Abstract
ELK transcription factors are expressed in brain, but it is unknown whether they are expressed in the peripheral nervous system. We show by RT-PCR that the previously described Elk1, Elk3/Elk3b/Elk3c and Elk4 mRNAs are expressed in adult dorsal root ganglia (DRG), together with the novel alternatively spliced isoforms Elk1b, Elk3d and Elk4c/Elk4d/Elk4e. These isoforms are also expressed in brain, heart, kidney and testis. In contrast to Elk3 protein, the novel Elk3d isoform is cytoplasmic, fails to bind ETS binding sites and yet can activate transcription by an indirect mechanism. The Elk3 and Elk4 genes are overlapped by co-expressed Pctk2 and Mfsd4 genes, respectively, with the potential formation of Elk3/Pctaire2 and Elk4/Mfsd4 sense-antisense mRNA heteroduplexes. After peripheral nerve injury the Elk3 mRNA isoforms are each upregulated ~2.3-fold in DRG (P<0.005), whereas the natural antisense Pctaire2 isoforms show only a small increase (21%, P<0.01) and Elk1 and Elk4 mRNAs are unchanged.
Keywords: Elk, Transcription factor, Dorsal root ganglia, Alternative splicing, Antisense mRNA, Axotomy
Introduction
The ETS (E twenty-six) family of transcription factors each contain an ETS domain of about 85 amino acids which can bind to a ~ 10-basepair sequence element with the highly conserved central core sequence 5′-GGA(A/T)-3′ (Mo et al., 2000; Oikawa and Yamada, 2003; Shore and Sharrocks, 1995). The family consists of 28/29 members in mouse and human, respectively, which can be assigned into nine subfamilies on the basis of homology and domain content (Kobberup et al., 2007). For example, members of the ELK (ETS-like) subfamily contain an N-terminal ETS DNA-binding domain and a B-box domain that allows the formation of a ternary complex with serum response factor (SRF) dimer, and so are also referred to as ternary complex factors (Buchwalter et al., 2004). The ELK subfamily comprises of the three proteins Elk1, Elk3 (also called Net, Erp or Sap2) and Elk4 (also called Sap1) (Buchwalter et al., 2004; Kobberup et al., 2007), which can have different potential protein-protein interactions (Buchwalter et al., 2004; Ducret et al., 1999; Ducret et al., 2000; Fitzsimmons et al., 1996; Maira et al., 1996), can respond differently to mitogen-activated protein (MAP) kinase signalling pathways (Buchwalter et al., 2004; Ducret et al., 2000), and Elk1 and Elk4 are known to have similar but distinct extended DNA-binding site sequence specificities (Mo et al., 2000; Shore and Sharrocks, 1995; Treisman et al., 1992).
A number of ETS-family transcription factors are expressed within the central nervous system (CNS; reviewed in (Oikawa and Yamada, 2003); (Hollenhorst et al., 2004; O’Leary et al., 2005)), including Elk1 and Elk3 mRNAs in adult mouse brain and Elk4 mRNA in human brain (Cesari et al., 2004; Giovane et al., 1994; Hollenhorst et al., 2004; Lopez et al., 1994; Price et al., 1995), whereas little is known about their expression in adult sensory neurons within dorsal root ganglia (DRG) of the peripheral nervous system (PNS; (Luo et al., 1999)). In contrast to neurons of the adult mammalian CNS, those of the PNS are capable of axonal regeneration following peripheral nerve transection (axotomy), and this is associated with altered gene expression within the cell bodies of the DRG (reviewed in (Navarro et al., 2007)). For example, the expression of neuropeptide galanin mRNA is upregulated around 80-fold in adult mouse DRG following axotomy, and recently we have implicated an 18 basepair (bp) sequence within the promoter/enhancer region as being critical for this response (Bacon et al., 2007; Kerr et al., 2008). This sequence contains overlapping putative Ets, Stat and Smad transcription factor binding sites (Bacon et al., 2007), without a nearby putative SRF binding site (CArG box). Therefore, we were interested in which ETS-family transcription factors are expressed in adult DRG.
