Abstract
Biomethylation is the major pathway for the metabolism of inorganic arsenic (iAs) in many mammalian species, including the human. However, significant interspecies differences have been reported in the rate of in vivo metabolism of iAs and in yields of iAs metabolites found in urine. Liver is considered the primary site for the methylation of iAs and arsenic (+3 oxidation state) methyltransferase (As3mt) is the key enzyme in this pathway. Thus, the As3mt-catalyzed methylation of iAs in the liver determines in part the rate and the pattern of iAs metabolism in various species. We examined kinetics and concentration-response patterns for iAs methylation by cultured primary hepatocytes derived from human, rat, mice, dog, rabbit, and rhesus monkey. Hepatocytes were exposed to [73As]arsenite (iAsIII; 0.3, 0.9, 3.0, 9.0 or 30 nmol As/mg protein) for 24 hours and radiolabeled metabolites were analyzed in cells and culture media. Hepatocytes from all six species methylated iAsIII to methylarsenic (MAs) and dimethylarsenic (DMAs). Notably, dog, rat and monkey hepatocytes were considerably more efficient methylators of iAsIII than mouse, rabbit or human hepatocytes. The low efficiency of mouse, rabbit and human hepatocytes to methylate iAsIII was associated with inhibition of DMAs production by moderate concentrations of iAsIII and with retention of iAs and MAs in cells. No significant correlations were found between the rate of iAs methylation and the thioredoxin reductase activity or glutathione concentration, two factors that modulate the activity of recombinant As3mt. No associations between the rates of iAs methylation and As3mt protein structures were found for the six species examined. Immunoblot analyses indicate that the superior arsenic methylation capacities of dog, rat and monkey hepatocytes examined in this study may be associated with a higher As3mt expression. However, factors other than As3mt expression may also contribute to the interspecies differences in the hepatocyte capacity to methylate iAs.
Introduction
Inorganic arsenic (iAs) is biomethylated in a sequence of enzymatically catalyzed reactions that produce methylarsenic (MAs), dimethylarsenic (DMAs), and trimethylarsenic (TMAs) species, which contain either trivalent arsenic (AsIII) or pentavalent arsenic (AsV) (Thomas et al., 2004; Waters et al., 2004a). The extent of methylation and the valency of As determine the reactivity and toxicity of these metabolites of iAs. Because some toxic or carcinogenic effects attributed to iAs are likely mediated by its methylated metabolites, understanding the pathway of methylation of iAs is critical to assessing risk associated with exposure to this metalloid. Rates and patterns of in vivo methylation of iAs have been extensively examined in laboratory studies with various species, including humans. These studies show that most mammalian species rapidly convert iAs to MAs and DMAs. In rodents, TMAs metabolites are also found. Notably, urinary profiles of metabolites of iAs indicate differences among species in capacity to form and excrete various arsenicals. For example, MAs represents a greater fraction of urinary arsenic in humans and rabbits than in other species, e.g., rat or mice (Vahter 1994, 1999). In addition, methylated arsenicals were not detected in urine of iAs-treated marmoset and tamarin monkeys (Vahter and Marafante, 1985; Zakharyan et al., 1996;), chimpanzees (Vahter et al., 1995a), and guinea pigs (Healy et al., 1997), suggesting that these species do not methylate iAs. Tissue distribution of As methylation activity in the methylating species has not been systematically examined. Methylation of arsenic increases the rate of whole body clearance; hence, whole body retention of arsenic is higher in non-methylating species than in species that rapidly methylate arsenic. In the mouse, the highest specific As methylation activity was found in testes, followed by kidney, liver, and lung (Healy et al., 1998). However, because of its place in portal circulation and its mass, the liver is generally regarded as the major site for the methylation of orally administered iAs.
Arsenic (+3 oxidation state) methyltransferase (As3mt) catalyzes all reactions in the pathway that convert iAs into MAs, DMAs, and TMAs (Waters et al., 2004 a, b; Thomas et al., 2007). We have previously shown that As3mt is expressed in cultured primary rat and human hepatocytes (Drobna et al., 2004) and that a knockdown of AS3MT expression by RNA interference in human hepatocellular carcinoma (HepG2) cells is associated with a proportional loss of the capacity to methylate iAs (Drobna et al., 2006). Heterologous expression of rat As3mt in a human uroepithelial cell line that cannot constitutively methylate iAs confers the capacity to produce the methylated metabolites of iAs (Drobna et al., 2005). Other studies have shown that sequence polymorphisms of As3mt affect profiles for iAs metabolism in in vitro methylation systems (Wood et al, 2006), in cell cultures (Drobna et al., 2004), and in human subjects (Hernandez et al., 2008; Schlawicke Engstrom et al., 2007; Meza et al., 2005). Similarly, point mutations targeting key cysteine residues altered rates and yields of methylation of iAs by recombinant mouse, rat, and human As3mt (Li et al., 2005; Fomenko et al. 2007, Song et al., 2009). In As3mt knockout mice, diminished methylation of orally administrated arsenate (iAsV) is associated with high retention of arsenic in tissues and a reduced rate of whole body clearance of arsenic (Drobna et al., 2009). Taken together, these data suggest that As3mt expression and polymorphism might be key factors affecting rates and patterns of iAs methylation and, consequently, the distribution and retention of As.
Intracellular reductants that support or modify As3mt activity are likely to be among the factors that affect capacity of the liver to methylate iAs. Several NADPH-dependent enzymatic systems that support in vitro activity of recombinant As3mt have been previously identified, including: (i) thioredoxin (TRx) and TRx reductase (TR); (ii) glutaredoxin, glutathione (GSH) reductase, and GSH, and (iii) dihydrolipoic acid and TR (Waters et al., 2004a,b; Thomas et al., 2007). The TRx/TR/NADPH coupled system supported the highest rates of in vitro methylation of iAsIII by As3mt. GSH was not essential for As3mt activity, but addition of GSH to a reaction mixture containing As3mt and TRx/TR/NADPH stimulated the overall rate of iAs methylation (Waters et al., 2004b; Thomas et al., 2007). The roles of TR and TRx in in vitro methylation of iAsIII were further examined in the recombinant As3mt assay mixture, in which exogenous TRx, TR and NADPH were replaced with freshly prepared guinea pig liver cytosol. Although guinea pig liver is a good source of endogenous reductants, including TRx and TR, iAsIII was not methylated in an in vitro assay containing guinea pig liver cytosol (Healy et al., 1997). However, iAsIII was methylated in an in vitro assay system containing guinea pig cytosol and rat recombinant As3mt (Waters et al., 2004a). Addition of aurothioglucose (ATG), a specific inhibitor of Se-cysteine-containing enzymes, including TR, sharply reduced MAs and DMAs production in this reaction mixture. These results indicated that TR (with or without TRx), but not other reductants present in the guinea pig liver cytosol, was a key co-factor for the As3mt-catalyzed methylation of iAsIII.
In the work reported here, we have examined differences in the metabolism of iAs in primary cultured hepatocytes from six species (human, rat, mouse, rabbit, dog, and macaque rhesus). The use of cultured hepatocytes is consistent with role of the liver as a primary site for the methylation of arsenicals. This work examined the roles of As3mt expression and protein structure, TR activity, and GSH concentration in differences among species in capacity of hepatocytes for iAs methylation. We found that hepatocytes from rat, macaque, and dog had much greater capacities to methylate iAs than do hepatocytes from human, mouse or rabbit. Neither TR activity nor GSH concentration in hepatocytes differed significantly between these two groups of species. Differences in the methylation capacities of hepatocytes from different species could not be rationalized by differences in the predicted amino acid sequences of this protein. However, As3mt protein levels were higher in hepatocytes from the high-methylator group than the lower methylator group. Thus, As3mt expression might be a key factor in interspecies differences in iAs metabolism by primary hepatocytes examined in this study.
Materials and Methods
Chemicals
A carrier-free [73As]arsenous acid (Brookhaven National Laboratory, Upton, NY) was reduced to arsenite ([73As]iAsIII) in a reaction with a metabisulfate-thiosulfate reagent (Reay and Asher, 1977). Sodium arsenite, 99% pure (Sigma; St. Louis, MO), was used as a carrier for [73As]iAsIII solutions. All other chemicals and reagents were of the highest grade available.
