Abstract
To achieve more accurate and consistent registration in an image population, a novel hierarchical groupwise registration framework, called Atlas Building by Self-Organized Registration and Bundling (ABSORB), is proposed in this paper. In this new framework, the global structure, i.e., the relative distribution of subject images is always preserved during the registration process by constraining each subject image to deform only locally with respect to its neighbors within the learned image manifold. To achieve this goal, two novel strategies, i.e., the self-organized registration by warping one image towards a set of its eligible neighbors and image bundling to cluster similar images, are specially proposed. By using these two strategies, this new framework can perform groupwise registration in a hierarchical way. Specifically, in the high level, it will perform on a much smaller dataset formed by the representative subject images of all subgroups that are generated in the previous levels of registration. Compared to the other groupwise registration methods, our proposed framework has several advantages: 1) It explores the local data distribution and uses the obtained distribution information to guide the registration; 2) The possible registration error can be greatly reduced by requiring each individual subject to move only towards its nearby subjects with similar structures; 3) It can produce a smoother registration path, in general, from each subject image to the final built atlas than other groupwise registration methods. Experimental results on both synthetic and real datasets show that the proposed framework can achieve substantial improvements, compared to the other two widely used groupwise registration methods, in terms of both registration accuracy and robustness.
Keywords: Groupwise registration, hierarchical registration, atlas building, self-organized registration, image bundling
1. Introduction
Image registration is one of the most important techniques in the field of medical image analysis due to its significance in both scientific researches and clinical applications (Crum et al., 2004). A large proportion of registration methods have been developed for pairwise image registration (Christensen and Johnson, 2001; Johnson and Christensen, 2002; Klein et al., 2009; Shen and Davatzikos, 2002; Vercauteren et al., 2009) where each subject image is registered independently with a selected template by a separately estimated spatial transformation. To better understand and analyze the group similarity and variation within a population, it is important to accurately and consistently register all images in the population. However, the pairwise registration can inevitably introduce bias to the registration, due to the bias in the selection of the template for registration. Accordingly, many groupwise registration methods have been recently proposed to achieve more accurate and consistent registration among the population by simultaneously registering all images within a single registration framework, thus facilitating the better investigation of the group similarity and variation in the population (Hajnal et al., 1995; Hill et al., 2001; Holden et al., 2000; Maintz and Viergever, 1998; Sabuncu et al., 2009; Sabuncu et al., 2007; Zitová and Flusser, 2003).
One way to achieve groupwise registration is based on the pairwise methods. For example, in (Park et al., 2005), an image that is the closest to the geometrical mean of a population is selected as a template by Multi-Dimensional Scaling (MDS) (Cox and Cox, 2000) and then all other images are registered to the selected template for achieving the least bias. Specially, the geometrical mean is estimated based on the registration results of all image pairs. As mentioned above, this type of groupwise registration is limited due to the selection of a particular image as a template for registration, which can inevitably introduce bias to the final registration. Another method proposed by Seghers et al. (Seghers et al., 2004) implements the pairwise registration on all pairs of images in the population, and each image is deformed by the average deformation field over the deformation fields estimated between this image and all other images. The atlas is thus built by averaging all the deformed images. However, the high computational load limits its application, especially when the number of images to be registered is large.
To avoid the potential bias in the registration, many other groupwise registration methods are proposed to directly register all images simultaneously by formulating groupwise registration as an optimization problem, with a global cost function particularly defined on all aligned images. For example, in the congealing registration method proposed in (Learned-Miller, 2006; Zitová and Flusser, 2003), an objective function based on the pixel stack entropy is defined over all aligned images in the dataset, to solve the groupwise registration problem by a gradient-based stochastic optimizer. The results by this congealing registration method can provide an estimated deformation field for each image and also generate an atlas through the averaging of all aligned images. The congealing registration method has been recently extended by Balci et al. (Balci et al., 2007a; Balci et al., 2007b) to perform non-rigid registration using B-Splines based deformation representation (Bhatia et al., 2004), and by Wang et al. (Wang et al., 2009b) to use the attribute vector (instead of the image intensity only) for guiding the registration and achieving more robust and accurate registration results. However, the curse of dimensionality from the huge number of variables involved in the global cost function poses challenges to the optimizer which is vulnerable to local minima.
Compared to the above-mentioned methods, Joshi et al. (Joshi et al., 2004) proposed to solve the groupwise registration in an iterative manner within the framework of diffeomorphism. Specifically, an interim atlas is first built by averaging all images after affine registration, and then all the images are registered to this interim atlas by diffeomorphic registration. (Note that this interim template is also called the group mean image in this paper.) After this first round of registration, the interim atlas is updated based on the newly registered images and a second round of registration is performed subsequently. By iteratively performing the steps of (1) the registration to the atlas and (2) the atlas updating, this method can provide an unbiased way to build the atlas, and also can converge fast with a few iterations. However, the registration process of this method could be misled by trying to register sharp individual images (with clear anatomical structures) to a blurry group mean image (with no clear anatomical structures) especially in the first rounds of registration.
It is generally difficult to achieve good registration in one step by simply registering each image to an explicit (or implicit) template directly, especially when anatomical variations are large across images within the group. To this end, Wang et al. (Wang et al., 2009a) proposed to perform groupwise registration by clustering all the images hierarchically into several small-scale subgroups, and thus the images in each subgroup can be better registered since they are more similar to each other. Specifically, a tree of images is first constructed based on the clustering results, and then the registration process starts from the subgroups on the leaf nodes and ends at the root. However, the problem of how to perform groupwise registration within each subgroup in a consistent and robust way remains untouched.
On the other hand, some other algorithms have been proposed to register the individual image to the template with help of intermediate templates (Baloch and Davatzikos, 2009; Baloch et al., 2007; Tang et al., 2009). These intermediate templates are selected from the dataset to build a connection path between each individual image and the template. The final registration result can be obtained for each individual image by deforming it along its respective connection path to the template. This idea can be applied to the groupwise registration by building a minimum spanning tree (MST) (Kruskal, 1956) where each node corresponds to one image and each edge weights the distance between two connected nodes (Munsell et al., 2009). The root node for the MST can be determined by selecting a node that has the minimal edge length to all other nodes. In (Hamm et al., 2009), after learning the intrinsic manifold from the whole dataset, the pseudo-geodesic median image is determined as the template since it minimizes the total path length from each image to the template. The corresponding geodesic paths between individual images and the template are computed to construct a tree on the learned manifold. Since a fixed image (i.e., the root image) is used as the final template to register with all other images, the bias is unavoidable in this scenario (as other pairwise-registration based groupwise registration methods as mentioned above), although the registration error could be reduced since each time only the nearby similar images need to be registered.
In this paper, a new framework for groupwise registration, termed as Atlas Building by Self-Organized Registration and Bundling, or ABSORB for short, is proposed to address the problems mentioned above, with the basic idea illustrated in Fig. 1. We resolve the groupwise registration problem in an iterative manner by warping each image in the population to the final atlas step by step on the learned manifold, and, at the same time, maintain the global distribution of the population. To achieve this goal, two new strategies, namely self-organized registration and image bundling, are proposed. Specifically, the self-organized registration is introduced to deform each image towards a subset of its neighbors that are closer to the global center (estimated in each iteration) and thus condense the distribution of image set on the learned manifold gradually. Note that the global center is updated iteratively and is used only to guide the selection of neighbors, thereby no fixed template is directly used for population registration. After several iterative registrations, some nearby subjects become close enough to each other and are thus bundled together spontaneously into a subgroup. By using these two strategies, ABSORB can perform groupwise registration from the lower level to the higher level hierarchically; particularly, in the higher level, the registration is performed on a much smaller dataset, which consists of the representative images of all subgroups formed in the previous registration steps. As the result of this hierarchical registration process, a pyramid of images is built automatically and the atlas image can be generated eventually once the registration arrives at the upmost level.
Figure 1.