In this study we focussed on expression of the ELK subfamily, as oligonucleotide microarrays had shown us that Elk3 mRNA was potentially upregulated in adult mouse DRG following axotomy (see below). We demonstrate that both known and novel ELK mRNA isoforms are expressed in adult DRG and other tested tissues, together with natural antisense mRNAs to Elk3 and Elk4. Alternative splicing of Elk3 mRNA results in protein isoforms with distinct functional properties, and quantitative RT-PCR demonstrates that the different Elk3 mRNA isoforms (but not Elk1 or Elk4) are upregulated after peripheral nerve injury.
Results
The expression of ELK mRNA isoforms in dorsal root ganglia (DRG)
We first investigated which ELK subfamily transcription factors and their isoforms are expressed in adult mouse DRG. The genes encoding the ELK subfamily proteins Elk1, Elk3 and Elk4 each have a large exon 3 (respectively, 447, 801 and 870 nucleotides (nt)), within which alternative splicing of Elk1 and Elk3 mRNAs is known to occur (Buchwalter et al., 2004). Therefore, gene-specific primers were designed to span this exon, and in the cases of Elk3 and Elk4 to prime from within this exon in order to accommodate the range of possible RT-PCR product sizes.
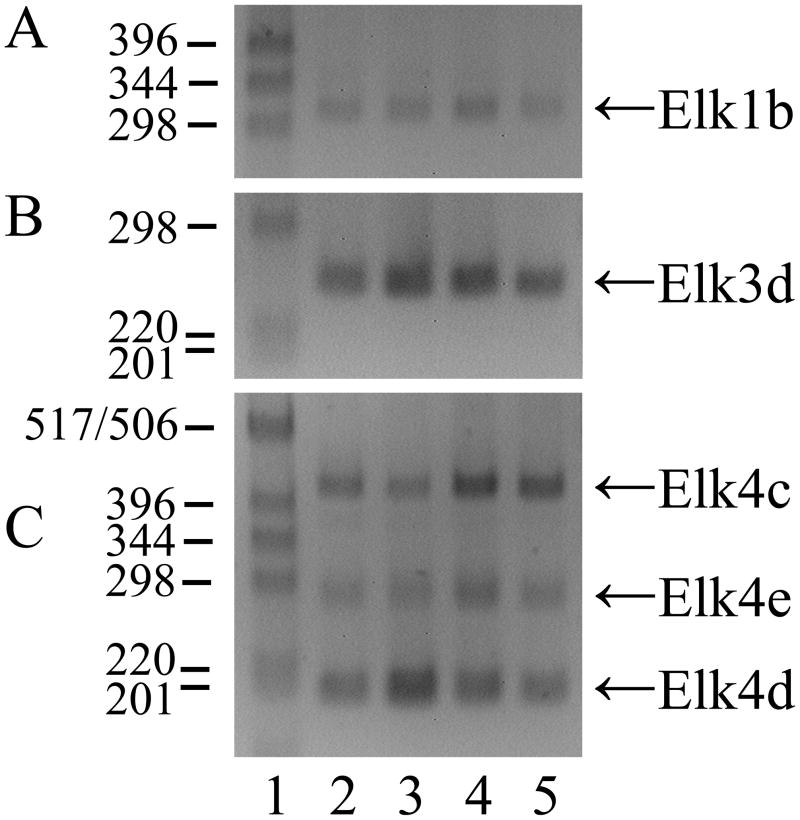
An Elk1 mRNA product of the expected size and sequence (n = 10) was detected in adult mouse DRG using primers to exons 2 and 4 (Fig. 1A), together with a minor, novel alternatively spliced Elk1b isoform of 314 bp that results from the deletion of 355 nt from the 5′ end of exon 3 (n = 10). There was no evidence of a ~240 bp product equivalent to the previously described human ΔElk1 mRNA which has a deletion of 429 nt from the 3′ end of the corresponding exon ((Rao and Reddy, 1993); M25269). Of relevance to the studies below on Elk3 and Elk4, there was no additional Elk1 product resulting from in-frame skipping of both exons 3 and 4 (that correspond to the single exon 3 of Elk3/Elk4; NM_007922) using primers to exons 2 and 5 (data not shown).