Cell culture
Freshly prepared suspensions of primary hepatocytes isolated from livers of a 7-week old male CD-1 mouse, 10-week old male Sprague-Dawley rat, 9-month old male Beagle dog, 4-month old female New Zealand White rabbit, 5-year old male Rhesus macaque, and a 56-year old woman were obtained from CellzDirect (Pittsboro, NC). Following the CellzDirect instructions, hepatocytes were plated on collagen-I coated 12- and 6-well plates (Becton-Dickinson) at respective densities of 0.75 and 1.5 × 106 viable cells/well for all animal species except mouse. The plating densities for mouse hepatocytes were 0.5 and 1.5 × 106 viable cells/well for 12- and 6-well plate, respectively. Human hepatocytes that are smaller in size than hepatocytes for the other species were plated in 24-well plates at 0.5 × 106 viable cells per well. The plating medium was William’s medium E (Sigma) containing 10% certified fetal bovine serum (FBS; Gibco), 2 mM glutamine (Sigma), 100 units/mL penicillin (Sigma), 100 µg/mL streptomycin (Sigma), 0.5 µM dexamethasone (Sigma), 5 µg/ml bovine insulin (Sigma), 5 µg/ml human transferrin (Sigma), and 5 ng/ml sodium selenite (Sigma). The same medium without FBS was used to culture attached hepatocyte monolayers for 3 to 5 days at 37 °C in a 95% air/5% CO2 atmosphere. The medium was changed daily.
Treatment
Hepatocytes from each species were used in two experimental designs. The first design focused on time course of MAs and DMAs production from iAsIII over a 24-hour incubation period; the second examined 24-hour yields of MAs and DMAs in response to various concentrations of iAsIII. To control for differences among species in number of cultured cells, concentrations of iAsIII were adjusted for cellular protein. Protein concentrations were determined shortly before the treatment with iAsIII using hepatocytes cultured in the same plates that were prepared for the treatment. The average protein concentration for 3 wells was used to determine the amount of iAsIII needed for the treatment. For time course experiments, cells were exposed to 0.3 nmol [73As]iAsIII per milligram of protein and media and cells were harvested after 3, 6 and 24 hours. For concentration-response experiments, cells were exposed to 0.3, 0.9, 3.0, 9.0 or 30 nmol [73As]iAsIII per milligram of protein and media and cells were harvested after 24 hours. Hepatocytes cultured in 12-well plates (24-well plates for human donor) were used in both types of experiments.
Analysis of iAs metabolites
Radioactivity associated with harvested cells and culture media were measured using Minaxi 5000 gamma counter (Packard Instruments Co., Downers Grove, IL). Radiolabeled metabolites of [73As]iAsIII were analyzed in CuCl extracts from cells and media by ion-exchange thin-layer chromatography (TLC) (Styblo et al., 1995a, 1995b, 1996). Radioactivity on TLC plates was monitored with a FLA 2000 imaging system (FujiFilm, Stamford, CT). Because CuCl extracts were oxidized with hydrogen peroxide before TLC separation, this technique could not determine the original oxidation states of As in separated metabolites. Thus, the As species detected by TLC were referred to generically as iAs (i.e., iAsIII + iAsV), MAs (i.e., MAsIII + MAsV), and DMAs (i.e., DMAsIII + DMAsV).
RT-PCR, DNA sequencing and multiple protein sequence alignment
As3mt mRNA levels were determined by touch-down PCR as previously described (Drobna et al., 2004). Briefly, total RNA was extracted using TRIzol reagent (Life Technology Inc., Gaithersburg, MD). Extracted RNA was treated with DNase (Promega, Madison, WI) and purified using an RNaesy Minikit (Qiagen, Valencia, CA). One microgram of RNA was reverse transcribed using SuperScript II reverse transcriptase and random primers (Invitrogen, Carlsbad, CA). An aliquot of cDNA representing 50 ng of RNA and As3mt primers specific for a conserved as3mt sequence were used for the PCR step. The primer sequences and the complementary nucleotide sequences in As3mt mRNA from rat, mouse, human, dog, and macaque are shown in Table 1. For all species examined, the PCR step amplified a 477-bp DNA fragment. The DNA fragment for each species was sequenced in both forward and reverse directions at the UNC Chapel Hill Automated DNA Sequencing Facility. The sequences of the DNA fragments were then blasted against as3mt sequences available from GenBank, including that for rat (Rattus norvegicus: NM_080890), human (Homo sapiens: NM_020682;), and mouse (Mus musculus: NM_020577), and also against dog and rhesus as3mt sequences (Canis familiaris: XM_543995; Macaca mulatta: XM_001113391) predicted by the automated computational analysis GNOMON. ExPasy proteomics tools (Swiss Institute of Bioinformatics; www.us.expasy.org) were used to translate the as3mt nucleotide sequences into the corresponding amino acid sequences. A multiple protein sequence alignment was generated using a EMBL-EBI ClustalW program (European Bioinformatics Institute, European Molecular Biology Laboratory; www.ebi.ac.uk/clustalw).
Table 1.
PCR primers and the corresponding nucleotide sequences in As3mt mRNA from rat, mouse, human, dog, and macaque*.
| FORWARD | REVERSE | |
|---|---|---|
| PCR Primers | ATTTTGGATCTGGGCAGTGGGAGT | TCTGCCCTCCACGTTTGGTCACT |
| Rat as3mt | ATTTTGGATCTGGGCAGTGGGAGT | TCTGCCCTCCACGTTTGGTCACT |
| Mouse as3mt | ATTTTGGATCTGGGCAGTGGGAGT | TCTGCCCTCCACGTTTGGTCACT |
| Human as3mt | ATTTTGGATCTGGGTAGTGGAAGT | TCTGCCCTCCACGTTTGGTCACT |
| Dog as3mt | ATTTTGGACCTGGGCAGTGGAAGT | TCTGCCCTCCACGTTTAGTCACT |
| Macaque as3mt | ATTTTGGATCTGGGTAGTGGAGGT | TCTGCCCTCCACGTTTGGTCACT |
| Rabbit as3mt | N/A** | N/A |
Mismatched nucleotides in as3mt sequences are underlined and bolded.
Nucleotide sequence of rabbit as3mt is not available.
Immunoblot analysis
Protein extracts for immunoblot analyses were prepared from hepatocytes cultured in 6-well plates (24-well plates for the human cells). Cells were lysed in RIPA buffer containing 50 mM Tris-HCl (pH 7.4), 1% NP-40, 150 mM NaCl, 0.25% Na-deoxycholate, 1 mM NaF, 1 mM EDTA, and protease and phosphatase inhibitors (Drobna et al., 2005). Cell lysates were spun at 10,000 × g and supernatants were stored at −80° C until analyzed. Proteins in cell lysates were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride Immobilon-P membranes (Millipore Corp., Bedford, MA). For As3mt analysis, the membranes were probed with a rabbit anti-serum raised against the recombinant rat As3mt (Alpha Diagnostic International, San Antonio, TX) or with a rabbit polyclonal antibody directed against a 20-amino acid fragment (341–360) of human AS3MT (Wood et al, 2006) (gift from Professor R.M. Weinshilboum, Mayo Clinic College of Medicine, Rochester, MN). β-actin was detected using a mouse monoclonal antibody (Abcam, Cambridge, MA). The antigen-antibody complexes were visualized after incubation with the corresponding HRP-conjugated antibody (Santa Cruz Biotechnology, Santa Cruz, CA) by enhanced chemiluminescence detection (Amersham, Biosciences, Pittsburgh, PA) using a computerized GeneGnome imaging system (Syngene, Frederick, MD).
Biochemical analyses
Intracellular concentrations of GSH and GSH disulfide (GSSG) were determined in cell lysates prepared in 5% sulfosalicylic acid, using a GSSG reductase-coupled recycling assay (Styblo et al. 1997). TR activity was measured in cell lysates prepared in 0.1 M Na/K phosphate-0.5% Triton ×100 buffer (pH 7.4), using 5,5’-dithiobis(2-nitrobenzoic acid) as a substrate (Lin at al. 1999). Bicinchoninic acid assay kit (Sigma) was used to determine protein concentrations in hepatocyte monolayers before treatment with [73As]iAsIII and in hepatocyte lysates or extracts prepared for various biochemical analyses. Calibration curves for this assay were prepared using bovine serum albumin as a standard.
Statistical analysis
One-way ANOVA and Tukey’s multiple comparison test were used to analyze differences between species. Differences between hepatocytes from species with low (human, mouse and rabbit) and high (rat, macaque and dog) capacities to methylate iAs were analyzed using unpaired t-test (GraphPad Software Inc., San Diego, CA). Nonparametric Spearman correlation was used to analyze associations between the rate or pattern of iAs methylation and the concentration or activity of the As3mt co-factors in cultured hepatocytes. Differences or correlations with p<0.05 were considered statistically significant.