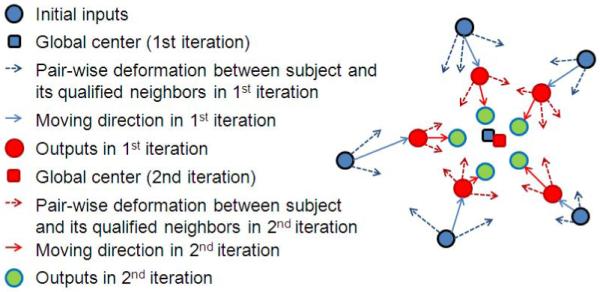
The framework for the proposed ABSORB algorithm. In each iteration, several deformation fields (designated by the dashed arrows) between a subject and some of its neighbors are combined to deform the subject to a new location by the average deformation field (designated by the solid arrows). The global center in each iteration is shown with the squares. This figure is best viewed in color printing.
Similar to the approaches that solve the groupwise registration in an iterative way (Hamm et al., 2009; Joshi et al., 2004; Munsell et al., 2009), the complete path from each individual image to the final atlas built by the proposed ABSORB method is composed of a series of small segments, connecting neighboring images. But the proposed ABSORB method is inherently different from those methods in three ways. First, in ABSORB, there are no intermediate templates used for all images in any iteration. Instead, the movement of each individual image on the manifold is driven only by a selected set of its neighboring images, not by a common explicit or implicit template. Second, in this proposed framework, the number of neighboring images that could have effects on the current image is adaptively determined according to the data structure learned online, and the complete path generated from each image to the final atlas on the manifold is generally smoother and more conservative as ABSORB always moves one image to its nearby location, instead of the global mean location. In contrast, in (Hamm et al., 2009; Joshi et al., 2004; Munsell et al., 2009), the direction and the amount of deformation for each image in each iteration are determined by only one image, i.e., the selected image used as the tentative template, which can often result in a zigzag path if the selected template cannot represent the data distribution very well, as will be demonstrated in the experimental result section. Finally, in the proposed ABSORB method, the registration path for each image is not pre-determined before the actual registration starts. In other words, it is a fully data-driven groupwise registration method. In contrast, the tree in (Hamm et al., 2009; Munsell et al., 2009) is built in the pre-processing step and fixed during the whole registration. The performance of ABSORB is evaluated on both synthetic and real image sets, showing that this novel framework of groupwise registration can significantly improve the registration accuracy and the quality of the built atlas, compared to other two groupwise registration methods (Joshi’s iterative groupwise registration method (Joshi et al., 2004) and the tree-based groupwise registration method (Hamm et al., 2009)).
The rest of this paper is organized as follows. The proposed groupwise registration framework (ABSORB) is detailed in Section 2. In Section 3, extensive experiments on both synthetic and real datasets, as well as the comparison with other two groupwise registration methods, are provided to demonstrate the performance of the ABSORB registration method. We conclude and discuss the future work in Section 4.
2. Method
In this section, a new framework for simultaneous registration of an image population, termed as ABSORB, is presented. We assume that a population I = {I1, I2, … , IN} with N different subject images has already been pre-processed under intensity normalization and affine registration, i.e., the global shape differences among subject images, such as translation, rotation, shearing and scaling, have been removed, e.g., by a groupwise affine registration method (Zöllei et al., 2005). The registered image of subject Ii at the beginning of iteration t is denoted as , where i = 1, 2, … , N and t ≥ 1. The goal of the proposed framework at iteration t is to further align all images and obtain an updated image set in the end of iteration t, so that all the images in It+1 are distributing more compactly to each other in the data space than they are in It. Here, we set I1 = I. It is worth noting that Nt denotes the current number of representative subjects under consideration, which can change dynamically with the registration. We will make it clear in Section 2.3.
In Fig. 1, the basic idea of how the registration process evolves in the proposed framework is illustrated. To move a subject towards a common space at iteration t, a self-organized registration is first performed. Specifically, a subset of neighbors of within It are selected based on some criteria that will be specified in Section 2.1, and then the deformation field defined to warp the current subject to is calculated by combining all the deformation fields between and each of its selected neighbors. Note that the deformation field between and each of its selected neighbors can be obtained by any pairwise registration methods, although in this paper we choose the diffeomorphic demons method1 because it is available in ITK (Ashburner, 2007; Vercauteren et al., 2009). After performing the self-organized registration on all subjects in the current iteration (t), we bundle some nearby subjects into respective subgroups if they have been registered very close to each other, and then update the deformed image set It with It+1. The same (self-organized registration and image bundling) steps are repeated on It+1 in the next iteration until the algorithm converges.
In the following, we first introduce in Section 2.1 a strategy for selection of neighboring subjects for the self-organized registration step. In Section 2.2, the detail of how to combine the deformation information from the neighborhood for guiding the movement of each subject is presented. Finally, the hierarchical structure of the whole registration framework is depicted in Section 2.3.
2.1. Selection of neighboring subjects
Selection of neighboring subjects is critical to self-organized registration. We design a particular procedure to adaptively choose a subset of neighbors for each subject by considering both local and global information. Specifically, a metric is first defined to measure the distance between any two subjects on the data manifold, and then a graph is built and updated iteratively to help locate the global center. Finally, the selection of neighboring subjects is performed for each subject and its deformation field is later calculated based on the qualified neighbors.
We first define a distance measurement between any two subjects in the dataset. In this paper, the intensity difference between two images and is used to define a metric, , due to its simplicity and fast calculation, i.e., , where M is the total number of voxels in the image and is the intensity value of the p-th voxel in the image . It is worth noting that other metrics, such as those defined in (Hamm et al., 2009; Munsell et al., 2009; Seghers et al., 2004), can also be applied. To better measure the distance between two subjects on the manifold that captures the intrinsic structure of the image space in the dataset, we can go one step further to pursue a distance defined on the manifold. Following the procedure described in the isomap algorithm (Tenenbaum et al., 2000), a k-NN isomap based on the pairwise distance can be constructed. The distance between and can be updated as , where is the shortest distance between and on the k-NN isomap.
To ensure that the registration process within the population is on the learned manifold, the interactions between different subjects in a single iteration are constrained to be within a local neighborhood. That means, for any subject image , its movement at iteration t is determined locally. This is extremely important in the early phase of registration (in order to achieve more accurate registration results), because it is always much easier to register two nearby subjects with similar structures precisely than to register two subjects far-away from each other. The local movement in each iteration will be used to warp one subject towards the global center step by step. One of the simple and straightforward ways to implement the above idea is to consider the movements to the local and global center separately. However, since the direction of the local movement is not necessarily consistent with that of the global movement, the overall registration path to the final atlas could be more like a zigzag. In this way, the whole population could converge slowly and possibly undermine the registration accuracy.
To solve this problem, we propose a different strategy to combine both the global and local information in one step, by embedding the global information into the selection of neighboring subjects. For example, when selecting the qualified neighbors of one subject for registration, we choose only subjects that are closer to the global center than the subject under consideration. It is worth noting that, here, the global center is different from the final atlas in two ways. First, the global center is not served as a template, to which each subject should be registered directly. Second, the global center is iteratively updated (rather than being fixed in many registration algorithms), and it is used only to provide general information for the selection of qualified neighbors. As depicted in Fig. 2, the neighborhood of subject is defined within a hyper-sphere in the high-dimensional space (i.e., the small disc in red). The global center at the current iteration is located at and another hyper-sphere with the radius equal to the distance from to is also calculated, i.e., the large disc in blue. Here, c(t) is the index of the subject selected as the global center at the current iteration t. Then, only those subjects within the intersection of two discs, i.e., subject (the solid triangle point), are chosen as the qualified neighbors of subject . Subject (denoted by the solid square) is not qualified as it is not closer to than , although is in the neighborhood of .
Figure 2.
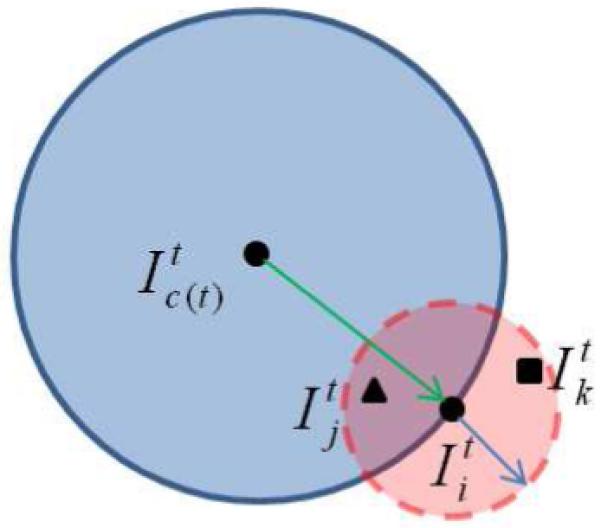
Selection of neighboring subjects. The small red disc with a dashed boundary defines the neighborhood of subject , and the intersection of the large blue disc and the small red disc defines a region including all points that are closer to than . Subject is one of the qualified neighbors of , while is not.