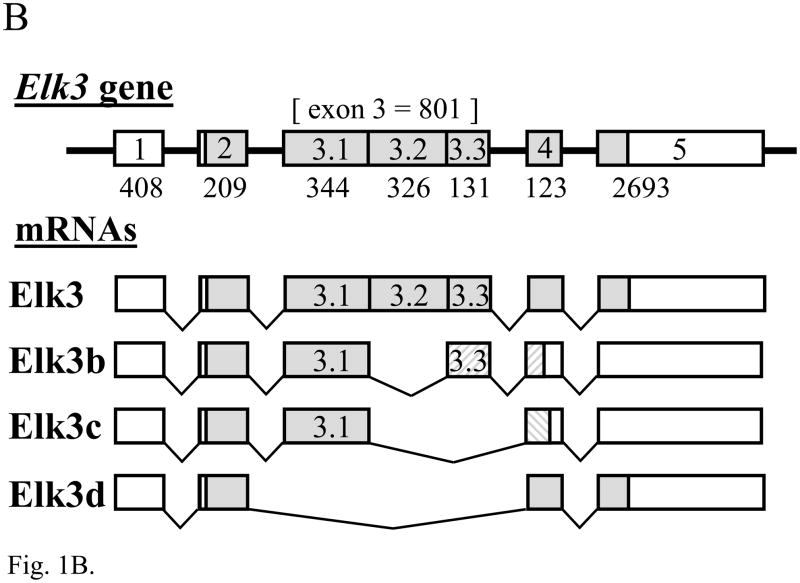
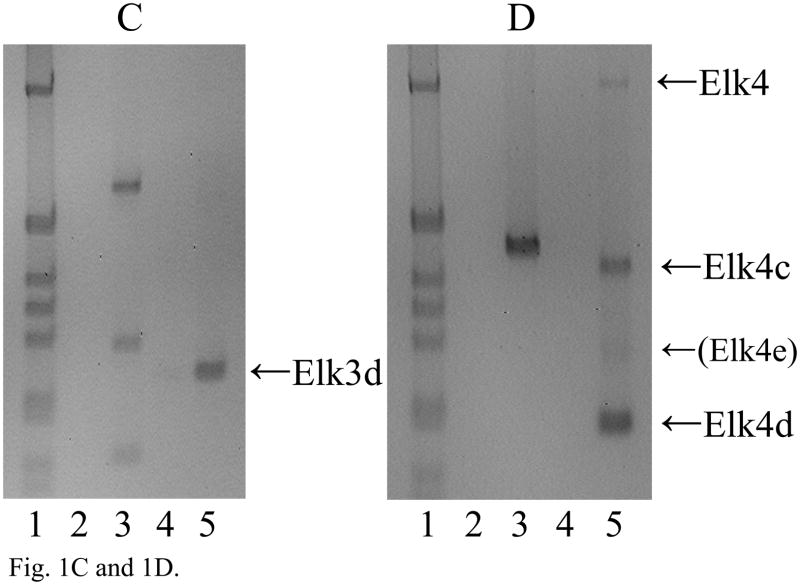
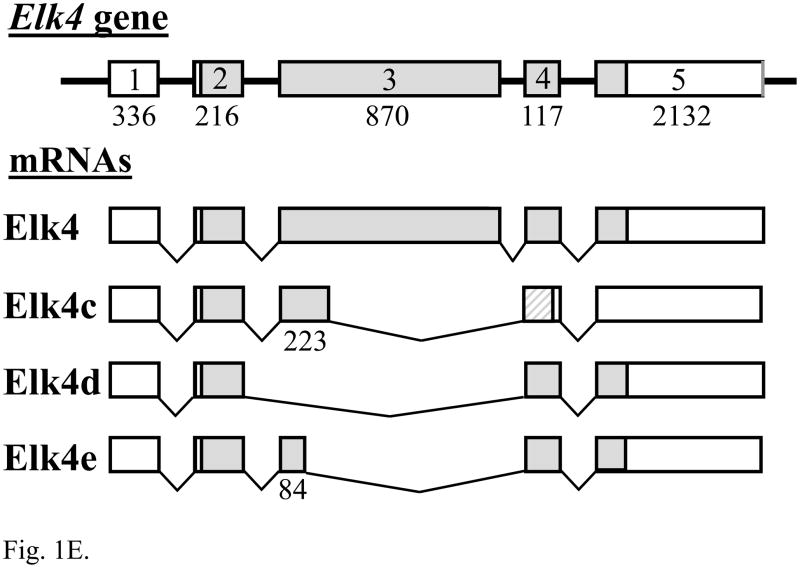
Fig. 1.