Results
Kinetics of iAs methylation: MAs and DMAs yields
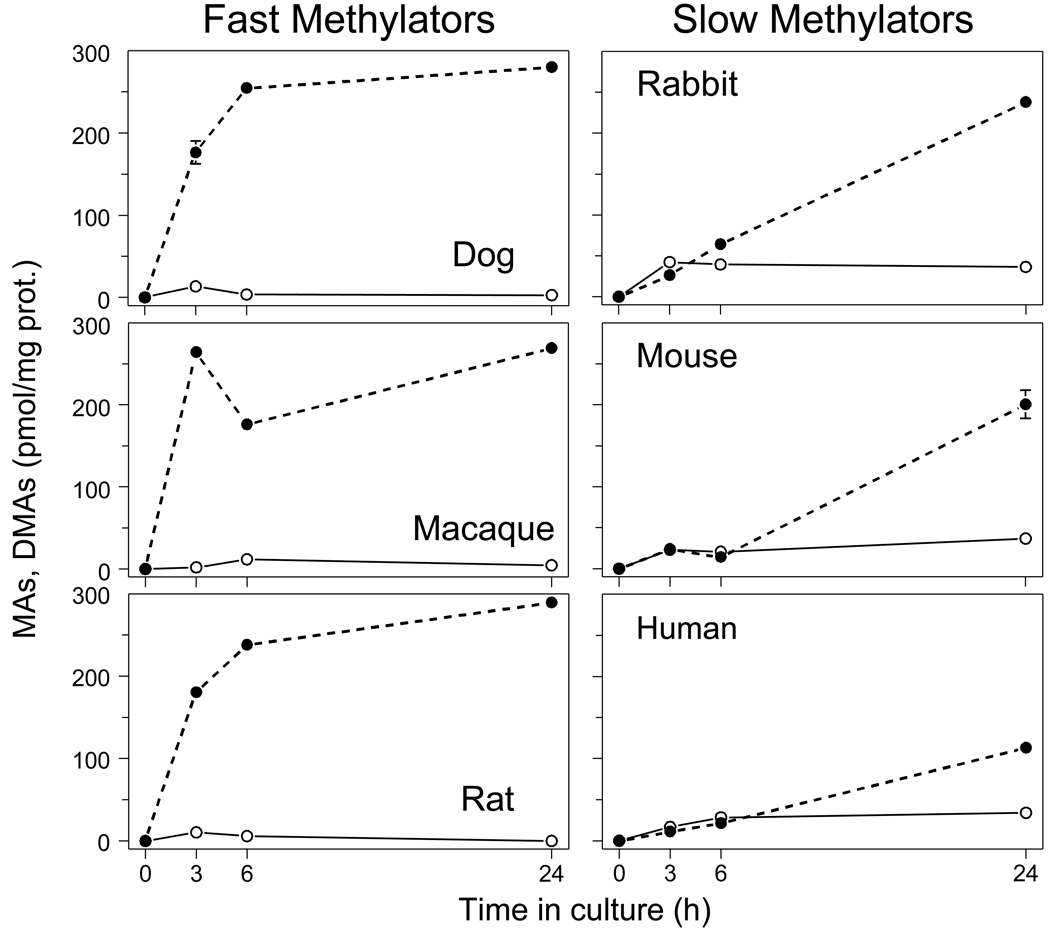
To examine interspecies differences in rates and patterns of iAs methylation, we exposed primary cultures of hepatocytes isolated from human, rat, mouse, dog, rabbit, and macaque to a low concentration of [73As]iAsIII (0.3 nmol/mg prot.) and quantified radiolabeled MAs and DMAs in cells and in medium after 3-, 6- or 24-hours of exposure (Fig. 1). The kinetics of iAsIII methylation by dog, macaque, and rat hepatocytes were characterized by fast production of DMAs over the first 3 to 6 hours. After 24 hours, 90 to 97% of total As in these cultures was DMAs; MAs was a minor metabolite (0.5 to 1.5% of total As). In contrast, the rates of DMAs production were much lower in cultures of rabbit, mouse, and human hepatocytes. Time courses for DMAs production for these three species were almost linear over the 24-hour interval. At 24 hours, DMAs accounted for 38 to 79% of total As in the cultures. During the first 6 hours, MAs yields were similar to those for DMAs. After 24 hours, 11 to 12% of total As in cultures was MAs. For all time points, DMAs/MAs ratios were significantly higher in cultures of dog, macaque, and rat hepatocytes compared to cultures of hepatocytes from rabbit, mouse, or human. After 24 hours, DMAs/MAs ratios in faster methylating hepatocyte cultures ranged from 60 (macaque) to 193 (rat) and in slower methylating cultures the ratios ranged from 3.4 (human) to 6.7 (rabbit).
Figure 1.
Kinetics of iAsIII methylation by primary cultures of hepatocytes isolated from dog, macaque, rat, rabbit, mouse, and human. The yields of MAs (○) and DMAs (●) produced by hepatocytes exposed to [73As]iAsIII (0.3 nmol/mg prot.) for 3, 6 and 24 hours (Mean ± SD, n = 3).
Kinetics of iAs methylation: Cellular retention of As species
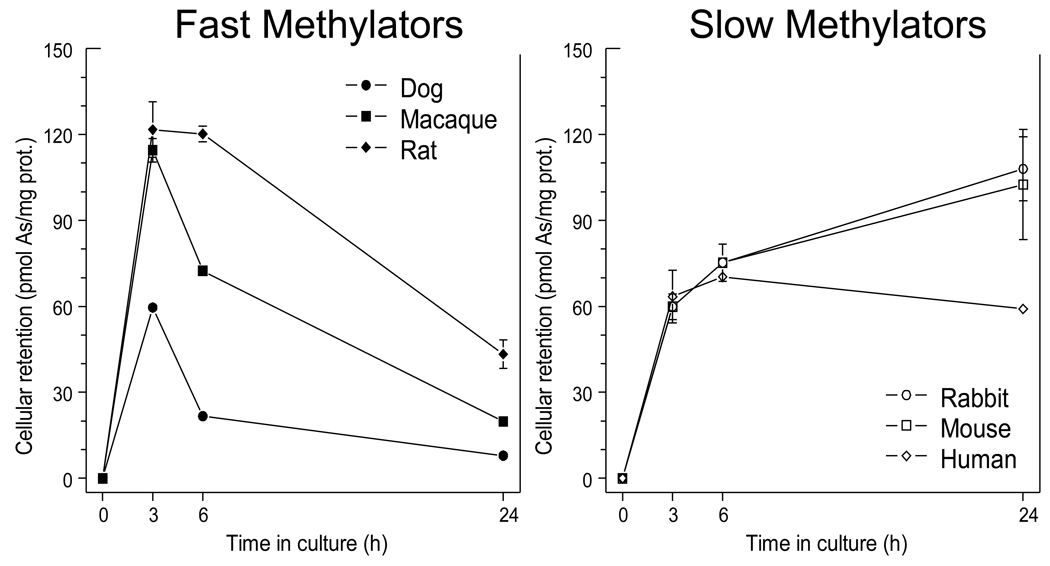
The sum of all arsenicals (iAs + MAs + DMAs) retained in hepatocytes exposed to 0.3 nmol [73As]iAsIII/mg protein significantly changed over the 24-hour treatment period. Notably, the dynamics of the cellular retention differed between faster methylating dog, macaque and rat hepatocytes and slower methylating rabbit, mouse and human hepatocytes (Fig. 2). Hepatocytes from all six species rapidly accumulated 73As during the first 3 to 6 hours of the exposure. Between 3 and 24 hours, levels of 73As in slower methylating hepatocytes remained high or further increased. In contrast, 73As initially accumulated in hepatocytes from faster methylating group was released into the medium. After 24 hours, the faster methylating hepatocytes retained from 8 to 43 pmol As/mg prot. as compared to 59 to 90 pmol As/mg prot. for the slower methylating hepatocytes.
Figure 2.
Cellular retention of total arsenic by dog, macaque, rat, rabbit, mouse, and human hepatocytes as a function of the exposure time. Hepatocytes were exposed to [73As]iAsIII (0.3 nmol/mg prot.) and the radioactivity associated with cells was measured after 3, 6 and 24 hours (Mean ± SD, n = 3).