As we have discussed above, the global information is embedded into the selection of local qualified neighbors by choosing only those closer to the global center. So the determination of global center is also critical to the performance of the algorithm. In (Joshi et al., 2004), the group mean image in the Euclidean space is adopted as the global center. However, this mean image is usually very fuzzy, which could lead to an inaccurate registration because of registering two images with different contrast, i.e., subject images are sharp with clear anatomical structures while the initial estimated group mean image is fuzzy with unclear anatomical structures. On the contrary, we select the median subject on the learned manifold as the global center due to its robustness to the outliers as shown in (Hamm et al., 2009). The median subject in a dataset is defined as the subject that minimizes the overall distance from that subject to all other subjects. To achieve this, an undirected graph, whose edges are assigned with distances between connected subjects, can help compute the median subject. Instead of building a fixed graph throughout the registration process, we propose a dynamic graph which is updated by the dataset composed of all registered images after each iteration. The global center is then updated accordingly on the manifold. This helps subjects adjust their moving directions more adaptively. However, if the graphs in different iterations are generated independently, the determined global center may change dramatically, especially in the early stage of registration, and thus the estimated deformation for each subject would lack smoothness in the perspective of registration process. A stable global center can provide a consistent direction to guide the registration of subjects and alleviate unnecessarily zigzagged paths. To obtain such a consistent but not fixed global center, we build an Iterative Neighborhood Graph (ING), , from k-NN isomap. The weight assigned to the edge connecting two images and in is defined as
| (1) |
where the current edge weight is calculated as a weighted summation of , the distance between and , and , the cumulative distance applied in the previous iteration. Here, we use α to include the distance information in previous iterations. We set α ∈ (0,1) so that the earlier iteration will have less effect on the current weight. Based on the weight assigned to each edge in , the median image (or the global center here) at the current iteration t can be selected by
| (2) |
According to Eq. 1, we can see that the information from different manifolds generated in all previous iterations (from 1 to t – 1) has been integrated together iteratively in graph . The ING can thus regulate the graph topologies built on the registered image set It in different iterations and can assure a gentle shift of the global center.
With the constructed and the selected global center , we can determine those qualified neighbors for each subject in its neighborhood as illustrated in Fig. 2. For the given subject at iteration t, we sort all other subjects in the ascending order of bilateral distances, . And a subscript set is built to include the top indices of the sorted subjects which are the closest to . We build another set containing the subscripts whose corresponding subjects are closer to the current global center than the subject under consideration The intersection between and , , with size , is exactly the set where each element corresponds to a qualified subject index that we expect to get. Note that could be zero since, for some subjects , the common area in Fig. 2 might not contain any other subject in , and is only true either when is the global center or when all of its nearest neighbors are farther to the global center. All the qualified neighbors form a set as and only they will guide the warping of at iteration t.
Such a neighboring subject selection strategy as we detailed above can successfully embed the global information into the selection of the local neighbors, which is a desirable property of the self-organized registration. It is worth noting that the global center is defined on the registered image set at each iteration, and is updated iteratively as the registration proceeds. Therefore, the global center is not necessarily the same in different iterations. After determining those qualified neighbors, the self-organized registration can be performed on each subject, which is detailed in the next section.
2.2. Averaging over dense deformation fields
In this section, we define a mechanism to warp the current subject by all its qualified neighbors.
Different from those pairwise-registration based groupwise registration methods (Joshi et al., 2004; Seghers et al., 2004; Wang et al., 2009a), we do not average over current qualified neighbor images to generate a local mean image for registration, since this local mean image could lose some key anatomical structures and thus mislead the registration as the group mean image does. Instead, we move the subject along an average deformation direction on the manifold according to the selected neighbors. In this way, the undesired registration to blurry images can be completely bypassed. With the subset of selected neighboring subjects , we can perform pairwise registration between and each of its qualified neighbor subject. In particular, we choose the diffeomorphic demons algorithm (Vercauteren et al., 2009) to register one subject to another.
The schematic illustration of how to combine different deformation fields is shown in Fig. 3. Here, the superscript t and the subscript i, indicating the iteration index and subject index, respectively, are dropped to demonstrate the process in a more general situation. In Fig. 3, I is the current subject to be registered and Ic the global center. All qualified neighbors selected by the procedure in Section 2.1 form a set J = {J1, J2, … , Jm}, m ≥ 1. Our goal is to move I to a new location I’ which should be closer to its destination in the end of registration. It can be achieved by averaging the warping directions from I to each Js, where s = 1,2, … , m. For each pair of subject I and Js, the dense deformation field Gs (i.e., the green solid arrow) is first estimated by Diffeomorphic Demons. Then, its inverse (i.e., the brown dotted arrow) are calculated using the method in (Christensen and Johnson, 2001; Shen and Davatzikos, 2002). Therefore, the average direction can be calculated based on the inversed deformation field (since they are defined on the same image space of I) by (the red dashed line). To emphasize the effect of those neighbors which are much close and similar to the center subject I, we can go one step further by weighting different based on the distance d(I, Js),
| (3) |
where is the Gaussian function and the variance σ is adaptively set as the median value of {d(I, Js)|s = 1,2, …, m}.
Figure 3.
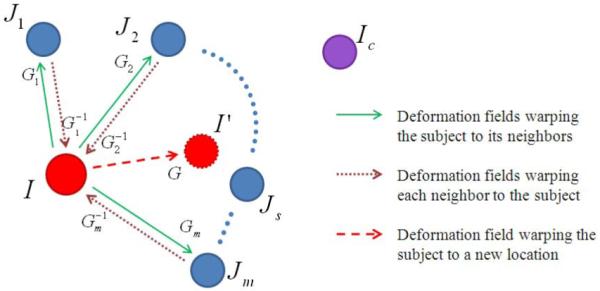
Illustration of the combination of multiple deformation fields. The current subject I has a total of m qualified neighboring subjects, J1, J2, … , Jm. The deformation field G as defined in Eq. 3 can deform I along the average direction to I’ (a red dashed circle), approaching closer to the global center Ic, indicated by a purple circle on the top right. This figure is best viewed in color printing.
For each in iteration t, we apply the above procedure, generate the deformation field by Eq. 3, and deform it following , thus moving closer to the current global center. Note, it is possible that qualified neighboring subjects of do not exist (i.e., ) because either neighbors of are farther away from the global center or is the global center. In any of these two cases, the subject under consideration will be assigned with an identity transformation temporarily, thus . In the following iteration, the distribution of the dataset in the image space becomes much denser, and this self-organized registration can proceed again as described above.
2.3. Hierarchical registration structure
We have described the details of self-organized registration which combines deformation fields from multiple qualified neighboring subjects to guide the movement of each subject in each iterative registration step. As the registration proceeds, it is possible that several nearby subjects converge spontaneously, thus partitioning the image set and obtaining subgroups of images. Each subgroup can be stable and compact, as the member subjects in each subgroup are very close to each other while different subgroups can be far-away. In this case, the selection of qualified neighbors for any subject will be restricted within the subgroup it belongs to. In order to break this spontaneous partition and to further refine the groupwise registration result, we employ a new strategy to perform registration across different subgroups. It is worth noting that each subgroup could have a bundle of aligned images, or just a single warped image.
Given a set of registered images, a clustering method (e.g., Affinity Propagation (AP) (Frey and Dueck, 2007)) can be adopted to bundle those aligned subjects (with very close distance to each other) into subgroups. Then, the representative image of each subgroup, determined automatically by the clustering method, forms a much smaller size of a new dataset. The same processing in Sections 2.1 and 2.2 can be applied to the new dataset (at a higher level), to further register subgroups of images together.