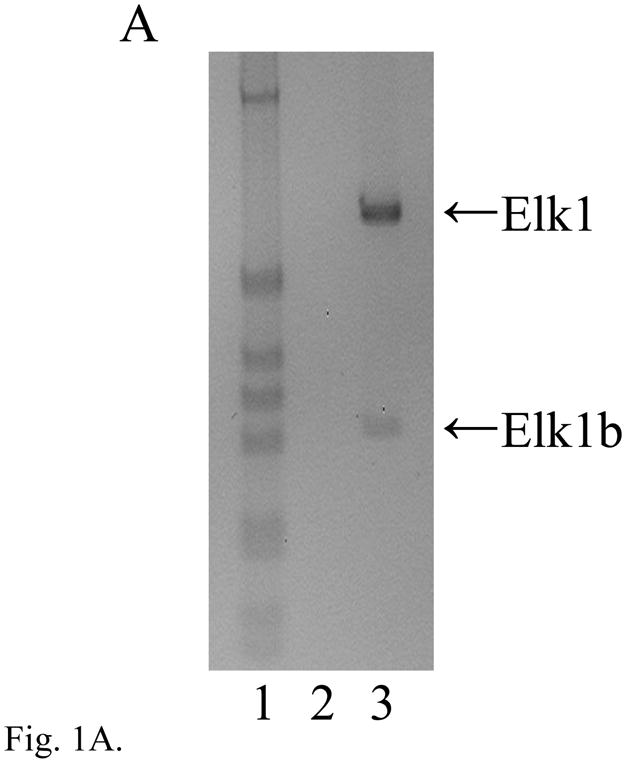
The expression of Elk1, Elk3 and Elk4 mRNA isoforms in adult DRG. (A) RT-PCR analysis showing expression of Elk1 (669 bp) and a minor, novel Elk1b isoform (314 bp; lane 3). In each gel image, lane 1 is a 1 kb DNA marker (Invitrogen) showing bands of 1018, 517/506, 396, 344, 298, 220/201 and 154/134 bp; other odd-numbered lanes are RT-PCRs using reverse transcribed RNA; and even-numbered lanes are corresponding reactions using RNA that had not been reverse transcribed (RT-minus controls), in which products were not detected. (B) Schematic diagram of the Elk3 gene with five exons (lengths in nt) and the transcribed full-length Elk3 mRNA, Elk3b isoform with an internal relative deletion of 326 nt within exon 3 (Δ exon 3.2), Elk3c isoform with a relative deletion of the 3′ 457 nt of exon 3 (Δ exons 3.2 and 3.3) ((Giovane et al., 1997); FN434119, FN434120) and the novel Elk3d isoform (see Fig. 1C). Coding regions are in gray, with those resulting from relative frameshifts shown in diagonal bars. (C) Expression of full-length Elk3, Elk3b and Elk3c isoforms (618, 292 and 161 bp, respectively) detected using primers to exons 3.1 and 4/5 (lane 3), whereas primers to exons 2 and 4 detected a novel Elk3d isoform (253 bp) under conditions in which the potential larger isoforms (597–1054 bp) were no longer detected (lane 5). (D) Expression of Elk4 detected using primers to exons 3 and 4 (457 bp; lane 3), whereas primers to exons 2 and 4 detected the now-larger Elk4 (1069 bp) plus the novel, major products Elk4c and Elk4d (422 and 199 bp, respectively) together with the minor product Elk4e (283bp; lane 5). (E) Schematic diagram of the Elk4 gene with five exons and the transcribed full-length Elk4 mRNA, together with the novel mRNA isoforms Elk4c, Elk4d and Elk4e (details as in (B)).
