Introduction
Inositol can be obtained through the diet or synthesized by some cells from glucose-6-phosphate (Daughaday et al., 1955; Holub, 1986). Through the actions of kinases and phosphatases, inositol can be phosphorylated and dephosphorylated to form a large number of intermediates in a complex metabolic pathway. With six positions available for phosphorylation there are roughly 70 potential intermediates, including both the lipid phosphatidylinositols and the soluble forms of inositol, several of which can contain pyrophosphate bonds (Irvine et al., 2001).
Inositol, 1,3,4-trisphosphate 5/6-kinase (ITPK1, E.C. 2.7.1.159) plays a pivotal role in inositol metabolism, is conserved from plants to humans, and is even found in Entamoeba histolytica (Field et al., 2000; Wilson et al., 1996; Wilson et al., 1997). The known phosphoryl transferase activities of ITPK1 are shown diagrammatically in Fig. 1. The substrate inositol 1,3,4-trisphosphate (Ins(1,3,4)P3) sits at a major branch point in the inositol metabolic pathway. It can be phosphorylated by ITPK1 in either the D5 or the D6 position of the inositol ring to form inositol 1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4 or inositol 1,3,4,6-tetrakisphosphate (Ins(1,3,4,6)P4), respectively (Fig. 1A). Either tetrakisphosphate (IP4) product can also serve as a substrate in an isomerization reaction (Ho et al., 2002; Miller et al., 2005). In addition, ITPK1 has also been shown to reversibly phosphorylate another IP4 isomer, inositol 3,4,5,6-tetrakisphosphate (Ins(3,4,5,6)P4), to form inositol pentakisphosphate (IP5) (Fig. 1B) (Chamberlain et al., 2007; Ho et al., 2002; Miller et al., 2005; Yang et al., 2000).
Fig. 1.
Activities of ITPK1. (A) Phosphorylation of Ins(1,3,4)P3 and isomerization of IP4 isomers and (B) reversible phosphorylation of Ins(3,4,5,6)P4.
The formation of Ins(1,3,4,6)P4 by ITPK1 represents the rate-limiting step in the formation of the higher phosphorylated forms of inositol in mammalian cells (Verbsky et al., 2005) including IP5, IP6, and the pyrophosphates diphosphoinositol-pentakisphosphate (IP7) and bisdiphosphoinositol tetrakisphosphate (IP8), shown diagrammatically in Fig. 2.
Fig. 2.
The pathway for the synthesis of the higher phosphorylated forms of inositol in mammalian cells.
Functions of IP6
In recent years, a number of functions for the more highly phosphorylated forms of inositol have come to light. IP6 has been shown to be an integral component of three proteins: adenosine deaminase (E.C.3.5.4.4) (Macbeth et al., 2005), arrestin (Milano et al., 2006), and the receptor for the plant hormone auxin (Tan et al., 2007). In addition to these structural roles, IP6 has been implicated in a wide range of cellular processes, including mRNA export from the nucleus (Alcazar-Roman et al., 2006; Feng et al., 2001; Weirich et al., 2006; York et al., 1999), regulation of transcription (Odom et al., 2000), increasing the ATPase activity of the dead box protein Dbp5 (E.C.3.6.1.-) (Alcazar-Roman et al., 2006), binding to the clathrin assembly proteins AP2 and AP180, inhibiting clathrin cage assembly in vitro (Chadwick et al., 1992; Norris et al., 1995; Theibert et al., 1991; Ye et al., 1995), inhibiting serine and threonine protein kinases (Larsson et al., 1997), stimulating protein kinases including casein kinase-2 (E.C.2.7.11.1) (Hilton et al., 2001; Solyakov et al., 2004), regulating nonhomologous DNA end joining of double strand breaks (Hanakahi et al., 2000; Hanakahi et al., 2002), and endocytosis (Hoy et al., 2002). Most recently, IP6 has been shown to activate the cysteine protease domains of both the Vibrio cholera RTX (repeats in toxin) (Lupardus et al., 2008; Prochazkova et al., 2008) and the Clostridium difficile A and B toxins (Pruitt et al., 2009).