Concentration dependence of MAs and DMAs yields
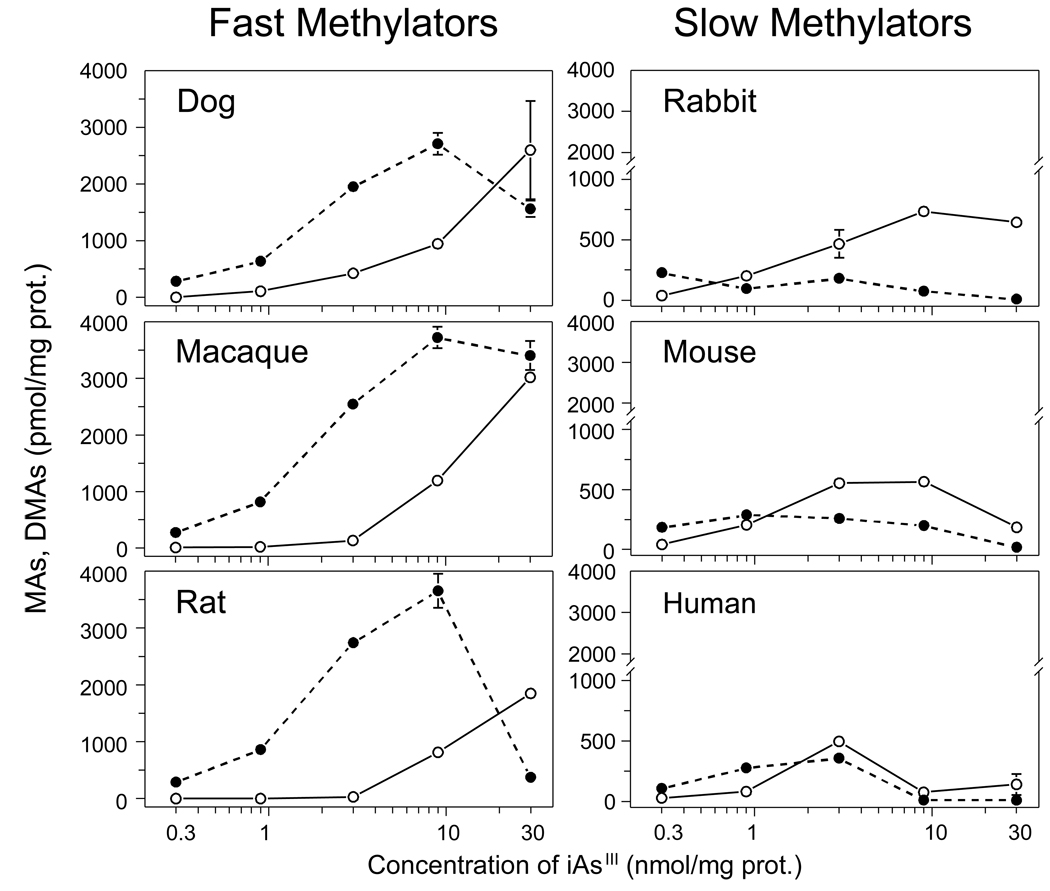
To examine for the concentration dependency of iAs methylation, hepatocyte cultures were exposed to [73As]iAsIII at final concentrations of 0.3, 0.9, 3.0, 9.0, or 30 nmol/mg protein. MAs and DMAs yields were determined in cells and in medium after a 24-hour exposure (Fig. 3). In cultures of faster methylating hepatocytes from dog, macaque, and rat, DMAs was the major methylated metabolite over the range of iAsIII concentrations of 0.3 to 9.0 nmol/mg prot. However, DMAs production was reduced in cultures exposed to 30 nmol iAsIII. At the highest exposure level of 30 nmol/mg prot., average MAs yield equaled DMAs yield in hepatocytes from macaque and was higher than DMAs yield in hepatocytes from dog and rat. DMAs was also the major methylated metabolite in slower methylating hepatocytes from rabbit, mouse, and human exposed to low concentrations of 0.3 or 0.9 nmol iAsIII/mg prot. At higher iAsIII concentrations, MAs was the major metabolite as DMAs production decreased or remained unchanged. Production of both MAs and DMAs was inhibited in cultures of human hepatocytes exposed to 9 or 30 nmol iAsIII/mg prot. and in cultures of mouse hepatocytes exposed to 30 nmol iAsIII/mg protein. Human hepatocytes were the least efficient methylators of iAsIII at any exposure level.
Figure 3.
The concentration-response patterns for iAsIII methylation by dog, macaque, rat, rabbit, mouse, and human hepatocytes. The yields of MAs (○) and DMAs (●) after 24-hour exposures to [73As]iAsIII at the initial concentrations of 0.3, 0.9, 3, 9, and 30 nmol iAsIII/mg protein (Mean ± SD, n = 3).
Concentration dependence of iAs methylation rates
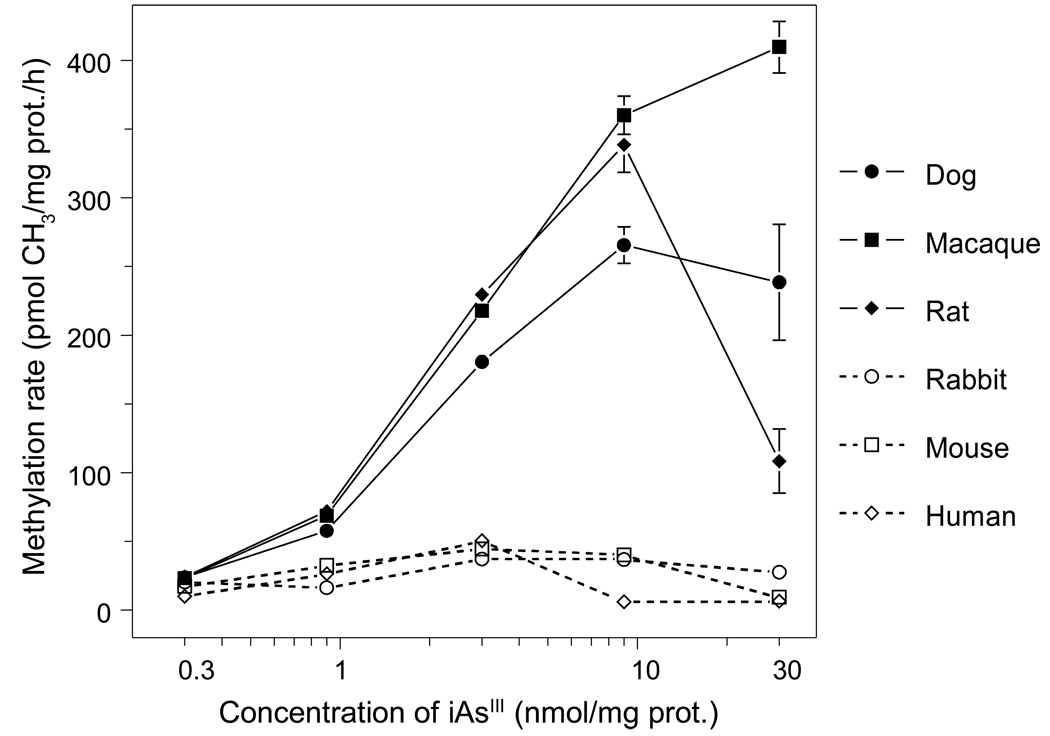
Average rates of iAsIII methylation were calculated for each exposure level as pmol of methyl groups transferred to iAsIII per hour per milligram of protein, using MAs and DMAs yields after a 24-hour exposure (Fig. 4). For each exposure level, methylation rates were consistently higher in cultured dog, macaque, or rat hepatocytes than in cultured rabbit, mouse, or human hepatocytes. Unlike other species, rabbit hepatocytes displayed a low and unchanged rate of iAs methylation at all exposure levels. Methylation rates increased proportionally with the initial concentration of iAsIII for dog, macaque, or rat hepatocytes exposed to up to 9 nmol iAsIII/mg protein; rates were proportional for rabbit, mouse, or human hepatocytes exposed to up to 3 nmol iAsIII/mg protein. Initial slopes for concentration-methylation rate curves for dog, macaque, or rat hepatocytes were greater than those for rabbit, mouse, or human hepatocytes. Exposure of rat hepatocytes to 30 nmol iAsIII/mg protein sharply decreased the methylation rate, but did not affect the methylation rate for dog hepatocytes. The iAsIII methylation rate decreased from peak values in cultured human hepatocytes exposed to 9 or 30 nmol iAsIII/mg protein and in cultured mouse hepatocytes exposed to 30 nmol iAsIII/mg protein. Rates of iAs methylation by macaque hepatocytes rose proportionally over the entire range of iAsIII concentrations.
Figure 4.
The rates of iAsIII methylation by dog, macaque, rat, rabbit, mouse, and human hepatocytes as a function of the initial concentration of [73As]iAsIII in the culture. The methylation rates (pmol CH3/mg protein per hour) are shown for cells exposed for 24 hours to 0.3, 0.9, 3, 9, and 30 nmol iAsIII/mg protein (Mean ± SD, n = 3).