The detail of the hierarchical registration structure is illustrated in Fig. 4. Initially, all the subjects in the population are placed on the bottom level (Level 1), where self-organized registration is performed on each of them, including the selection of neighboring subjects (in Section 2.1) and the combination of multiple deformations to qualified neighboring subjects (in Section 2.2). We then apply the AP clustering method to detect whether the registered images have fallen into a stable partition. If the clustering results on It and It+1 (t ≥ 1) are not exactly the same, both the self-organized registration on It+1 and the clustering of the whole population are repeated. If at some iteration t* the clustering results on datasets It* and It+1* do not change, i.e. both the subgroup partition and the representative images are exactly the same, the proposed groupwise registration framework will go to the next level and initiate a new image set containing all representative images. The same procedures are repeated on this new image set, and the iterative registration will terminate once the representative images are clustered into a single group, or when the registration reaches the top of the hierarchical structure.
Figure 4.
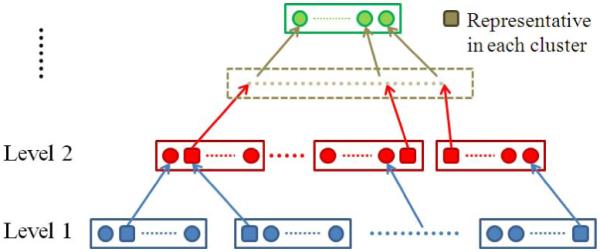
The hierarchical structure built during the groupwise registration. The center image (the squared one) in each cluster (i.e., all subjects within each box) is selected as a representative image and sent to the higher level for registration. When the maximal number of levels is reached or all representative images are clustered into a single group, the pyramid reaches its top level and the groupwise registration can be completed.
It is worth noting that the registration accuracy will not decrease as the registration procedure moves upwards in the hierarchical structure. Also, we will have no problem on the smoothness of the estimated deformation fields, with reasons justified next. First, the images in each subgroup have been well aligned together to each other when the AP clustering result becomes stable. Therefore, it is reasonable of using the representative image to select a common set of new qualified neighbors for all (non-representative) images in the same cluster. Second, each (representative or non-representative) image will always have a chance to be registered individually with its new qualified neighbors (selected by its representative image). This indicates that the deformation field estimated for the representative image will be not directly applied onto non-representative images as their new registration results. Therefore, there is no problem on decrease of registration accuracy on the non-representative images, since all of them will be separately registered with the new qualified neighbors (selected from the representative images of other clusters). Third, the previous estimated deformation field for each image will be used as a good initialization for the current registration of the same image, therefore there is no problem on accumulated discontinuities since we will not try to connect separate deformations (estimated from different phase of registration) together. Fourth, when we register each image to its new qualified neighbors in each iteration, we always enforce the smoothness of the deformation field. Therefore, the estimated deformation field can be always smooth after each registration, and we have no problem on discontinuities of deformation field. Based on all of these four novel designs, it shows that our proposed hierarchical registration strategy will not affect the registration accuracy on the whole dataset.
In the upmost level, all subjects are registered very close to each other on the learned manifold, therefore we can take their average image as the final atlas. The average image is no longer fuzzy, but of clear anatomical structures and sharp boundaries as will be demonstrated in the experiments section.
It is worth noting that the way of employing the clustering method in our framework is quite different from that in (Wang et al., 2009a). Clustering is intrinsic to our framework where spontaneous partition naturally happens during registration. Subjects that are similar in the population will be more likely to converge by self-organized registration. In many cases, they arrive at a local common space before the reaching global center. The clustering result depicts the consequence of the self-organized registration, and also helps trigger the registration in a higher level, rather than a mandated pre-processing step to guide the subsequent registration as in (Wang et al., 2009a). Note that, in (Wang et al., 2009a), the clustering technique is applied before the actual registration process starts, to hierarchically partition the entire population into several subgroups at different levels. Then, the groupwise registration is solved based on small-scale registrations on the subgroups of images, and the final atlas is synthesized from different subgroups hierarchically. The clustering procedure is used to alleviate the complexity of groupwise registration on a population with a large number of images.
Another difference between our method and the method in (Wang et al., 2009a) is on how to send an image for registration to the higher level. In our algorithm, the cluster is described by the representative image which is derived from AP clustering. The groupwise registration on the higher level is performed on the set of representative images. However, in (Wang et al., 2009a), the mean image of each cluster contributes to the registration at the higher level, which can suffer from the same problem of using fuzzy mean image as a template for registration as in (Joshi et al., 2004). On the contrary, in our method, the representative image of each cluster is the warped individual image that contains abundance of key anatomical structures. Thus, the groupwise registration accuracy at a higher level can be guaranteed.
2.4. Summary of ABSORB registration method
To summarize the framework of the proposed ABSORB registration method, we enumerate all steps in the following:
0. Set the input dataset (after linear registration) of the 1st iteration I1 to be the original dataset I. Set initial parameters including iteration index t = 1, level index g = 1, maximum level index gmax, and the weighting factor α in Eq. (1).
1. Build the ING on dataset It and find the current global center image .
- 2. For each subject in the dataset It:
- 2a. Select qualified neighbors within the -nearest neighborhood that is closer to than , and perform pairwise registration between and each qualified neighbor.
- 2b. Calculate the average deformation field and warp to .
- 2c. If g > 1 and there are other non-representative images in the subgroup with the representative image , follow the same procedure in Step 2(a-b) and warp each of them separately to its new neighboring images determined by its representative image.
- 3. Apply affinity propagation clustering method on the current deformed image set It+1.
- 3a. If the clustering result of It+1 is different from that of It, t ← t + 1 and go back to Step 1.
- 3b. If the clustering results are the same for It and It+1, the number of clusters are more than one and the level index g < gmax, reset It+1 with all representative images. t ← t + 1 and go back to Step 1.
- 3c. Otherwise (i.e., the level index g = gmax or the number of clusters is one), go to Step 4.
4. Average all the registered images to generate the final atlas.
Note that when the registration process arrives at Step 4, the image population has been aligned very well. We can average all registered images to obtain the atlas since the mean image of a well-aligned dataset is sharp and keeps all major anatomical structures as will demonstrated in our experiments below.
2.5. Implementation issues
Several implementation issues about the parameter settings and computation complexity are discussed in this section.
In the proposed framework, there are several parameters related to the selection of the qualified neighbors for each subject to drive its deformation. Before we construct the ING , the k-NN isomap is built on the registered image set at iteration t first. Later on, the neighborhood of is defined by a hyper-sphere in which the number of neighbors is pre-specified as . Different settings for k or may result in different and different selected neighbors. However, it is worth noting that as long as is satisfied, the registration process will not change. This is because the edge weight in a graph built on k1-NN isomap is not larger than the corresponding edge weight in a graph built on k2-NN isomap if k1 > k2, and thus the neighbor selection within a neighborhood of size is the same. Therefore, in this paper, we set .
One of the key parameters in affinity propagation is the self-similarity, which is highly related to the clustering results. Since the self-similarity measures the possibility for a subject to be chosen as a center image (or a representative image) in a subgroup, all subjects are assigned with a common self-similarity in order to ensure that they have equal chances to be selected as a representative image. Different selections of self-similarity will lead to different clustering results. When the median (or minimum) of all pairwise similarity values is selected as the self-similarity, more (or fewer) subgroups will be generated (Frey and Dueck, 2007). In the lower levels of our hierarchical registration structure, a relatively large number of clusters can help reduce the distance between different clusters and thus make it easier for the cluster representative images to be registered with each other in the next higher level. But as the registration reaches a higher level, subjects are aligned to be similar to each other, so a smaller number of clusters will help register all subjects together and also speed up the convergence of registration without undermining the registration accuracy. Therefore, in the proposed ABSORB method, we choose to use the median as the self-similarity in Levels 1 and 2 (see Fig. 4), and the minimum as the self-similarity in all higher levels.
The pairwise image registration serves as a basic operation at each level in the proposed framework. Specifically, in each iteration, one subject is involved in no more than k pairwise registrations. If we assume that the total number of iterations is T, the computation complexity of the proposed ABSORB method is O(kNT). In our experiments, k ⪡ N and T ⪡ N.
3. Experiments
In this section, extensive experiments on both synthetic and real datasets are performed to demonstrate the performance of the proposed ABSORB registration method. For comparison, the results from other two groupwise registration methods are also provided. The first groupwise registration method under comparison is the group mean method proposed in (Joshi et al., 2004), where all subjects are registered to the group mean image, and the group mean image is updated upon the tentatively aligned images during the registration procedure. The second groupwise registration method is the tree-based registration method (Hamm et al., 2009). In this method, after building a tree and locating the root node, all other images are registered to the root subject by deforming along the path from each subject to the root.