To underscore the importance of IP6 in cells, deletion of the murine genes encoding the two other kinases leading to the synthesis of IP6 results in early embryonic lethality, suggesting that IP6 is essential for life (Frederick et al., 2005; Verbsky et al., 2005; Wilson et al., 2009).
Results and Discussion
Generation of an ITPK1-deficient mouse
We have generated a genetically engineered mouse harboring a hypomorphic allele in Itpk1 (Wilson et al., 2009). The β-galactosidase (βgal) containing gene trap cassette is inserted in the second intron of the Itpk1 gene, and by alternative splicing produces both an Itpk1-βgal fusion transcript as well as a wild type, full-length transcript. RT-PCR using mRNA extracted from brain tissue of wild type animals as well as those heterozygous and homozygous for the gene trap allele show that all animals produce mRNA for Itpk1 (Fig. 3). RT-PCR of L19, a ribosomal protein, is shown as a loading control. The levels of Itpk1 mRNA vary in both the heterozygotes and homozygotes, consistent with varying levels of ITPK1 protein produced by these animals (Wilson et al., 2009).
Fig. 3.

RT-PCR of Itpk1 (upper panel) and L19 (lower panel) from 2 animals each heterozygous for the gene trap allele (A,B), homozygous for the gene trap allele (C,D), or wild type (E,F). The ribosomal protein L19 is used as a loading control.
Expression of ITPK1 in mouse tissues
We have previously reported that Itpk1 is widely expressed throughout the nervous system in developing embryos and in adult mice (Wilson et al., 2009). Using Xgal staining, we have also analyzed Itpk1expression in other mouse tissues. Expression was seen in the vascular smooth muscle cells in arterioles of kidney (Fig. 4A-C), thorax (Fig. 4D), and gut (Fig. 4E-F). In the digestive tract, expression is seen in serosal cells (Fig. 4E), visceral smooth muscle cells (Fig. 4F-H), and crypt cells (Fig. 4I). Expression was also evident in the smooth muscle cells in the vas deferens (Fig. 4J), neuroepithelial derivatives including brainstem neurons (Fig. 4K), and pia mater (Fig. 4L).
Fig. 4.
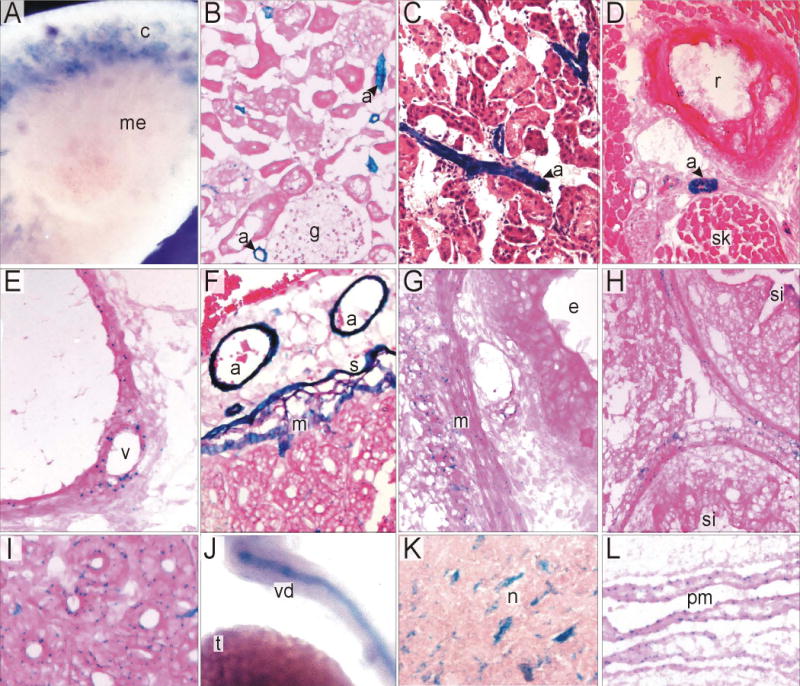
Expression of Itpk1 in mouse tissues. Tissue sections from mice containing the Itpk1-βgal gene trap allele were subjected to Xgal staining. (A) Whole mount of bisected kidney showing expression in vasculature within the renal cortex (c) but not medulla (me). (B,C) Expression in vascular smooth muscle of renal arterioles (a), including those adjacent to glomeruli (g). (D) Expression in an intercostal arteriole (a) between a rib (r) and skeletal muscle (sk). (E) Expression in smooth muscle cells of the vasa vasorum. (F) Expression in arterioles (a), serosal cells (s) and visceral smooth muscle (m) of the small intestine. (G,H) Expression in visceral smooth muscle (m) of the esophagus (e) and small intestine (si). (I) Weak expression in duodenal crypts. (J) Whole mount showing expression in vas deferens (vd) but not testis (t). (K) Expression in brain stem neurons (n). (L) Expression in pia mater (pm) of the brain.