Concentration dependence of arsenical distribution between hepatocytes and medium
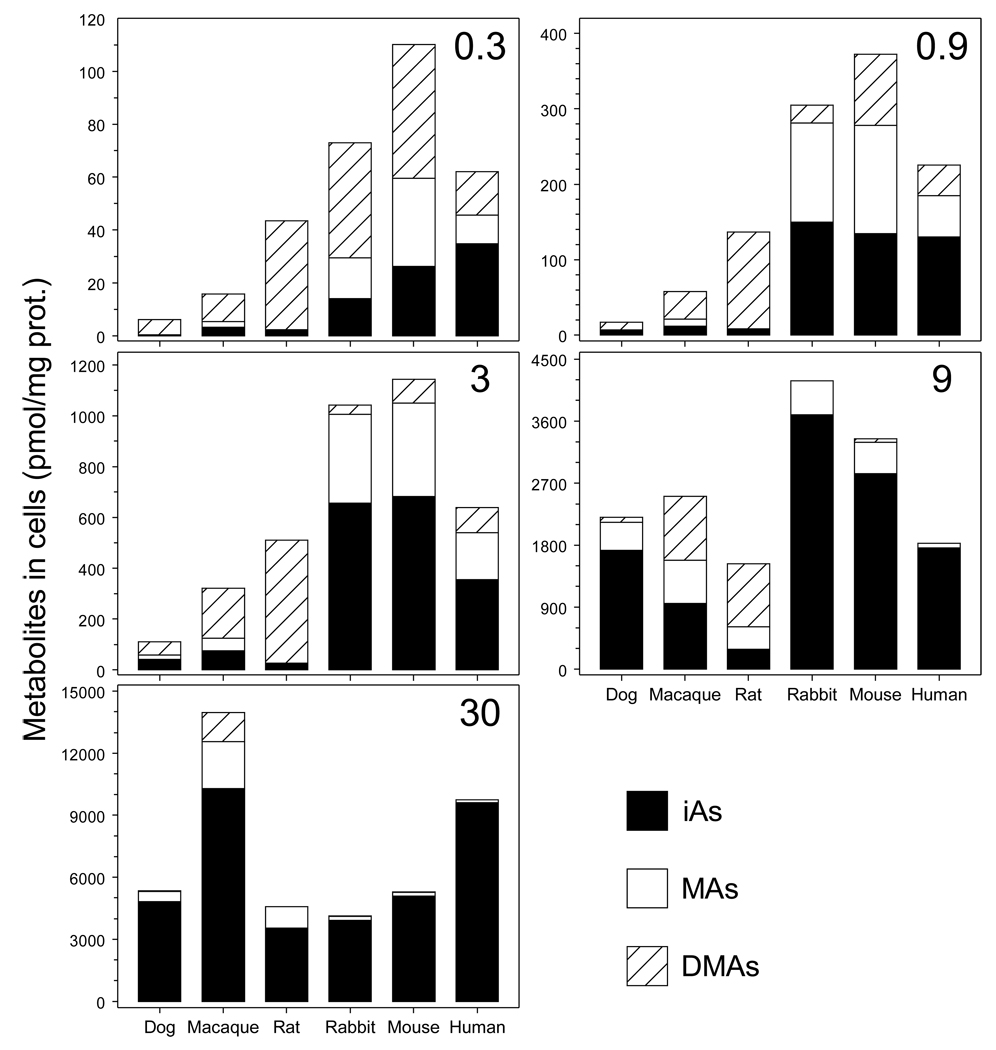
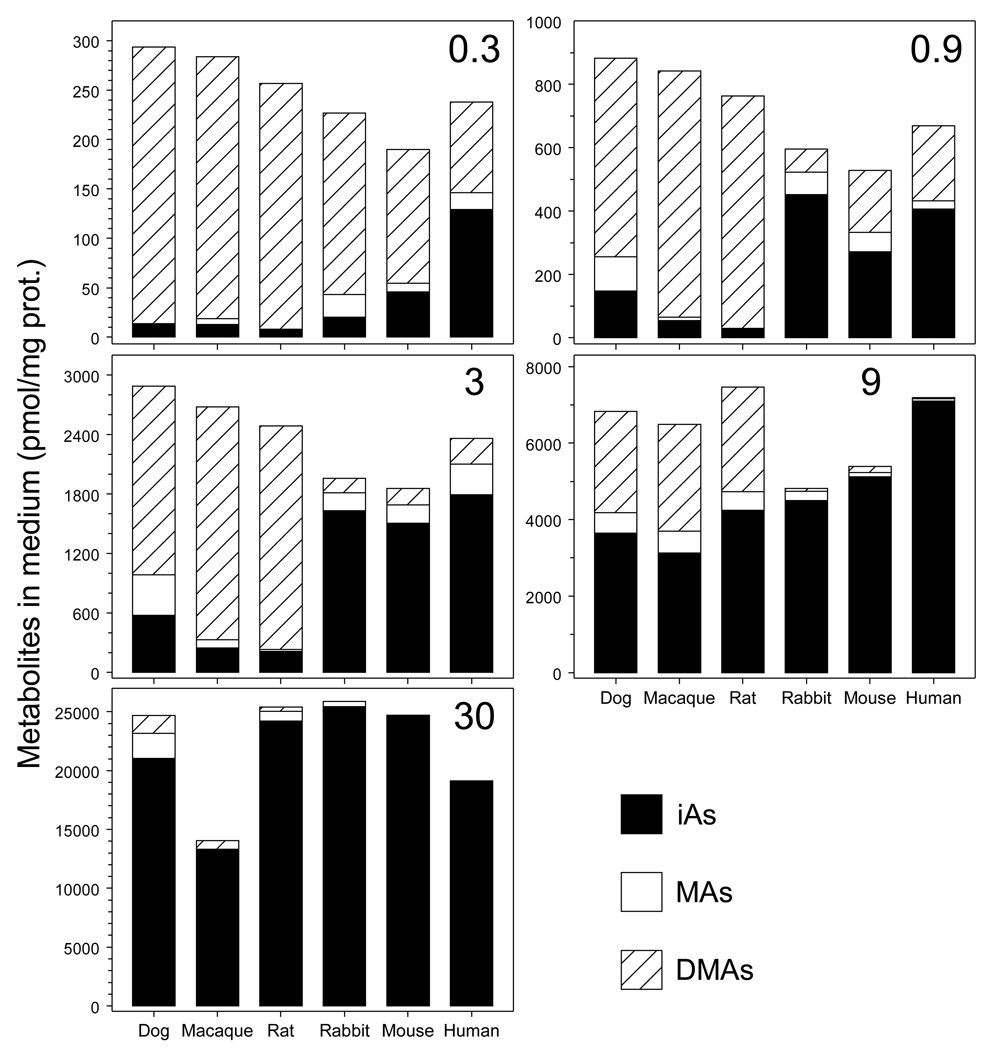
The level of iAsIII in medium affected both concentrations and proportions of As species retained by hepatocytes after 24-hour exposures (Fig. 5). At low exposures levels (0.3 to 3 nmol iAsIII/mg protein), DMAs was the major arsenical in dog, macaque, or rat hepatocytes, accounting for 46 to 95% of total cellular As. Over this range, MAs accounted for less than 16%. In contrast, MAs accounted for 18 to 42% of total As retained by rabbit, mouse, or human hepatocytes and iAs accounted for 19 to 63%. At these exposure levels, dog, macaque, or rat hepatocytes retained less total As (i.e., iAs + MAs + DMAs) than did rabbit, mouse, or human hepatocytes. At higher exposure levels (9 and 30 nmol iAsIII/mg protein) concentration of DMAs in dog, macaque, or rat hepatocytes decreased and concentrations of MAs and iAs increased. Rabbit, mouse, or human hepatocytes exposed to 9 or 30 nmol iAsIII/mg protein retained almost exclusively iAs; the concentration and percentage of MAs decreased in a concentration-dependent manner. At the lowest exposure level (0.3 nmol iAsIII/mg protein), DMAs was the major arsenical found in media of all cultures (Fig. 6). At the higher exposure levels, DMAs concentrations in media gradually decreased. However, DMAs concentrations in medium were consistently higher in dog, macaque and rat hepatocyte cultures as compared to the cultures of hepatocytes from rabbit, mouse, or human.
Figure 5.
Intracellular concentrations of the metabolites of iAsIII in dog, macaque, rat, rabbit, mouse, and human hepatocytes as a function of the initial concentration of [73As]iAsIII in the culture. The intracellular concentrations of iAs, MAs, and DMAs normalized for cellular protein (Mean, n = 3) are shown for cells exposed for 24 hours to 0.3, 0.9, 3, 9, and 30 nmol iAsIII/mg protein.
Figure 6.
Concentrations of the metabolites of iAsIII in culture medium of dog, macaque, rat, rabbit, mouse, and human hepatocytes as a function of the initial concentration of [73As]iAsIII in the culture. The concentrations of iAs, MAs, and DMAs normalized for cellular protein (Mean, n = 3) are shown for cells exposed for 24 hours to 0.3, 0.9, 3, 9, and 30 nmol iAsIII/mg protein.