Four datasets are used to evaluate the performance of all three methods, i.e., one synthetic dataset with 61 images and three real brain image datasets, including 18 elderly brain subjects (Resnick et al., 2000), NIREP brain image dataset (Christensen et al., 2006) and LONI LPBA40 dataset (Shattuck et al., 2008). We first evaluate three methods on a synthetic data set in which the three different types of structure for gyri and sulci are synthesized. The converging process on the synthetic data proves the efficacy and the ability of ABSORB to perform registration on the manifold. For the real brains, we present the mean image and the overlapping ratio of the registered brain images on different tissues, i.e., white matter (WM), grey matter (GM), and ventricle (VN), or on different regions of interest (ROIs). In all experiments, we use the same set of parameters, i.e., the maximum level of the built hierarchical structure gmax = 4, and the weight factor α = 0.5 in Eq. 1. Note that adaptively learning these parameters from each individual dataset may help improve the performance of ABSORB.
3.1. Synthetic dataset
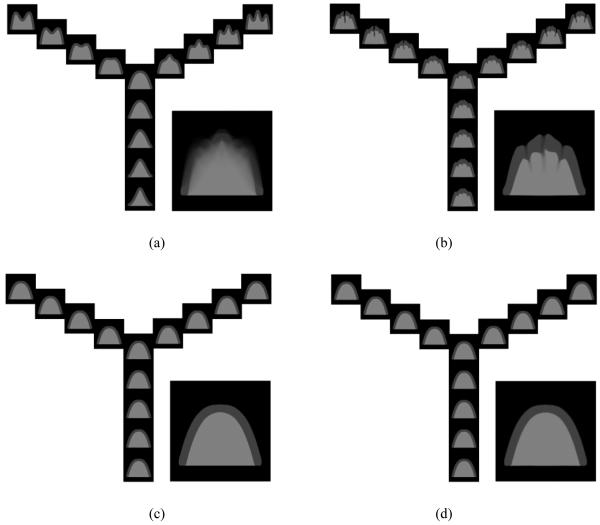
The proposed algorithm is first evaluated on a synthetic dataset, demonstrating the effect of self-organized registration and image bundling strategies. The synthetic dataset simulates the sulci and gyri around cortical region in MR brain image, as shown in Fig. 5(a). Each image has the size of 256×256. From the central image with a single wide gyrus, three different types of images are generated. Each type has 20 images with four of them being shown in each branch of the Y-shape structure in Fig. 5(a). There are totally 61 (20×3+1) images in the dataset. These three types of synthetic images are, respectively: i) a single synthetic gyrus changes from wide to narrow (i.e., a branch from the center to the bottom); ii) one gyrus splits into two gyri and the newly generated sulcus becomes deeper and deeper (i.e., a branch from the center to the top-left corner); and iii) one gyrus splits into three gyri with deeper and deeper sulci (i.e., a branch from the center to the top-right corner). The areas with different gray values are specified to represent the background, GM and WM, respectively. The built atlases and the registration accuracy are compared among the group mean method, the tree-based method, and the ABSORB method.
Figure 5.
The experimental results on the synthetic dataset. The original synthetic images and their blurry mean image (before registration) are shown in (a). The registration results for (b) the group mean method, (c) the tree-based method, and (d) the proposed ABSORB method are demonstrated together with their corresponding atlas images. The images on the same location of different Y-shape structures correspond to the same subject before and after the registration. The results for ABSORB and the tree-based method are visually similar, and their final built atlas images are much shaper and more reasonable than that of the group mean method.
Registered images and the built atlases
After performing the groupwise registration with the group mean method, we can obtain the registered images, which are placed in the same location of the Y-shape structure in Fig. 5 (b). Although those images with a single gyrus seem aligned well with each other, all other images with two or three gyri are not registered together completely since the bottom part of the sulcus is lagging behind the motion of its neighboring anatomies during the registration and thus a deep fissure is formed with a similar depth at the corresponding position of each sulcus. This is resulted mainly because the group mean image is initially very blurry (i.e., the one shown on the bottom right of Fig. 5(a)), and it is very difficult for those sulcal parts to warp towards the right direction consistently. It is also seen that the built atlas (as shown on the bottom right of Fig. 5(b)), with three vague cracks and a bumpy surface, does not look like any of the initial images. In Fig. 5(c) and 5(d), the results of the tree-based method and the proposed ABSORB method are shown together with the final built atlas. The results of the tree-based method are visually similar to those of ABSORB because the root node is selected to be very close to the geometrical mean and the underlying data space is well sampled by the current dataset. We can see that all the images with different number of sulci and different depth of each sulcus have been well registered onto the final atlas, which is very close to the geometrical mean.
Registration process
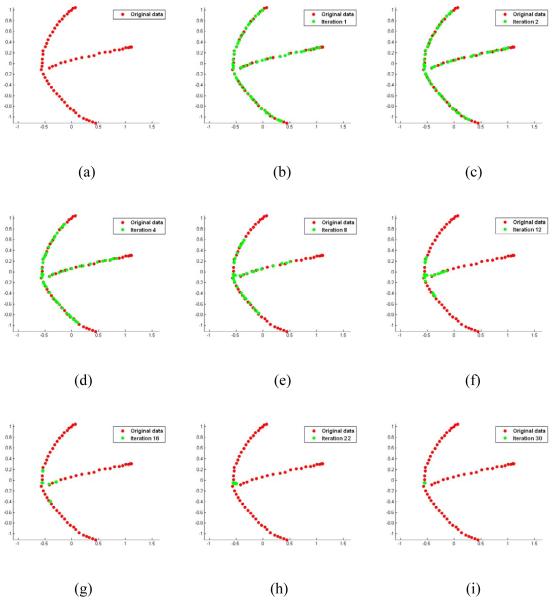
We illustrate the registration process of the proposed ABSORB method on the synthetic dataset in Fig. 6. The original dataset is projected onto a two dimensional (2D) space which is spanned by the two eigen-vectors corresponding to the largest two eigen-values after applying Principal Component Analysis (PCA) on the original dataset (Fig. 6(a)), and the updated image set in all later iterations is projected onto the same 2D space to visualize the converging process. The learned manifold can be clearly seen as a Y-shape in Fig. 6(a). In Fig. 6 (b-g), the whole population is converging, with the moving direction of each subject determined by both its qualified neighbors and the (tentatively estimated) global center. After 16 iterations, it reaches a stable distribution of the population, so all representative images are automatically chosen to form a new dataset and the registration process moves up to the second level in the pyramid (Fig. 6(g)). On the second level, the same procedure is applied to a much smaller population, and all other non-representative images follow the movement of their representatives. Finally, the whole registration process arrives at the top level where all images are clustered into a single group and registered together (Fig. 6(i)).
Figure 6.
The illustration of the registration process of ABSORB on the synthetic dataset. The original dataset is projected onto a 2D PCA space (a), and the same projection is applied to all later registered images. All subjects converge to the global center on the learned manifold during iteration 1 till 16 (b-g). After applying AP clustering method on the results of iteration 16 (g), all points converge together to form several subgroups, and each point in (g) represents a bundle of aligned images. The registration continues at higher levels (h-i), and all subjects are finally registered together (i). This figure is best viewed in color printing.
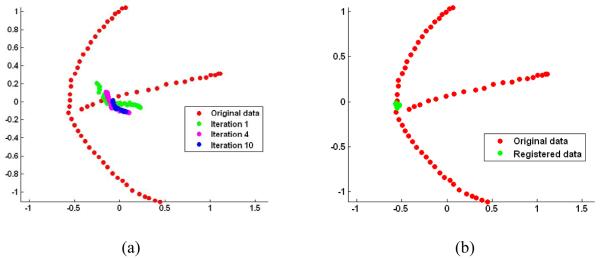
For comparison, the registration results by the group mean method and the tree-based method on the same 2D projection space are shown in Fig. 7. It can be observed that the registration results of ABSORB are much more compact than those of the other two methods. In Fig. 7 (a), we can see that, after the first iterative registration by the group mean method, the warped dataset travels away from the manifold (i.e., the data structure represented by three branches) because of the use of initially very fuzzy mean image as shown in Fig. 5(a). Since the registration process is misled in the very first step, the following registrations can be refined only around the new but wrong center of images in all later steps, although it converges very fast using only about 4 iterations. In contrast, the registration process of ABSORB is constrained strictly on the manifold as shown in Fig. 6, and also the topological structure of the dataset is well preserved until all subjects reach the final atlas. In Fig. 7 (b), the registration result of the tree-based method is also shown, which seems not as compact as that of ABSORB (see Fig. 6(i)).