Neural tube defects in Itpk1 hypomorphic embryos
Between embryonic day (E) 9.5 and E11.5, 12% of embryos resulting from mating animals homozygous for the gene trap allele have some form of neural tube defect (NTD), either spina bifida, exencephaly, or both (Wilson et al., 2009). The severity of the NTDs is quite variable, ranging from the mild caudal defects (Fig 5D), to moderately severe cranial defects (Fig. 5A-B) to cases of exencephaly that would be incompatible with life (Fig. 5C). The animals used in these experiments are of a mixed genetic background (C57Bl/6 × 129(P2)Ola). Animals are currently being backcrossed to pure genetic backgrounds representing each of the parental strains to determine whether genetic background plays a role in the incidence of NTDs, as has been shown to be the case for NTDs in the curly tail mouse, a mouse model for folic acid-resistant NTDs (Letts et al., 1995; Neumann et al., 1994; Van Straaten et al., 2001). Defects in neural tube development have also been shown to occur in mice harboring deletion of PIP5KIγ (Wang et al., 2007) further establishing the role of inositol signaling in neuronal development.
Fig. 5.
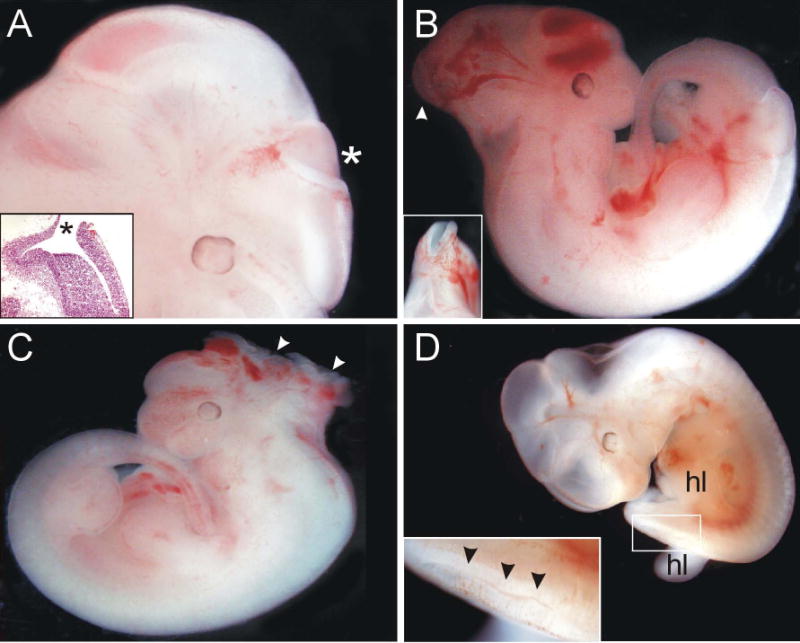
NTDs in Itpk1 hypomorphic mouse embryos. (A) Exencephaly in an E12.5 Itpk1 hypomorph. The inset shows an H&E stained section through the open NTD (*). (B) Exencephaly in an E12.5 Itpk1 hypomorph. The inset shows a dorsal view of the open NTD (arrowhead). (C) Severe exencephaly (arrowheads) in an E11.5 Itpk1 hypomorph. (D) Splayed hindlimbs (hl) in conjunction with a caudal NTD in an E12.5 Itpk1 hypomorph. The inset shows a higher magnification of the boxed region. Note the open and kinked neural tube (arrowheads).