As3mt expression
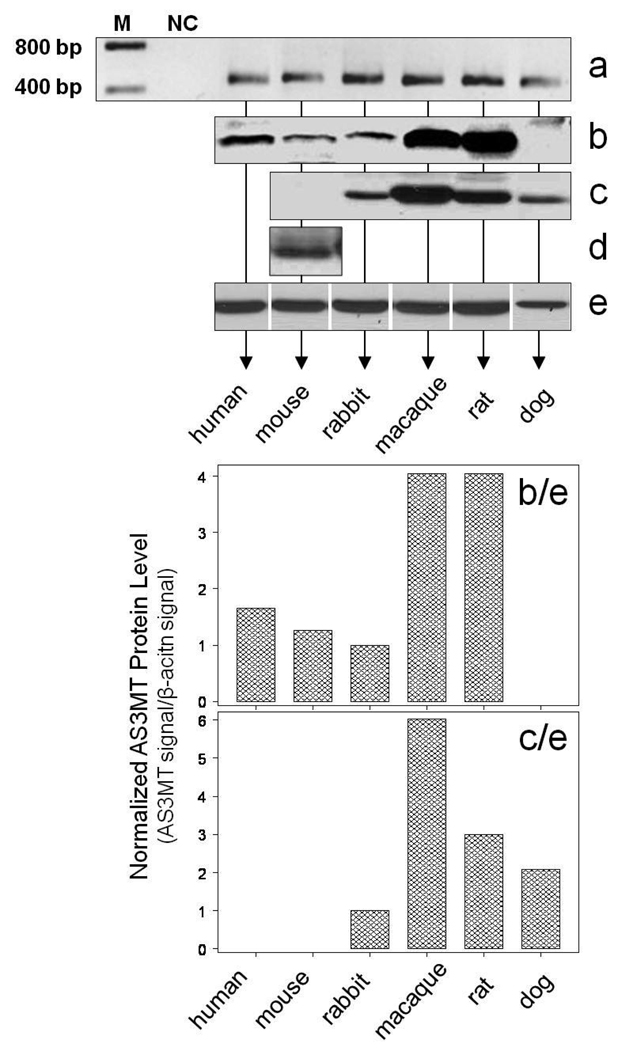
Expression of As3mt in cultured hepatocytes before exposure to iAsIII was examined at the levels of mRNA and protein (Fig. 7). As3mt mRNA was detected by RT-PCR in hepatocytes from all six species (Fig. 7a). The first immunoblot analysis using rabbit anti rat recombinant As3mt antiserum detected immunoreactive protein in human, mouse, rabbit, macaque, and rat hepatocytes but not in dog hepatocytes (Fig. 7b). The second immunoblot analysis used a rabbit polyclonal anti-human AS3MT antibody (1:5,000 dilution). Here, As3mt protein was detected in hepatocytes from rabbit, macaque, rat and dog, but not in hepatocytes from mouse (Fig. 7c). This antibody was not used for analysis of AS3MT in human hepatocytes due to a limited number of the cells available for this study. To increase assay sensitivity, the immunoblot analysis of As3mt in mouse hepatocytes was repeated using the rabbit anti-human AS3MT antibody diluted 1 to 2,000. Under these conditions, a weak signal for As3mt protein was detected (Fig. 7d). The As3mt immunoblot signals in hepatocytes from the six species were then normalized to the corresponding immunoblot signals of β-actin (Fig. 7e). The normalized values for the two different anti-AS3MT antibodies show that the macaque and rat and, possibly, the dog hepatocytes contained higher levels of As3mt than did the hepatocytes from rabbit, mouse or human (Fig. 7b/e and Fig. 7c/e).
Figure 7.
Expression of As3mt in cultured primary hepatocytes from dog, macaque, rat, rabbit, mouse, and human: Analysis of As3mt mRNA by RT-PCR (a); As3mt protein detected by rabbit anti-serum against recombinant rat As3mt (b); As3mt protein detected by rabbit polyclonal antibody against human AS3MT diluted 1:5,000 (c) and 1:2,000 (d); immunoblot analysis of β-actin (e); As3mt protein level detected by the rabbit anti-serum normalized for β-actin (b/e); As3mt protein levels detected by the rabbit polyclonal antibody normalized for β-actin (c/e).
TR activity and GSH and GSSG concentrations
Mono- and dithiol compounds are modulators of the catalytic activity of As3mt. TR is required for As3mt catalysis of iAs methylation in in vitro assays and GSH modulates MAs and DMAs yields in in vitro TR-dependent As3mt-catalyzed methylation of iAsIII (Waters et al., 2004a,b; Thomas et al., 2007). Here, TR activity and GSH and GSSG concentrations were determined in cultured hepatocytes from the six species before exposures to iAsIII (Table 2). No statistically significant correlations were found between rates of iAsIII methylation and TR activity, GSH, or GSSG concentration regardless of the exposure level (data not shown). Categorizing hepatocytes into faster and slower methylation groups as described above found no correlation between methylation rate and TR activity or between methylation rate and GSH or GSSG concentrations (data not shown).
Table 2.
Protein and GSH concentrations and TR activity in cultured primary hepatocytes from six mammalian species (mean ±SD, n = 3 or 4).
| SPECIES | Plating density (cells/ well) |
Protein | GSH (nmoles/ mg protein) |
GSSG (nmoles/ mg protein) |
GSH/ GSSG Ratio |
TR activity (U/mg/ protein) |
|
|---|---|---|---|---|---|---|---|
| mg/well | mg/106 cells |
||||||
| *Human | 0.25 ×106 | 0.12 ± 0.02 |
0.49 | 70.7 ± 2.74 |
1.48 ± 0.01 |
48 ± 2.2 |
5.1 ± 0.08 |
| Rabbit | 0.75 ×106 | 0.72 ± 0.20 |
0.96 | 135.1 ± 4.55 |
0.78 ± 0.03 |
173 ± 11.9 |
2.6 ± 0.03 |
| Mouse | 0.5 ×106 | 0.41 ± 0.04 |
0.81 | 101.5 ± 1.40 |
1.98 ± 0.01 |
51 ± 0.9 |
5.5 ± 0.05 |
| Macaque | 0.75 ×106 | 0.45 ± 0.05 |
0.60 | 152.7 ± 2.64 |
0.52 ± 0.03 |
292 ± 20.2 |
2.0 ± 0.05 |
| Rat | 0.75 ×106 | 0.52 ± 0.00 |
0.70 | 33.6 ± 4.35 |
0.26 ± 0.09 |
128 ± 30.8 |
3.6 ± 0.03 |
| Dog | 0.75 ×106 | 0.38 ± 0.04 |
0.51 | 115.2 ± 3.81 |
2.55 ± 0.13 |
45 ±3.7 |
1.8 ± 0.04 |
Human hepatocytes were cultured in 24-well plates; cells from all other species were cultured in 12-well plates.
Discussion
The capacity of cultured cells to methylate iAs can be characterized by the rate of conversion of iAs to MAs and DMAs and by relative yields of DMAs and MAs or by the DMAs/MAs ratio at a range of low to high exposures to iAs. Using these criteria, we have previously shown that at the same substrate concentrations cultured rat primary hepatocytes methylated iAsIII much faster than did primary human hepatocytes from several donors (Styblo et al., 1999). In addition, methylation of iAsIII by human hepatocytes, specifically DMAs production, was found to be sensitive to substrate inhibition, resulting in DMAs/MAs ratios < 1 even in hepatocytes exposed to relatively low initial concentrations of iAsIII. At equal iAsIII concentrations, rat hepatocytes produced DMAs as the major metabolite and DMAs/MAs ratios were consistently higher than those in human hepatocytes. We have also shown that the capacity of primary human hepatocytes to methylate iAsIII varies dramatically among donors (Drobna et al., 2004, 2010). The highest (maximal) methylation rates were usually recorded at iAsIII concentrations of 3 to 30 nmol/mg protein and ranged from about 2 to 30 pmol CH3 per milligram of protein per hour.