In addition to full-length Elk3 mRNA, the two alternatively spliced isoforms Elk3b (Net-b) and Elk3c (Net-c) have previously been described which have deletions of 326 or 457 nt within exon 3 ((Giovane et al., 1997); NM_013508), as shown in Fig. 1B. Three RT-PCR products of the expected sizes and sequences of Elk3, Elk3b and Elk3c (each n = 10) were detected in adult DRG using primers designed to amplify from exon 3.1 to the junction of exons 4 and 5 (Fig. 1C, lane 3). However, using additional primers to amplify Elk3 exons 2 to 4 allowed the detection of a novel Elk3d isoform (253 bp) under conditions in which the now much larger full-length Elk3 product (1054 bp) was variably detected and the Elk3b and Elk3c isoforms were no longer detected (Fig. 1C, lane 5, and data not shown). By DNA sequencing Elk3d was shown to result from the in-frame skipping of the whole of exon 3 (n = 10; Fig. 1B). By comparison to the Elk3 reference mRNA coding region (NM_013508) the sequenced clones contained the same nine substitutions as found in another cDNA sequence (BC005686), of which six also occur in the original cDNA sequences ((Giovane et al., 1994; Lopez et al., 1994); see Table S1). These differences may be due to strain-specific polymorphisms.
Two human ELK4 (SAP-1) cDNAs have previously been isolated, SAP-1a and SAP-1b (Dalton and Treisman, 1992), though SAP-1b expression was not apparent in eleven tested human tissues (Price et al., 1995) and the sequence has a 3′-extended exon 3 or possible retained intron sequence (NM_021795; NT_004487) that is not conserved in the corresponding mouse genomic sequence. A single Elk4 product (n = 10) was detected in adult mouse DRG using primers to exons 3 and 4 (Fig. 1D, lane 3), whereas primers to exons 2 and 4 detected the now much larger Elk4 together with the novel major products Elk4c and Elk4d, and the barely detected minor product Elk4e (Fig. 1D, lane 5; each product, n = 10). [These isoform designations were used to avoid potential confusion with the human isoforms SAP-1a and SAP-1b, recently termed ELK4 transcript variants ‘a’ and ‘b’.] As shown in Fig. 1E, the Elk4c isoform results from the deletion of ~75% of exon 3 (647/870 nt) which causes a frameshift, whereas the Elk4d isoform results from the in-frame skipping of the whole of exon 3 and so corresponds to the Elk3d isoform discussed above. Finally, the minor Elk4e isoform results from the in-frame deletion of ~90% of exon 3 (786/870 nt).
In summary, adult mouse DRG express the previously described Elk1, Elk3/Elk3b/Elk3c and Elk4 mRNAs, and the novel Elk1b, Elk3d and Elk4c/Elk4d/Elk4e mRNA isoforms. Transcripts corresponding to either the human ΔElk1 or ELK4/SAP-1 isoform SAP-1b were not detected, and each of the novel Elk1, Elk3 and Elk4 mRNA isoforms results from alternative splicing using canonical GT and AG dinucleotides at the 5′ and 3′ splice sites, respectively (Table S2).