Measurements of myo-inositol
The NTDs observed in the Itpk1 hypomorphs likely represent an example of folate-resistant NTDs. In humans, folic acid supplementation provides a 70% protective effect (Wald, 1991) leaving 30% which are folate-resistant. There are several animal models for folate-resistant NTDs. In some of these, the level of inositol affects the prevalence of the NTDs (Cockroft, 1988; Cockroft et al., 1992; Copp et al., 2003; Reece et al., 1997). In order to determine whether the NTDs seen in the Itpk1 hypomorphs result from a perturbation in inositol metabolism, animals will be fed diets containing varying levels of inositol and the prevalence of NTDs will be examined. To determine the level of inositol in the mouse chow as well as in the mice, to confirm that the levels of inositol are being altered, it is necessary to be able to assay for inositol concentrations. The enzyme inositol dehydrogenase (E.C.1.1.1.18) converts myo-inositol to scyllo-inosose in an NAD+-dependent oxidation reaction (Ramaley et al., 1979; Weissbach, 1965). Using purified solutions of inositol in a room temperature reaction containing 50 μg/ml of inositol dehydrogenase (Sigma Aldrich), the generation of NADH is measured by following the absorbance at 340 nm (Fig. 6A). Reactions reach their maximum absorbance by 70 minutes, as the A340 does not increase at time points beyond this time. To ensure that reactions go to completion, the A340 is read after 120 minutes. The assay is linear up to 0.25 mM inositol (Fig. 6B).
Fig. 6.
Measurement of myo-inositol. (A). Time course for inositol dehydrogenase reaction. (B). Concentration-dependence of inositol dehydrogenase.
For determination of inositol content in food (Harlan Teklad), samples were homogenized and extracted as described (Kawa et al., 2003). In addition to the standard mouse chow used by the Washington University Division of Comparative Medicine, two diet samples were obtained from Harlan Teklad (Madison, WI) for assay. Both test diets were based on the AIN-93 nutritionally complete rodent diet (Reeves et al., 1993). The level of inositol in the standard diet was 0.8 mg inositol/kg chow. A diet containing soybean oil (70 g/kg; TD.06706) and a diet containing 7% corn oil (TD.09146) both had reduced levels of inositol: 0.095 mg/kg and 0.072 mg/kg, respectively. We are currently assaying other test diets in order to obtain a relatively inositol free diet.
Summary
ITPK1 is the rate limiting enzyme in the pathway leading to formation of the highly phosphorylated inositol phosphates including IP6 and the inositol pyrophosphates. One or more of these metabolites are essential for life as deletion of either of the kinases that form IP5 or IP6 in mice results in embryonic lethality. We have produced mice harboring a hypomorphic allele for Itpk1, and mice homozygous for this gene trap allele produce low but detectable levels of active enzyme. We have studied the expression of Itpk1 in various tissues and found that the enzyme is highly expressed in smooth muscle of vessels and other tissues. In addition, these mice have neural tube defects in 12% of homozygous embryos. Since the levels of enzyme expression vary greatly in homozygous animals, we speculate that relative deficiency of one or more inositol phosphates accounts for these defects. We plan to feed an inositol deficient diet or one with supplemental inositol to animals to demonstrate altered prevalence of neural tube defects.