Figure 7.
The registration results of (a) the group mean method and (b) the tree-based method. The image registration process of the group mean method diverges away from the learned manifold (starting from the first iteration shown in green in (a). The registration result of the tree-based method (b) is better; however, it is not as compact as that by the ABSORB method. This figure is best viewed in color printing.
Registration accuracy
We also compare the registration accuracy in a quantitative way by measuring the concentration on the registered dataset and the landmarks. First, the registration accuracy is evaluated on the registered dataset. Specifically, the pairwise distance (i.e., intensity difference) between each registered image and the atlas (the mean image) is calculated, and the mean and standard deviation (std) of these distances are used to measure how concentrated the registered results are for all three methods. The mean of these distances for the group mean method, the tree-based method, and ABSORB are 3.5, 1.6 and 1.4, respectively, and the corresponding stds are 1.7, 0.4 and 0.2, respectively. Compared to the mean and std (16.3 and 4.0) for the original dataset, the registration accuracy by the tree-based method and ABSORB is significantly improved, compared to that by the group mean method; and also ABSORB is slightly better than the tree-based method.
Second, the registration accuracy is evaluated on the landmarks. As we know the ground truth of the transformation for generating the synthetic images, we can evaluate the groupwise registration results by the alignment of the corresponding landmarks. Totally, 193 landmark points are located in each image of this synthetic dataset, and thus we have 193 point sets, with each set having 61 correspondences from 61 synthetic images. All 61 correspondences in each point set should be close to each other after groupwise registration. In the original dataset, the mean and std of all pairwise distances from each of 61 correspondences to their center (over all 193 point sets) are 12.4 and 15.8, respectively. The mean/std for the group mean method, the tree-based method, and ABSORB are 6.9/7.2, 1.7/2.8 and 1.2/1.6, respectively. ABSORB ranks top over all registration methods.
3.2. Experiment on 18 elderly brains dataset
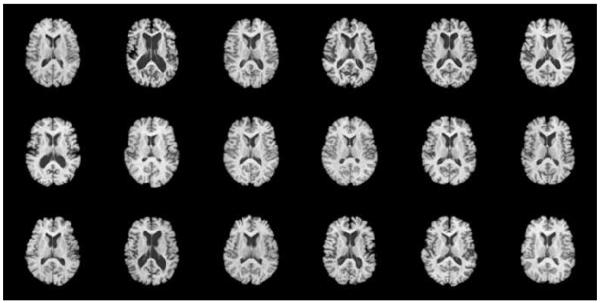
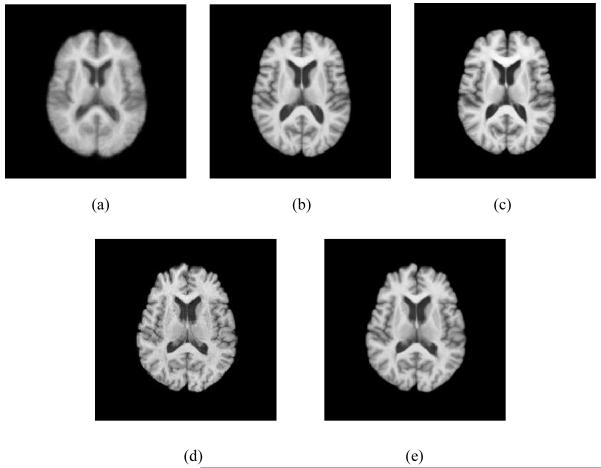
A brain MR image dataset with 18 elderly subjects is used to further evaluate the performance of ABSORB on real images. Fig. 8 indicates large variations of structures across 18 different brain images. Affine registrations have been performed to these 18 brain images to remove their global differences. In this section, we will continue to compare the performances among three different methods, namely the group mean method (Joshi et al., 2004), the tree-based method (Hamm et al., 2009), and ABSORB, on the registration accuracy both qualitatively and quantitatively.
Figure 8.
18 elderly brain images with large anatomical variations.
Registered images and the built atlases
First, we compare the atlases generated by three different methods visually in Fig. 9. It can be observed that the atlas image from ABSORB is slightly sharper than that of the group mean method, although they are visually similar to each other. The atlas from the tree-based method is biased to the selected root image, which may not represent the group mean very well, although the atlas is relatively sharper. From the result of the tree-based method on both synthetic and real datasets, we can see that, if the selected root image cannot represent the whole population, the bias could be introduced into the whole registration process, as well as the built atlas.
Figure 9.
The atlas images generated by three different methods. The mean image for the original dataset (a), the atlas image constructed by the group mean method (b), and the proposed ABSORB method (c) are shown in the top row from left to right. The atlas by ABSORB is visually similar to that of the group mean method. For the tree-based method, its selected root image and the corresponding atlas are shown in (d) and (e), respectively. It can be observed that the atlas image is very similar to the root image.
Registration accuracy
The improvement of ABSORB over the group mean method and the tree-based method can be examined more clearly by measuring the overlapping rates on different tissues and also the average entropy on the segmentations of registered images. Here, we use the Jaccard Coefficient metric (Jaccard, 1912) to measure the similarity between two registrations of the same region, which provides a similar but stricter definition of the overlapping rate than the popularly used Dice Coefficient (van Rijsbergen, 1979). For two registered region U and V, the Jaccard Coefficient is defined as
| (4) |
where |·| defines the area of region under consideration. To demonstrate the group overlapping on the registered segmentation images, tissue labels on each voxel first vote to obtain a common segmentation atlas. This is done by assigning each voxel with a tissue label that is the majority of all tissue labels at the same location from all the aligned images. Then, the Jaccard Coefficient between each of the registered segmentation images and the voted common segmentation atlas is calculated, with the average score listed in Table 1. Note that this is a very strict definition to measure the overlapping rate and thus the respective value seems low compared to other definitions. Our method achieves the best overlapping rates on all three different tissues, and the average increase over the other two methods is about 4.0%. The group mean method and the tree-based method have similar average overlapping rates. The average entropy of our method on the aligned segmentation images is 0.17, which is about 10% better than the group mean method and the tree-based method.
Table 1.
The overlapping rates and average entropy of the registered segmentation images by three different methods
| Overlapping rate (%) |
Entropy | |||
|---|---|---|---|---|
| GM | WM | VN | ||
| Before registration | 35.5% | 45.4% | 48.6% | 0.33 |
|
| ||||
| Group mean method | 49.0% | 68.0% | 72.6% | 0.19 |
|
| ||||
| Tree-based method | 51.7% | 61.9% | 74.6% | 0.19 |
|
| ||||
| ABSORB | 54.0% | 71.0% | 76.6% | 0.17 |
Robustness of registration
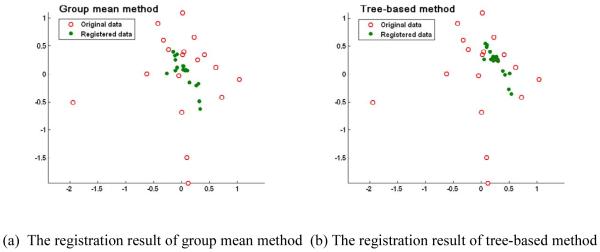
The robustness of registration to the outliers of different methods is also compared. The registration results of three methods are shown in Fig. 10 by projecting all the registered subjects onto the 2D PCA space. It can be observed that the final registered images of ABSORB and the tree-based method are more concentrated around the geodesic mean than those of the group mean method. In other words, the group mean method is easily to be distracted by the outliers, i.e., the left-most and the bottom-most points in Fig. 10 (a), and ABSORB and the tree-based method can obtain more robust registration results even some outliers exist in the dataset. It is worth noting that the registration results of ABSORB and the tree-based method are also more compactly distributed in the projective space than those of the group mean method.
Figure 10.