The novel ELK mRNA isoforms are expressed in multiple tissues
The tissue distributions of mouse full-length Elk1 and Elk3 mRNAs vary between previous reports (Giovane et al., 1994; Lopez et al., 1994; Rao et al., 1989) but taken together they include expression in brain, heart, kidney and testis, as confirmed here, while the expression of full-length Elk4 mRNA in these tissues (Fig. S1) is similar to human (Price et al., 1995). In Fig. 2 we show that the novel Elk1b, Elk3d and Elk4c/Elk4d/Elk4e mRNA isoforms identified in DRG are also expressed in adult brain, heart, kidney and testis. The identity of each RT-PCR product was confirmed by sequencing and no products were detected in control reactions using RNA which had not been reverse transcribed. Therefore, unlike some ‘DRG-specific’ genes such as the sodium channel Nav1.8/SNS (Akopian et al., 1996), the novel ELK mRNA isoforms are expressed in each of the tested tissues.
Fig. 2.
The expression of novel ELK mRNA isoforms is not DRG-specific. (A) Expression of Elk1b (314 bp) detected by RT-PCR in adult brain (lane 2), heart (lane 3), kidney (lane 4) and testis (lane 5). (B) Expression of Elk3d (253 bp), and (C) expression of Elk4c (422 bp), Elk4d (199 bp) and Elk4e (283 bp) in the same tissue panel. In each case, primers were to exons 2 and 4, and lane 1 is a 1 kb DNA marker with fragment sizes indicated in bp.
The predicted proteins encoded by the novel ELK mRNA isoforms
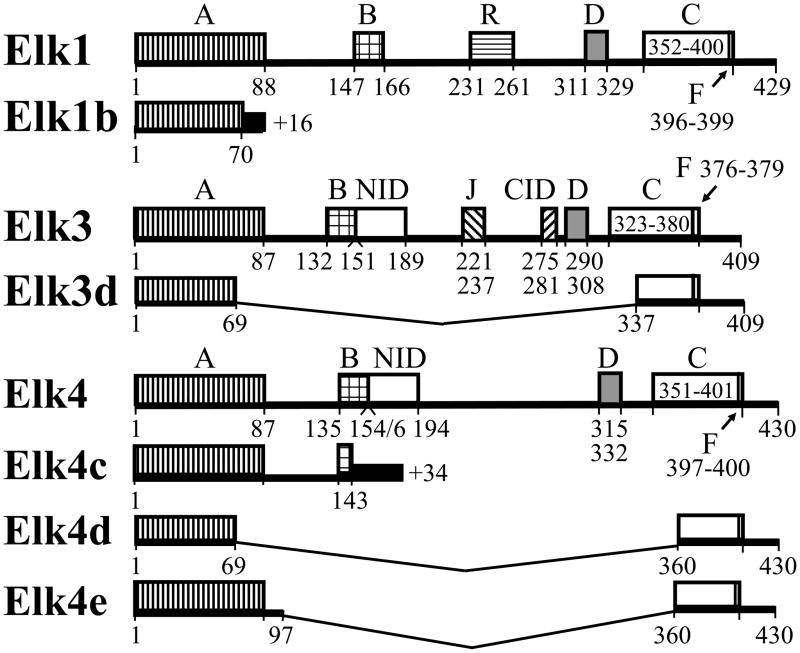
The Elk1, Elk3 and Elk4 proteins each include the five homologous domains A–D and F, as detailed in the legend of Figure 3. Elk1 protein has a total of six designated domains (Fig. 3; (Buchwalter et al., 2004; Yang et al., 2002)), whereas the frameshifted Elk1b mRNA encodes a predicted protein of 10.0 kDa with only a truncated N-terminal DNA-binding domain (70/88 residues of domain A) joined to 16 amino acids with no significant similarity to other mouse proteins.
Fig. 3.
