propose that components of the nuclear matrix such as matrin 3 and SAF-A play a role in the dynamic formation of these functional arrays and serve as a platform for the assembly of multiple repeating patterns of sites that define a functional neighborhood in the cell nucleus (C, D).
Further evidence supporting this view is provided by application of the minimal spanning tree (MST) algorithm (Dussert, et al. 1988; Dussert, et al. 1987). This pattern recognition imaging approach connects the discrete sites identified following segmentation into a network. The line path of the MST network follows aspects of nearest neighbor relations between the sites and a minimal distance for the entire pathway. The MST network generated for matrin 3 and SAF-A was in close alignment and demonstrated many sites of overlap (Malyavantham, et al. 2008b). In contrast, the MST patterns formed by pol II sites were spatially less closely aligned and showed no visible overlap with the network patterns of matrin 3 or SAF-A (Malyavantham, et al. 2008b). These findings further support the view that matrin 3 and SAF-A are components of an extensive nuclear matrix architecture in the cell nucleus and that the functional combinatorial arrays for DNA replication and transcribed are anchored to the matrix at a much more limited number of sites.
Summary
There is growing realization that the spatio-temporal positioning of functional sites and factors within the cell nucleus are critical determinants of genomic function and regulation within the interphase cell nucleus. Previous studies from our group identified higher order functional zones of replication and transcriptional activity in chromatin domains replicated in early S phase where actively transcribed genes are enriched. It was proposed that these higher order zones represent the modular basis for the coordinate regulation of genomic replication and transcription in the cell.
In this article we review our recent findings concerning the spatio-temporal dynamics of early S phase replicated chromatin domains, sites of gene transcription and associated protein factors. Our findings led us to propose that an important step in activation for transcription in the cell nucleus is the unfolding of regions within the discrete chromatin domains so that transcriptionally active chromatin can interact with the machinery for transcription such as RNA polymerase II and the multitude of additional factors involved in transcription and its regulation. Regulation of transcription from this global perspective likely involves temporal programming for the coordinate and differential unfolding of chromatin regions in accordance with the transcriptional programming of the cell. For example, active genes from different chromatin domains could undergo transcription at common sites or transcription factories. In addition, genes regions from within the same chromatin domains could be active at different transcription factories. All this is likely made possible by the spatial arrangement of the protein factors involved in the replicational and transcriptional processes which form combinatorial arrays that define the functional landscape of the cell nucleus.
Our results demonstrate a degree of precision for the higher order spatial arrangement of these combinatorial arrays of functional components. It is thus conceivable that alterations in the spatial organization of these arrays could lead to dysfunction or altered regulation of replicational or transcriptional processes at the global level. Studies of the nuclear matrix associated proteins matrin 3 and SAF-A further suggest that anchoring to the nuclear matrix architecture may be a critical determining factor in the combinatorial properties and for the higher order arrangement of these genomic functional complexes into 3-D network-like arrays.
Acknowledgments
Studies reported from the authors’ laboratory were funded by NIH Grant GM 072131-27
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belgrader P, Dey R, Berezney R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. The Journal of biological chemistry. 1991;266:9893–9899. [PubMed] [Google Scholar]
- Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Advances in enzyme regulation. 2002;42:39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- Berezney R, Malyavantham KS, Pliss A, Bhattacharya S, Acharya R. Spatio-temporal dynamics of genomic organization and function in the mammalian cell nucleus. Adv Enzyme Regul. 2005;45:17–26. doi: 10.1016/j.advenzreg.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Berezney R, Mortillaro M, Ma H, Meng C, Samarabandu J, Wei X, Somanathan S, Liou WS, Pan SJ, Cheng PC. Connecting nuclear architecture and genomic function. Journal of cellular biochemistry. 1996;62:223–226. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C223::AID-JCB10%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int Rev Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Berezney R, Wei X. The new paradigm: integrating genomic function and nuclear architecture. J Cell Biochem Suppl. 1998;30-31:238–242. [PubMed] [Google Scholar]
- Bhattacharya S, Acharya R, Pliss A, Malyavantham KS, Berezney R. Automated matching of genomic structures in microscopic images of living cells using an information theoretic approach. Conf Proc IEEE Eng Med Biol Soc. 2005;6:6425–6428. doi: 10.1109/IEMBS.2005.1615969. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Mahy NL, Chambeyron S. Do higher-order chromatin structure and nuclear reorganization play a role in regulating Hox gene expression during development? Cold Spring Harbor symposia on quantitative biology. 2004;69:251–257. doi: 10.1101/sqb.2004.69.251. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & development. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Current opinion in cell biology. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Current biology. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. Duplicating a tangled genome. Science. 1998;281:1466–1467. doi: 10.1126/science.281.5382.1466. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B. High-Resolution Mapping Reveals Links of HP1 with Active and Inactive Chromatin Components. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Functional architecture in the cell nucleus. The Biochemical journal. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussert C, Rasigni G, Llebaria A. Quantization of directional properties in biological structures using the Minimal Spanning Tree. J Theor Biol. 1988;135:295–302. doi: 10.1016/s0022-5193(88)80245-5. [DOI] [PubMed] [Google Scholar]