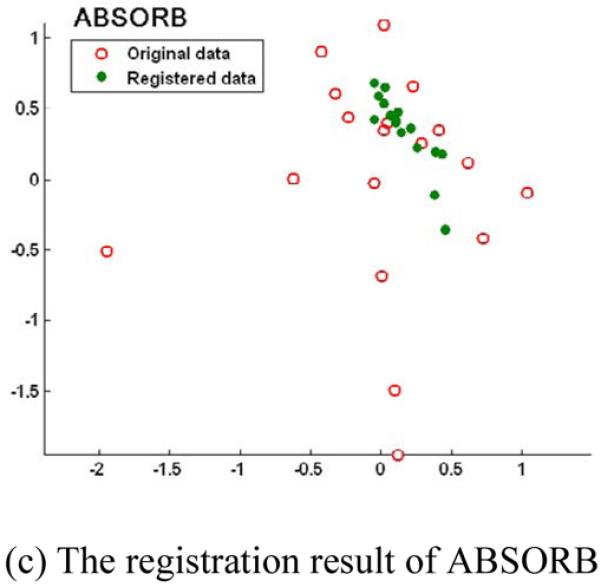
The registration results of the group mean method (a), the tree-based method (b), and the proposed ABSORB method (c) on 18 elderly brain images. The final registered images of the group mean method are distracted by the outlier images, i.e., the left-most and the bottom-most points, while the results by the other two methods are robust to the outliers and finally concentrate around the geodesic mean.
Smoothness on registration path
The registration paths of all three methods are examined in Fig. 11 to compare the smoothness of paths generated by different methods. We select three different subjects (labeled as Sub 1, Sub 2 and Sub 3 in Fig. 11), and the registration path for each of them is delineated by connecting all segments from its original position to the final position. For the tree-based method, its built tree has the height of 5, and thus the segments on each path are no more than 4. The selected three subjects are those of the longest distance to the root node on the built tree. It can be observed that the path generated from our method is always much smoother and more direct to the final location than that of the tree-based method, which is more twisted and devious. The main reason is that each deformation segment along the path of our method is the result of averaging several different moving directions, and also different segments on the same path share a similar direction to the global center. However, in the tree-based method, the path is pre-determined by a tree and a fixed root node (before image registration), without considering the dynamic change of overall distribution of the dataset after each iterative registration. Also, the moving directions of different segments on the same path are independently estimated in the tree-based method, thus potentially resulting in a more zigzag path and affecting the registration results. On the other hand, the paths generated by the group mean method become nearly unchanged after the first round of registration, which indicates that the group mean method could be easily trapped by the local minima.
Figure 11.
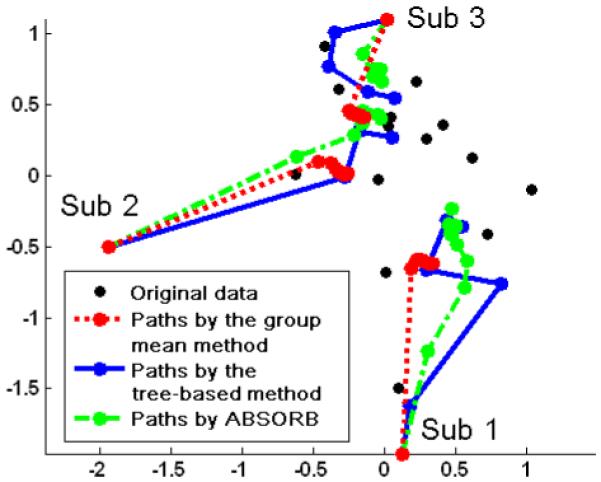
The registration paths produced by three different methods. The paths generated by ABSORB are much smoother than the tree-based method. The paths given by the group mean method shows that there is not much progress after the first iteration of registration.
3.3. Experiment on NIREP dataset
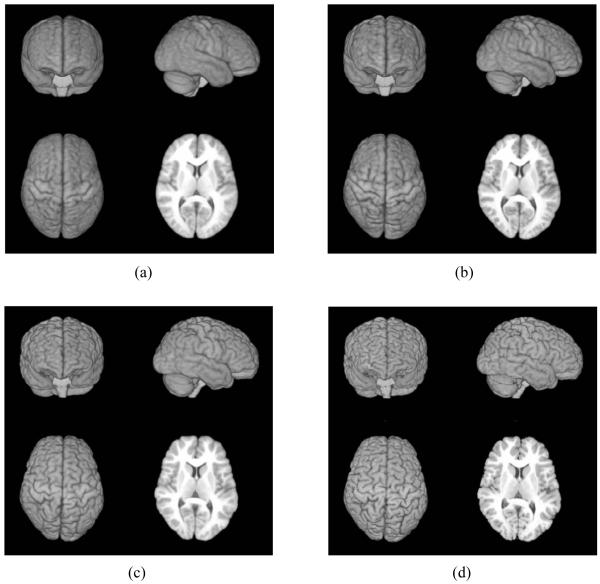
In this experiment, all three methods are tested on NIREP dataset including 16 brain subjects. The atlas images generated by the group mean method, the proposed ABSORB method and the tree-based method are displayed in Fig. 12, respectively. ABSORB can generate an atlas image with more anatomical details than the group mean method, especially on the cortical regions as indicated by red arrows in Fig. 12. As we have pointed out in previous experiments, the registration results given by the tree-based method can be biased by the selected fixed rood subject, although the built atlas is sharper.
Figure 12.
The atlas images of NIREP brain dataset built by the group mean method (a), ABSORB (b) and the tree-based method (c). The atlas generated by ABSORB can keep more anatomical details than that generated by the group mean method, especially on the cortical regions marked by red arrows. The atlas built by the tree-based method (c) is clearly biased to the root subject (d).
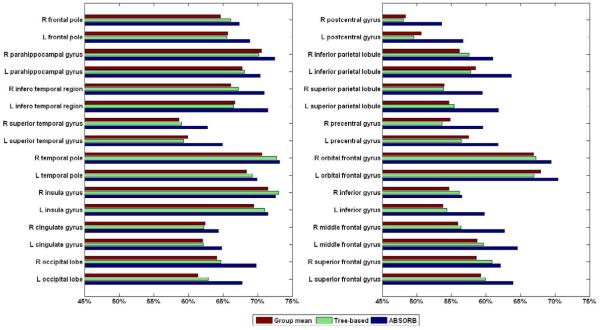
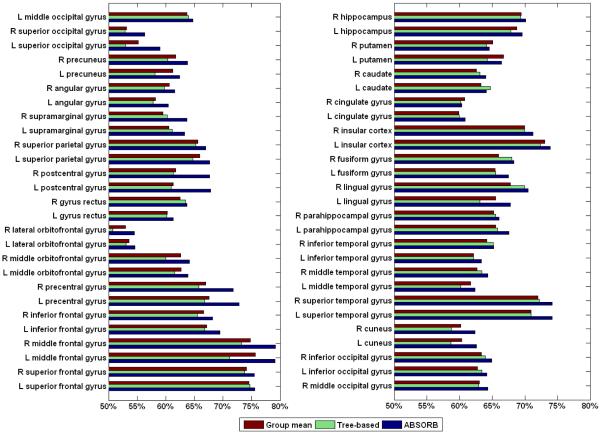
To demonstrate the performance of the proposed ABSORB framework quantitatively, the overlap rates are calculated based on the registered images, which have been manually labeled with 32 ROIs. The average overlap rate over all 32 ROIs in the original dataset is 46.21%. After the groupwise registration of ABSORB, the average overlap ratio increases to 65.31%, which is much higher than the results of the group mean method (61.25%) and the tree-based method (61.69%). We plot the overlap rates of all 32 ROIs in Fig. 13. It can be seen that ABSORB outperforms other two methods on 31 ROIs, except that the overlap ratio of ABSORB on R insula gyrus is only 0.5% lower than that of the tree-based method. The average entropy on the registered image dataset is also calculated. The average entropy on the original dataset is 0.801. The ABSORB method gives the smallest average entropy (0.505) among all three methods, which is much smaller than that of the group mean method (0.600) and the tree-based method (0.590).
Figure 13.
The overlap rates of 32 ROIs on the registered NIREP dataset by three different groupwise registration methods.