The domain structures of the mouse Elk1, Elk3 and Elk4 novel protein isoforms. The Elk1, Elk3 and Elk4 proteins each include the five homologous domains A–D and F that correspond to the ETS DNA-binding domain (A); the serum response factor (SRF) interaction domain (B); the transactivation domain (C); a docking site for MAP kinases (domain D); and the FXFP motif docking site for MAP kinases (domain F). These are shown schematically along with the Elk1-specific repression domain (R); the Net Inhibitory Domain (NID); and the Elk3-specific CtBP Inhibition Domain (CID) and docking site for c-Jun N-terminal protein kinase (JNK; domain J) (Buchwalter et al., 2004). The predicted proteins encoded by the novel ELK mRNA isoforms are shown aligned to the full-length proteins, with unrelated sequences resulting from frameshifts indicated by solid black boxes. The previously described Elk3b (Net-b) and Elk3c (Net-c) proteins (Giovane et al., 1997) are not shown. Amino acid numbering follows reference protein sequences (NP_031948; NP_038536; NP_031949), with domain boundaries following previous reports (domains A, D, F and CID in Figs. 2 and 5 of (Buchwalter et al., 2004)); B and C (Dalton and Treisman, 1992; Giovane et al., 1994); R (Yang et al., 2002); NID (Maira et al., 1996; Stinson et al., 2003); and J (Ducret et al., 2000)).
By comparison to the eight designated domains of full-length Elk3 protein (Fig. 3; (Buchwalter et al., 2004)), the predicted Elk3d protein of 16.1 kDa includes a truncated ETS DNA-binding domain (69/87 residues of domain A) linked to most of the transcriptional activation domain (44/58 residues of domain C) including the FXFP motif docking site (domain F) that is specific for MAP kinases of the extracellular signal-regulated kinase (ERK) subfamily (Buchwalter et al., 2004; Giovane et al., 1994; Jacobs et al., 1999).
Elk4 protein has six designated domains (Fig. 3; (Buchwalter et al., 2004; Stinson et al., 2003)), whereas the predicted Elk4d protein of 15.8 kDa has a structure similar to that described above for Elk3d, i.e. a truncated ETS DNA-binding domain (69/87 residues) linked to most of the transcriptional activation domain (42/51 residues) including the FXFP motif docking site (Buchwalter et al., 2004; Dalton and Treisman, 1992; Jacobs et al., 1999). By comparison to Elk4d, the predicted Elk4e protein of 19.1 kDa includes an additional 28 amino acids (in common with full-length Elk4) that results in a complete ETS DNA-binding domain (Fig. 3). Finally, the frameshifted Elk4c mRNA encodes a predicted 20.3 kDa protein with an intact ETS DNA-binding domain, but with a truncated SRF interaction domain (9/20 residues of domain B; (Giovane et al., 1994)) joined to a sequence of 34 amino acids with no significant similarity to other mouse proteins. Therefore, the predicted Elk4c and Elk4e proteins have intact ETS DNA-binding domains, whereas this domain is truncated in the predicted Elk1b, Elk3d and Elk4d proteins.
Elk3 protein expression and differences in subcellular localization between isoforms
We focussed on the expression of Elk3 protein since we had shown the mRNA to be upregulated in DRG after peripheral nerve injury (see section below). An antiserum raised against the C-terminal 25 residues of Elk3 has been shown previously to detect a protein doublet of ~50 kDa and a band of >80 kDa in mouse NIH-3T3 fibroblasts transiently transfected with full-length mouse Elk3 cDNA (Maira et al., 1996), and in tissues should also detect the predicted Elk3d isoform of ~16 kDa but not the protein isoforms encoded by the frameshifted Elk3b or Elk3c mRNAs which do not contain this epitope (see Fig. 1B).