The main goal of this study was to compare metabolic profiles for iAsIII in primary cultures of hepatocytes from a human and from several mammalian species, some of which have previously been used in studies examining either toxicity or metabolism of iAs. We found that rates and patterns of iAsIII methylation by primary hepatocytes from one group of species (Sprague-Dawley rat, Rhesus macaque and Beagle dog) differed markedly from those in the other group of species (human, CD-1 mouse, and New Zealand White rabbit). Methylation of iAsIII by hepatocytes from the first group of species was characterized by high methylation rates, high DMAs/MAs ratios, fast clearance of DMAs from cells, and relative insensitivity to the inhibition of methylation by high iAsIII concentrations. In contrast, the second group of species exhibited low methylation rates, low DMAs/MAs ratios, delayed clearance of As species, and marked inhibition of the methylation reactions, especially DMAs production, by moderate iAsIII concentrations. Notably, the rates of iAs methylation by human hepatocytes used in this study exceeded the methylation rates reported for human hepatocytes in our previous studies (Drobna et al., 2004, 2010). The maximal methylation rate of 49 pmol CH3/mg prot. per hour was found in human hepatocytes exposed to 3 nmol iAsIII/mg prot. However, consistent with our previous findings, even this relatively high methylation rate was almost 7 times lower that the maximal methylation rate recorded for rat hepatocytes exposed to 10 nmol iAsIII/mg prot: 340 pmol CH3/mg prot.
Complete predicted amino acid sequences for As3mt from human, mouse, rat and Rhesus monkey and the incomplete predicted amino acid sequence for dog As3mt were compared to see if sequences provide any insights into species differences in capacity or pattern of As methylation (Table 3). This comparison also included a portion of a predicted amino acid sequence for rabbit As3mt which we obtained by translating the nucleotide sequence of a 477-base DNA fragment generated in the present study by RT-PCR (Fig. 7a). The multiple protein sequence alignment of the full length of As3mt using Clustal-W program (www.ebi.ac.uk/clustalw) showed high degrees of sequence homology among all species. The highest degree of homology was found between the human and macaque As3mt sequences (Table 4). In contrast, the rate and pattern of iAs metabolism by primary hepatocytes from these two species were markedly different. Notably, sequences of motifs commonly found in non-DNA methyltransferases (Kagan and Clarke, 1994) were highly conserved in these six species. In addition, analogs of Cys157 and Cys207 that are essential for the activity of mouse As3mt (Fomenko et al., 2007) are found in As3mt sequences for the other 5 species examined in this study, including the fragment of rabbit As3mt. Thus, no major differences in predicted amino acid sequences in As3mt from these species can be invoked as a basis for differences in rates of formation or patterns of metabolites produced by hepatocytes exposed to iAsIII.
Table 3.
Clustal-W 2.0.5 multiple protein sequence alignment.
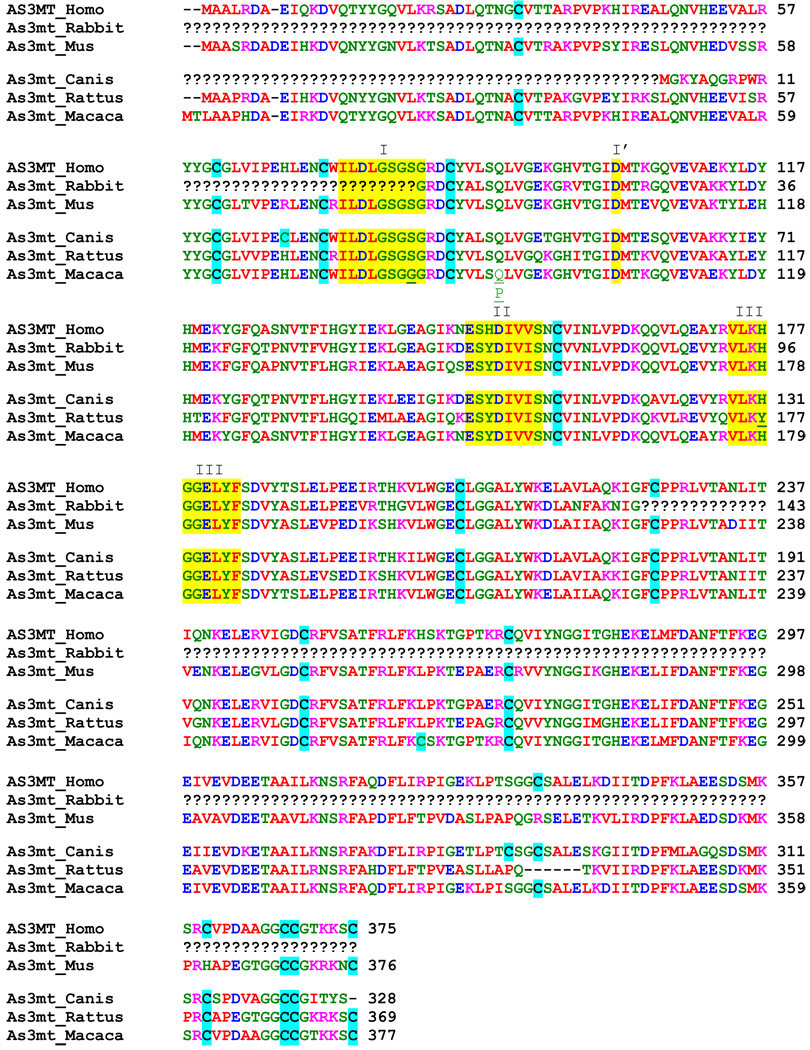 |
I, I’, II, and III: non DNA-methyltransferase sequence motifs.
Table 4.
Comparison of protein sequences of human, mouse, dog, rat, and macaque As3mt.
| SeqA | Name | Length(aa) | SeqB | Name | Length(aa) | Score* |
|---|---|---|---|---|---|---|
| 1 | AS3MT_Homo | 375 | 2 | As3mt_Mus | 376 | 75 |
| 1 | AS3MT_Homo | 375 | 3 | As3mt_Canis | 328 | 83 |
| 1 | AS3MT_Homo | 375 | 4 | As3mt_Rattus | 369 | 76 |
| 1 | AS3MT_Homo | 375 | 5 | As3mt_Macaca | 377 | 97(Q)/96(P)** |
| 2 | As3mt_Mus | 376 | 3 | As3mt_Canis | 328 | 75 |
| 2 | As3mt_Mus | 376 | 4 | As3mt_Rattus | 369 | 87 |
| 2 | As3mt_Mus | 376 | 5 | As3mt_Macaca | 377 | 75 |
| 3 | As3mt_Canis | 328 | 4 | As3mt_Rattus | 369 | 74 |
| 3 | As3mt_Canis | 328 | 5 | As3mt_Macaca | 377 | 82 |
| 4 | As3mt_Rattus | 369 | 5 | As3mt_Macaca | 377 | 76 |
Score represents % similarity between two As3mt protein sequences (SeqA and SeqB)
Macaque was a heterozygote for two variants of As3mt
As3mt plays a key role in the methylation of iAs in hepatic cells (Drobna et al., 2006). Therefore, the rate and pattern of iAsIII methylation by hepatocytes depend in part on As3mt expression. Results of our non-quantitative RT-PCR confirm that As3mt mRNA is present in hepatocytes from all six species examined in this study. We also performed a semiquantitative immunoblotting to compare As3mt protein levels in hepatocytes from different species. To account for probable interspecies variation in As3mt structures, we used two immunoreagents, an antiserum raised against recombinant rat As3mt and a polyclonal antibody against a 20-amino acid fragment (341–360) of human AS3MT (Wood et al., 2006), for detection and quantitation of As3mt. Notably, the human AS3MT sequence used for antibody production is highly conserved in rat, dog, macaque, and mouse As3mt (Table 3). (A corresponding sequence in rabbit As3mt has not been determined.) Results of two independent immunoblot analyses indicated that the hepatocytes from macaque, Sprague-Dawley rat, and possibly, from Beagle dog contained higher levels of As3mt protein than did hepatocytes from human, CD1 mouse or New Zealand White rabbit. Molecular masses of the dog, macaque and rabbit As3mt have not yet been determined; however, electrophoretic mobilities of the immunoreactive protein bands from the corresponding hepatocytes (Fig. 7c) suggests the molecular masses of dog, macaque, and rabbit As3mt to be similar to those of As3mt from the other species.
The metabolism of iAs in hepatocytes may also be affected by availability of intracellular reductants, specifically TR and GSH that are known to support or modulate in vitro activity of recombinant As3mts (Waters et al., 2004a, 2004b). However, results of this study found no statistically significant correlations between TR activity or GSH concentrations and rates of iAsIII methylation by primary hepatocytes from the six species. Thus, our data suggest the differences in rates and patterns of iAsIII methylation by hepatocytes from these species to be mainly due to differences in As3mt expression, specifically in the As3mt protein levels. In should be emphasized that hepatocytes from a single animal from each of the five animal species were examined in this study. However, inter-animal variations are possible within each of these animal species, even within the inbred mouse or rat strain. Thus, the patterns of iAs metabolism by primary hepatocytes as described in this study are not necessarily representative of the entire species. In addition, factors other than As3mt expression may also contribute to the interspecies differences in the capacity of primary hepatocytes to methylate iAsIII and to retain metabolites of iAs in cells. Identification of these factors is the goal of our future studies.