- Dussert C, Rasigni M, Palmari J, Rasigni G, Llebaria A, Marty F. Minimal spanning tree analysis of biological structures. J Theor Biol. 1987;125:317–323. doi: 10.1016/s0022-5193(87)80063-2. [DOI] [PubMed] [Google Scholar]
- Fackelmayer FO, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. European journal of biochemistry / FEBS. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- Fraser P. Transcriptional control thrown for a loop. Current opinion in genetics & development. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Hatton KS, Dhar V, Brown EH, Iqbal MA, Stuart S, Didamo VT, Schildkraut CL. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A, Iborra F. The balance sheet for transcription: an analysis of nuclear RNA metabolism in mammalian cells. Faseb J. 2000;14:242–254. [PubMed] [Google Scholar]
- Kipp M, Gohring F, Ostendorp T, van Drunen CM, van Driel R, Przybylski M, Fackelmayer FO. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Molecular and cellular biology. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Chubb JR, Hall CB, Capodieci P, Singer RH. The spatial order of transcription in mammalian cells. Journal of cellular biochemistry. 2007;102:609–617. doi: 10.1002/jcb.21495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Singer RH. Gene expression and the myth of the average cell. Trends in cell biology. 2003;13:4–6. doi: 10.1016/s0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. The Journal of cell biology. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. The Journal of cell biology. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Alonso WD, Acharya R, Berezney R. Spatiotemporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma. 2008a;117:553–567. doi: 10.1007/s00412-008-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Barbeitos M, Mukherjee L, Xu J, Fackelmayer FO, Berezney R. Identifying functional neighborhoods within the cell nucleus: proximity analysis of early S-phase replicating chromatin domains to sites of transcription, RNA polymerase II, HP1gamma, matrin 3 and SAF-A. Journal of cellular biochemistry. 2008b;105:391–403. doi: 10.1002/jcb.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes & development. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Nuclear matrins: identification of the major nuclear matrix proteins. Proc Natl Acad Sci U S A. 1991;88:10312–10316. doi: 10.1073/pnas.88.22.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez E, Kwon YS, Hutt KR, Hu Q, Cardamone MD, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell. 2008;132:996–1010. doi: 10.1016/j.cell.2008.01.051. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature genetics. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pliss A, Malyavantham K, Bhattacharya S, Zeitz M, Berezney R. Chromatin dynamics is correlated with replication timing. Chromosoma. 2009;118:459–470. doi: 10.1007/s00412-009-0208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Jones E, Iborra FJ, Kimura H, Sugaya K, Cook PR, Jackson DA. Specialized transcription factories within mammalian nuclei. Critical reviews in eukaryotic gene expression. 2000;10:21–29. [PubMed] [Google Scholar]
- Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS biology. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin SV, Iarovaia OV, Sjakste N, Sjakste T, Bagdoniene L, Rynditch AV, Eivazova ER, Lipinski M, Vassetzky YS. Chromatin domains and regulation of transcription. J Mol Biol. 2007;369:597–607. doi: 10.1016/j.jmb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol Cell Biol. 1994;72:177–185. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Singer RH. Gene expression within a dynamic nuclear landscape. The EMBO journal. 2006;25:3469–3479. doi: 10.1038/sj.emboj.7601226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli E, Levi M, Rossi E. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J Cell Sci. 1994;107(Pt 11):3097–3103. doi: 10.1242/jcs.107.11.3097. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, Choi JY, Vradii D, Zaidi SK, Pratap J, Young D. Organization of transcriptional regulatory machinery in nuclear microenvironments: Implications for biological control and cancer. Adv Enzyme Regul. 2007 doi: 10.1016/j.advenzreg.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, Zaidi SK, Young D, Choi JY, Gutierrez S, Pockwinse S. Nuclear microenvironments support assembly and organization of the transcriptional regulatory machinery for cell proliferation and differentiation. Journal of cellular biochemistry. 2004;91:287–302. doi: 10.1002/jcb.10777. [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi JY, Stein JL, Lian JB, Javed A. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends in cell biology. 2003;13:584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Strouboulis J, Wolffe AP. Functional compartmentalization of the nucleus. Journal of cell science. 1996;109(Pt 8):1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Belmont AS. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nature cell biology. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. Journal of cell science. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EH. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]