Acknowledgments
We thank Jun Zou, Jasna Marjanovic, and Marina Kisseleva for helpful discussions. This work was supported by grants from the National Institutes of Health (HL-16634-43 to P.W.M.; DK07618, to D.B.W.; and DK52574, to the Histology Core Facility) and the Children's Discovery Institute (MD-II-2009-174, to M.P.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–6. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Chadwick CC, Timerman AP, Saito A, Mayrleitner M, Schindler H, Fleischer S. Structural and functional characterization of an inositol polyphosphate receptor from cerebellum. J Biol Chem. 1992;267:3473–81. [PubMed] [Google Scholar]
- Chamberlain PP, Qian X, Stiles AR, Cho J, Jones DH, Lesley SA, Grabau EA, Shears SB, Spraggon G. Integration of inositol phosphate signaling pathways via human ITPK1. J Biol Chem. 2007;282:28117–25. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockroft DL. Changes with gestational age in the nutritional requirements of postimplantation rat embryos in culture. Teratology. 1988;38:281–90. doi: 10.1002/tera.1420380312. [DOI] [PubMed] [Google Scholar]
- Cockroft DL, Brook FA, Copp AJ. Inositol deficiency increases the susceptibility to neural tube defects of genetically predisposed (curly tail) mouse embryos in vitro. Teratology. 1992;45:223–32. doi: 10.1002/tera.1420450216. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–93. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- Daughaday WH, Larner J, Hartnett C. The synthesis of inositol in the immature rat and chick embryo. J Biol Chem. 1955;212:869–75. [PubMed] [Google Scholar]
- Feng Y, Wente SR, Majerus PW. Overexpression of the inositol phosphatase SopB in human 293 cells stimulates cellular chloride influx and inhibits nuclear mRNA export. Proc Natl Acad Sci U S A. 2001;98:875–9. doi: 10.1073/pnas.021558098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Wilson MP, Mai Z, Majerus PW, Samuelson J. An Entamoeba histolytica inositol 1,3,4-trisphosphate 5/6-kinase has a novel 3-kinase activity. Mol Biochem Parasitol. 2000;108:119–23. doi: 10.1016/s0166-6851(00)00197-3. [DOI] [PubMed] [Google Scholar]
- Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci U S A. 2005;102:8454–9. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC. Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell. 2000;102:721–9. doi: 10.1016/s0092-8674(00)00061-1. [DOI] [PubMed] [Google Scholar]
- Hanakahi LA, West SC. Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. Embo J. 2002;21:2038–44. doi: 10.1093/emboj/21.8.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton JM, Plomann M, Ritter B, Modregger J, Freeman HN, Falck JR, Krishna UM, Tobin AB. Phosphorylation of a synaptic vesicle-associated protein by an inositol hexakisphosphate-regulated protein kinase. J Biol Chem. 2001;276:16341–7. doi: 10.1074/jbc.M011122200. [DOI] [PubMed] [Google Scholar]
- Ho MW, Yang X, Carew MA, Zhang T, Hua L, Kwon YU, Chung SK, Adelt S, Vogel G, Riley AM, Potter BV, Shears SB. Regulation of Ins(3,4,5,6)P(4) signaling by a reversible kinase/phosphatase. Curr Biol. 2002;12:477–82. doi: 10.1016/s0960-9822(02)00713-3. [DOI] [PubMed] [Google Scholar]
- Holub BJ. Metabolism and function of myo-inositol and inositol phospholipids. Annu Rev Nutr. 1986;6:563–97. doi: 10.1146/annurev.nu.06.070186.003023. [DOI] [PubMed] [Google Scholar]
- Hoy M, Efanov AM, Bertorello AM, Zaitsev SV, Olsen HL, Bokvist K, Leibiger B, Leibiger IB, Zwiller J, Berggren PO, Gromada J. Inositol hexakisphosphate promotes dynamin I-mediated endocytosis. Proc Natl Acad Sci U S A. 2002;99:6773–7. doi: 10.1073/pnas.102157499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:327–38. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Kawa JM, Przybylski R, Taylor CG. Urinary chiro-inositol and myo-inositol excretion is elevated in the diabetic db/db mouse and streptozotocin diabetic rat. Exp Biol Med (Maywood) 2003;228:907–14. doi: 10.1177/153537020322800806. [DOI] [PubMed] [Google Scholar]
- Larsson O, Barker CJ, Sjoholm A, Carlqvist H, Michell RH, Bertorello A, Nilsson T, Honkanen RE, Mayr GW, Zwiller J, Berggren PO. Inhibition of phosphatases and increased Ca2+ channel activity by inositol hexakisphosphate. Science. 1997;278:471–4. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- Letts VA, Schork NJ, Copp AJ, Bernfield M, Frankel WN. A curly-tail modifier locus, mct1, on mouse chromosome 17. Genomics. 1995;29:719–24. doi: 10.1006/geno.1995.9946. [DOI] [PubMed] [Google Scholar]
- Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322:265–8. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–9. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J Biol Chem. 2006;281:9812–23. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- Miller GJ, Wilson MP, Majerus PW, Hurley JH. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell. 2005;18:201–12. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Neumann PE, Frankel WN, Letts VA, Coffin JM, Copp AJ, Bernfield M. Multifactorial inheritance of neural tube defects: localization of the major gene and recognition of modifiers in ct mutant mice. Nat Genet. 1994;6:357–62. doi: 10.1038/ng0494-357. [DOI] [PubMed] [Google Scholar]
- Norris FA, Auethavekiat V, Majerus PW. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J Biol Chem. 1995;270:16128–33. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–9. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Prochazkova K, Satchell KJ. Structure-function analysis of inositol hexakisphosphate-induced autoprocessing of the Vibrio cholerae multifunctional autoprocessing RTX toxin. J Biol Chem. 2008;283:23656–64. doi: 10.1074/jbc.M803334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Chagot B, Cover M, Chazin WJ, Spiller B, Lacy DB. Structure-Function Analysis of Inositol Hexakisphosphate-induced Autoprocessing in Clostridium difficile Toxin A. J Biol Chem. 2009;284:21934–40. doi: 10.1074/jbc.M109.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaley R, Fujita Y, Freese E. Purification and properties of Bacillus subtilis inositol dehydrogenase. J Biol Chem. 1979;254:7684–90. [PubMed] [Google Scholar]
- Reece EA, Khandelwal M, Wu YK, Borenstein M. Dietary intake of myo-inositol and neural tube defects in offspring of diabetic rats. Am J Obstet Gynecol. 1997;176:536–9. doi: 10.1016/s0002-9378(97)70543-x. [DOI] [PubMed] [Google Scholar]
- Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–51. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- Solyakov L, Cain K, Tracey BM, Jukes R, Riley AM, Potter BV, Tobin AB. Regulation of casein kinase-2 (CK2) activity by inositol phosphates. J Biol Chem. 2004;279:43403–10. doi: 10.1074/jbc.M403239200. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–5. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Theibert AB, Estevez VA, Ferris CD, Danoff SK, Barrow RK, Prestwich GD, Snyder SH. Inositol 1,3,4,5-tetrakisphosphate and inositol hexakisphosphate receptor proteins: isolation and characterization from rat brain. Proc Natl Acad Sci U S A. 1991;88:3165–9. doi: 10.1073/pnas.88.8.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten HW, Copp AJ. Curly tail: a 50-year history of the mouse spina bifida model. Anat Embryol (Berl) 2001;203:225–37. doi: 10.1007/s004290100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J, Lavine K, Majerus PW. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc Natl Acad Sci U S A. 2005;102:8448–53. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW. The pathway for the production of inositol hexakisphosphate in human cells. J Biol Chem. 2005;280:1911–20. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- Wald Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. MRC Vitamin Study Research Group Lancet. 1991;338:131–7. [PubMed] [Google Scholar]
- Wang Y, Lian L, Golden JA, Morrisey EE, Abrams CS. PIP5KI gamma is required for cardiovascular and neuronal development. Proc Natl Acad Sci U S A. 2007;104:11748–53. doi: 10.1073/pnas.0700019104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–76. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Weissbach A. myo-inositol. In: Bergmeyer HU, editor. Methods of Enzymatic Analysis. Vol. 1965. pp. 171–174. [Google Scholar]
- Wilson MP, Hugge C, Bielinska M, Nicholas P, Majerus PW, Wilson DB. Neural tube defects in mice with reduced levels of inositol 1,3,4-trisphosphate 5/6-kinase. Proc Natl Acad Sci U S A. 2009;106:9831–5. doi: 10.1073/pnas.0904172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MP, Majerus PW. Isolation of inositol 1,3,4-trisphosphate 5/6-kinase, cDNA cloning and expression of the recombinant enzyme. J Biol Chem. 1996;271:11904–10. doi: 10.1074/jbc.271.20.11904. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Majerus PW. Characterization of a cDNA encoding Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase. Biochem Biophys Res Commun. 1997;232:678–81. doi: 10.1006/bbrc.1997.6355. [DOI] [PubMed] [Google Scholar]
- Yang X, Shears SB. Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase. Biochem J. 2000;351(Pt 3):551–5. [PMC free article] [PubMed] [Google Scholar]
- Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995;270:1564–8. [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]