Acknowledgements
We thank Professor R.M. Weinshilboum (Mayo Clinic College of Medicine) for the generous gift of the rabbit anti human AS3MT antibody and Dr. David Paul (UNC Nutrition) for his help with quantitative analysis of immunoblot signals. This work has been supported in part by NIH grant 1R01 ES010845 to M.S. and a Clinical Nutrition Research Center Grant DK 56350. This manuscript has been reviewed in accordance with the policy of the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Drobna Z, Waters SB, Walton FS, LeCluyse EL, Thomas DJ, Styblo M. Interindividual variation in the metabolism of arsenic in cultured primary human hepatocytes. Toxicol. Appl. Pharmacol. 2004;201:166–177. doi: 10.1016/j.taap.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Drobna Z, Waters SB, Devesa V, Harmon AW, Thomas DJ, Styblo M. Metabolism and toxicity of arsenic in human urothelial cells expressing rat arsenic (+3 oxidation state)-methyltransferase. Toxicol. Appl. Pharmacol. 2005;207:147–159. doi: 10.1016/j.taap.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Xing W, Thomas DJ, Styblo M. shRNA silencing of AS3MT expression minimizes arsenic methylation capacity of HepG2 cells. Chem. Res. Toxicol. 2006;19:894–898. doi: 10.1021/tx060076u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Narenmandura H, Kubachka KM, Edwards BC, Herbin-Davis K, Styblo M, Le XC, Creed JT, Maeda N, Hughes MF, Thomas DJ. Disruption of the arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and retention of orally administered arsenate. Chem. Res. Toxicol. 2009;22:1713–1720. doi: 10.1021/tx900179r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Walton FS, Xing W, Paul DS, Thomas DJ, Styblo M. Metabolism of arsenic in human liver: The role of membrane transporters. Arch. Toxicol. 2010;84:3–16. doi: 10.1007/s00204-009-0499-7. [DOI] [PubMed] [Google Scholar]
- Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-Throughput Identification of Catalytic Redox-Active Cysteine Residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- Healy SM, Casarez EA, Ayala-Fierro F, Aposhian HV. Enzymatic methylation of arsenic compounds. V. Arsenite methyltransferase activity in tissues of mice. Toxicol. Appl. Pharmacol. 1998;148:65–70. doi: 10.1006/taap.1997.8306. [DOI] [PubMed] [Google Scholar]
- Healy SM, Zakharyan RA, Aposhian HV. Enzymatic methylation of arsenic compounds: IV. In vitro and in vivo deficiency of the methylation of arsenite and monomethylarsonic acid in the guinea pig. Mutat. Res. 1997;386:229–239. doi: 10.1016/s1383-5742(97)00014-8. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Xamena N, Sekaran C, Tokunaga H, Sampayo-Reyes A, Quinteros D, Creus A, Marcos R. High arsenic metabolic efficiency in AS3MT287Thr allele carriers. Pharmacogenet. Genomics. 2008;18:349–355. doi: 10.1097/FPC.0b013e3282f7f46b. [DOI] [PubMed] [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Li J, Waters SB, Drobna Z, Devesa V, Styblo M, Thomas DJ. Arsenic (+3 oxidation state) methyltransferase and the inorganic arsenic methylation phenotype. Toxicol. Appl. Pharmacol. 2005;204:164–169. doi: 10.1016/j.taap.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lin S, Cullen WR, Thomas DJ. Methylarsenicals and arsinothiols are potent inhibitors of mouse liver thioredoxin reductase. Chem. Res. Toxicol. 1999;12:924–930. doi: 10.1021/tx9900775. [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ. A novel S-Adenosyl-L-methionine: arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Meza MM, Yu L, Rodriguez YY, Guild M, Thompson D, Gandolfi AJ, Klimecki WT. Developmentally restricted genetic determinants of human arsenic metabolism: association between urinary methylated arsenic and CYT19 polymorphisms in children. Environ. Health Perspect. 2005;113:775–781. doi: 10.1289/ehp.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay PF, Asher CJ. Preparation and purification of 74As-labeled arsenate and arsenite for use in biological experiments. Anal. Biochem. 1977;78:557–560. doi: 10.1016/0003-2697(77)90117-8. [DOI] [PubMed] [Google Scholar]
- Schlawicke Engstrom K, Broberg K, Concha G, Nermell B, Warholm M, Vahter M. Genetic polymorphisms influencing arsenic metabolism: evidence from Argentina. Environ. Health Perspect. 2007;115:599–605. doi: 10.1289/ehp.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Geng Z, Zhu J, Li C, Hu X, Bian N, Zhang X, Wang Z. Structure-function roles of four cysteine residues in the human arsenic (+3 oxidation state) methyltransferase (hAS3MT) by site-directed mutagenesis. Chem. Biol. Interct. 2009;179:321–328. doi: 10.1016/j.cbi.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Styblo M, Delnomdedieu M, Hughes MF, Thomas DJ. Identification of methylated metabolites of inorganic arsenic by thin-layer chromatography. J. Chromatogr. B Biomed. Appl. 1995a;668:21–29. doi: 10.1016/0378-4347(95)00053-l. [DOI] [PubMed] [Google Scholar]
- Styblo M, Yamauchi H, Thomas DJ. Comparative in vitro methylation of trivalent and pentavalent arsenicals. Toxicol. Appl. Pharmacol. 1995b;135:172–178. doi: 10.1006/taap.1995.1220. [DOI] [PubMed] [Google Scholar]
- Styblo M, Hughes MF, Thomas DJ. Liberation and analysis of protein-bound arsenicals. J Chromatogr. B Biomed. Appl. 1996;677:161–166. doi: 10.1016/0378-4347(95)00490-4. [DOI] [PubMed] [Google Scholar]
- Styblo M, Serves SV, Cullen WR, Thomas DJ. Comparative inhibition of yeast glutathione reductase by arsenicals and arsenothiols. Chem. Res. Toxicol. 1997;10:27–33. doi: 10.1021/tx960139g. [DOI] [PubMed] [Google Scholar]
- Styblo M, Del Razo LM, LeCluyse EL, Hamilton GA, Wang C, Cullen WR, Thomas DJ. Metabolism of arsenic in primary cultures of human and rat hepatocytes. Chem. Res. Toxicol. 1999;12:560–565. doi: 10.1021/tx990050l. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Waters SB, Styblo M. Elucidating the pathway for arsenic methylation. Toxicol Appl Pharmacol. 2004;198:319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Styblo M. Arsenic (+3 oxidation state) methyltransferase and methylation of arsenicals. Exp. Biol. Med. 2007;232:3–13. [PMC free article] [PubMed] [Google Scholar]
- Vahter M, Marafante E. Reduction and binding of arsenate in marmoset monkeys. Arch. Toxicol. 1985;57:119–124. doi: 10.1007/BF00343121. [DOI] [PubMed] [Google Scholar]
- Vahter M. Species differences in the metabolism of arsenic compounds. Appl. Organomet. Chem. 1994;8:175–182. [Google Scholar]
- Vahter M, Couch R, Nermell B, Nilsson R. Lack of methylation of inorganic arsenic in the chimpanzee. Toxicol. Appl. Pharmacol. 1995;133:262–268. doi: 10.1006/taap.1995.1150. [DOI] [PubMed] [Google Scholar]
- Vahter M. Methylation of inorganic arsenic in different mammalian species and population groups. Sci Prog. 1999;82:69–88. doi: 10.1177/003685049908200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters SB, Devesa-Perez V, Del Razo LM, Styblo M, Thomas DJ. Endogenous reductants support catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem. Res. Toxicol. 2004a;17:404–409. doi: 10.1021/tx0342161. [DOI] [PubMed] [Google Scholar]
- Waters SB, Devesa V, Fricke MW, Creed JT, Styblo M, Thomas DJ. Glutathione modulates recombinant rat arsenic (+3 oxidation state) methyltransferase-catalyzed formation of trimethylarsine oxide and trimethylarsine. Chem. Res. Toxicol. 2004b;17:1621–1629. doi: 10.1021/tx0497853. [DOI] [PubMed] [Google Scholar]
- Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. J. Biol. Chem. 2006;281:7364–7373. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- Zakharyan RA, Wildfang E, Aposhian HV. Enzymatic methylation of arsenic compounds. III. The marmoset and tamarin, but not the rhesus, monkeys are deficient in methyltransferases that methylate inorganic arsenic. Toxicol. Appl. Pharmacol. 1996;140:77–84. doi: 10.1006/taap.1996.0199. [DOI] [PubMed] [Google Scholar]