3.4. Experiment on LONI LPBA40 dataset
Finally, we evaluate the proposed ABSORB method on a much larger dataset, LONI LPBA40, which has 40 brain images with 54 manually labeled ROIs. The average overlap rate over all 54 ROIs on the original dataset is 62.08%, since all 40 subjects are linearly aligned before performing manual delineations. The proposed ABSORB method increases the average overlap rate to 69.5%. The same measurements given by the group mean method and the tree-based method are 66.36% and 66.95%, respectively. We plot the overlap rates of all 54 ROIs in Fig. 14.
Figure 14.
The overlap rates of 54 ROIs on the registered LONI LPBA40 dataset by three different groupwise registration methods.
4. Conclusion
A new framework for groupwise registration, called Atlas Building by Self-Organized Registration and Bundling, or ABSORB, has been presented. In this new framework, the global structure of subject distribution on the data space is always preserved during the registration process, and the deformation of each subject is constrained locally along the learned image manifold. As the two novel strategies proposed in the ABSORB method, self-organized registration and image bundling are both employed to perform the groupwise registration hierarchically, by automatically building a pyramid of images during the registration procedure. An atlas can be finally built once the registration arrives at the top level. Extensive experiments have been conducted to evaluate the performance of the ABSORB registration method, which shows that ABSORB can perform the registration more accurately and consistently, compared to other two groupwise registration methods, namely the group mean method and the tree-based method. Specifically, the overlap rates of the same tissues across different subjects after registration are much higher than any of two methods under comparison. In the future, we will apply ABSORB to large clinical datasets with brain disorders such as Alzheimer’s disease or schizophrenia, to test its performance in detecting brain abnormalities.
Footnotes
Source code can be downloaded from http://www.insight-journal.org/browse/publication/154.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5. References
- Ashburner J. A Fast Diffeomorphic Image Registration Algorithm. Neuroimage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. 2007. [DOI] [PubMed] [Google Scholar]
- Balci SK, Golland P, Shenton M, Wells WM. Free-Form B-spline Deformation Model for Groupwise Registration; Med Image Comput Comput Assist Interv. MICCAI 2007; 2007a. [Google Scholar]
- Balci SK, Golland P, Wells W. Non-rigid Groupwise Registration using B-Spline Deformation Model; Open Source and Open Data for MICCAI; 2007b.pp. 105–121. [Google Scholar]
- Baloch S, Davatzikos C. Morphological appearance manifolds in computational anatomy: Groupwise registration and morphological analysis. Neuroimage. 2009;45:S73–S85. doi: 10.1016/j.neuroimage.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloch S, Verma R, Davatzikos C. An Anatomical Equivalence Class Based Joint Transformation-Residual Descriptor for Morphological Analysis. Information Processing in Medical Imaging. 2007:594–606. doi: 10.1007/978-3-540-73273-0_49. [DOI] [PubMed] [Google Scholar]
- Bhatia KK, Hajnal JV, Puri BK, Edwards AD, Rueckert D. Consistent groupwise non-rigid registration for atlas construction; IEEE Int. Symp. Biomed. Imag.: Macro Nano; 2004.2004. pp. 908–911. [Google Scholar]
- Christensen GE, Geng X, Kuhl JG, Bruss J, Grabowski TJ, Pirwani IA, Vannier MW, Allen JS, Damasio H. Introduction to the Non-rigid Image Registration Evaluation Project (NIREP) 3rd International Workshop on Biomedical Image Registration; 2006.pp. 128–135. [Google Scholar]
- Christensen GE, Johnson HJ. Consistent Image Registration. IEEE Transactions on Medical Imaging. 2001;20:568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- Cox T, Cox MAA. Multidimensional Scaling. second ed. Chapman & Hall; London: 2000. [Google Scholar]
- Crum WR, Hartkens T, Hill DLG. Non-rigid Image Registration: Theory and Practice. British Journal of Radiology. 2004;77:S140–153. doi: 10.1259/bjr/25329214. [DOI] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. Clustering by Passing Messages Between Data Points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- Hajnal JV, Saeed N, Soar EJ, Oatridge A, Young IR, Bydder GM. A Registration and Interpolation Procedure for Subvoxel Matching of Serially Acquired MR Images. Journal of Computer Assisted Tomography. 1995;19:289–296. doi: 10.1097/00004728-199503000-00022. [DOI] [PubMed] [Google Scholar]
- Hamm J, Davatzikos C, Verma R. Efficient large deformation registration via geodesics on a learned manifold of images; MICCAI 2009; London, UK. 2009; [DOI] [PubMed] [Google Scholar]
- Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical Image Registration. Physics in Medicine and Biology. 2001;46:R1–R45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- Holden M, Hill DLG, Denton ERE, Jarosz JM, Cox TCS, Rohlfing T, Goodey J, Hawkes DJ. Voxel Similarity Measures for 3-D Serial MR Brain Image Registration. Medical Imaging, IEEE Transactions on. 2000;19:94–102. doi: 10.1109/42.836369. [DOI] [PubMed] [Google Scholar]
- Jaccard P. The distribution of flora in the alpine zone. New Phytologist. 1912;11:37–50. [Google Scholar]
- Johnson HJ, Christensen GE. Consistent landmark and intensity-based image registration. IEEE Transactions on Medical Imaging. 2002;21:450–461. doi: 10.1109/TMI.2002.1009381. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased Diffeomorphic Atlas Construction for Computational Anatomy. Neuroimage. 2004;23:S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang M-C, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal JB. On the Shortest Spanning Subtree of a Graph and the Traveling Salesman Problem. Proceedings of the American Mathematical Society. 1956;7:48–50. [Google Scholar]
- Learned-Miller EG. Data Driven Image Models through Continuous Joint Alignment. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2006;28:236–250. doi: 10.1109/TPAMI.2006.34. [DOI] [PubMed] [Google Scholar]
- Maintz JBA, Viergever MA. A Survey of Medical Image Registration. Medical Image Analysis. 1998;2:1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- Munsell BC, Temlyakov A, Wang S. Fast Multiple Shape Correspondence by Pre-Organizing Shape Instances; IEEE Conference on CVPR; 2009.pp. 840–847. [Google Scholar]
- Park H, Bland PH, Hero AO, Meyer CR. Least Biased Target Selection in Probabilistic Atlas Construction; MICCAI; 2005; pp. 419–426. [DOI] [PubMed] [Google Scholar]
- Resnick SM, Goldszal A, Davatzikos C, Golski S, Kraut MA, Metter EJ, Bryan RN, Zonderman AB. One-year age changes in MRI brain volumes in older adults. Cerebral Cortex. 2000;10:464–472. doi: 10.1093/cercor/10.5.464. [DOI] [PubMed] [Google Scholar]
- Sabuncu MR, Balci SK, Shenton ME, Golland P. Image-Driven Population Analysis Through Mixture Modeling. IEEE Transactions on Medical Imaging. 2009;28:1473–1487. doi: 10.1109/TMI.2009.2017942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Shenton ME, Golland P. Joint Registration and Clustering of Images; MICCAI 2007; Springer, Heidelberg, Germany. 2007.pp. 47–54. [Google Scholar]
- Seghers D, D’Agostino E, Maes F, Vandermeulen D, Suetens P. Construction of a Brain Template from MR Images Using State-of-the-Art Registration and Segmentation Techniques; MICCAI; 2004.pp. 396–703. [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: Hierarchical Attribute Matching Mechanism for Elastic Registration. IEEE Trans. on Medical Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Tang S, Fan Y, Shen D. RABBIT: Rapid Alignment of Brains by Building Intermediate Templates. Neuroimage. 2009;47:1277–1287. doi: 10.1016/j.neuroimage.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum JB, Silva V.d., Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- van Rijsbergen CJ. Information Retrieval. 2nd ed. Butterworth-Heinemann; London: 1979. [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009;45:S61–72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chen L, Shen D. Group-wise registration of large image dataset by hierarchical clustering and alignment; Medical Imaging 2009. SPIE.; 2009a. [Google Scholar]
- Wang Q, Yap P-T, Wu G, Shen D. Attribute Vector Guided Groupwise Registration; MICCAI 2009; London, UK. 2009b; [DOI] [PubMed] [Google Scholar]
- Zöllei L, Learned-Miller E, Grimson E, Wells W. Efficient Population Registration of 3D Data; Computer Vision for Biomedical Image Applications (ICCV); 2005.pp. 291–301. [Google Scholar]
- Zitová B, Flusser J. Image Registration Methods: A Survey. Image and Vision Computing. 2003;21:911–1000. [Google Scholar]