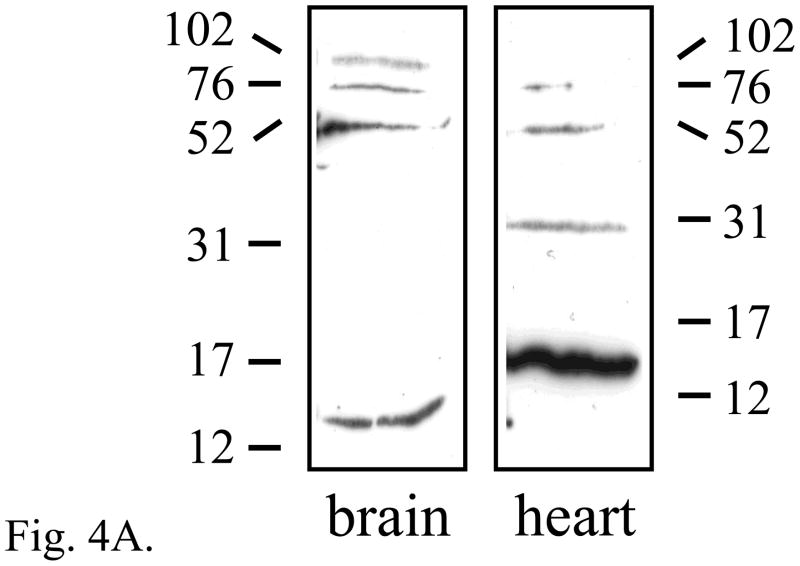
In Western blots of whole-cell tissue lysates from adult brain, the major immunoreactive proteins detected with affinity-purified antiserum had relative molecular weights (Mr) of approximately 14, 50, 75 and 95 kDa (Fig. 4A). The bands of ~14 and ~50 kDa are consistent, respectively, with the size of protein detected in COS-7 cells transfected with an Elk3d expression vector (data not shown) and with full-length Elk3 (Giovane et al., 1997), while the larger bands may correspond to the sumoylated conjugates of Elk3 detected in COS-7 cells transfected with both Elk3 and SUMO-1 cDNAs (Wasylyk et al., 2005). In adult heart, an additional band of ~29 kDa of unknown identity was also detected (Fig. 4A). Western blot analysis of Elk3 from DRG was not performed due to the small size of mouse ganglia, which would require the sacrifice and dissection of ~35 animals.
Fig. 4.
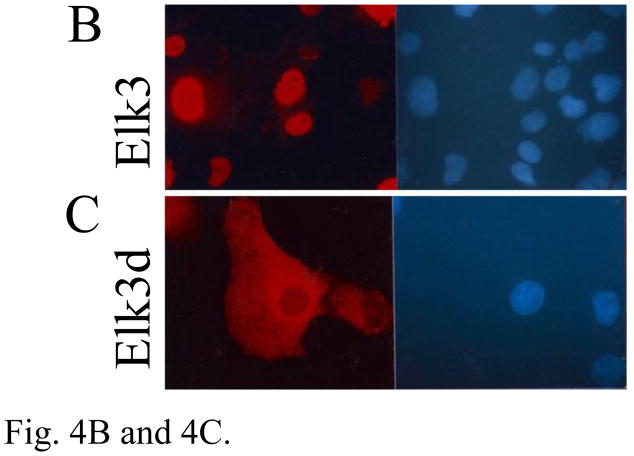
The expression of Elk3 proteins and differences in subcellular localization between isoforms. (A) Western blots showing expression of proteins of the expected sizes for Elk3d (~14 kDa) and full-length Elk3 (~50 kDa) in adult mouse brain and heart, together with a heart-specific band of unknown identity (~29 kDa). (B) COS-7 cells transfected with an Elk3 expression vector expressed Elk3 immunoreactivity (red, left panel) that co-localized with Hoeschst nuclear staining in the same field (blue, right panel). (C) COS-7 cells transfected with an Elk3d expression vector expressed Elk3d immunoreactivity that is predominantly cytoplasmic (red, left panel), as compared to Hoeschst nuclear staining in the same field (blue, right panel).
To address the possibility of differences in intracellular localization between Elk3 and Elk3d proteins, COS-7 cells were transiently transfected with the corresponding expression vectors. Elk3 was localized mainly in the nucleus with a minor detectable amount in the cytoplasm (Fig. 4B) whereas Elk3d was predominantly cytoplasmic (Fig. 4C). This cytoplasmic localization was observed in several cell types expressing different levels of Elk3d protein (data not shown). The affinity-purified antiserum specifically detected the introduced proteins, since there was no signal with the non-expressing cells in the transfected cell population (Fig. 4), with non-transfected cells or with pre-immune sera (data not shown). In summary, we provide evidence of Elk3 and Elk3d protein expression in brain and heart, and have shown that the two proteins appear to have different subcellular localizations.
Elk3d protein does not form complexes with DNA containing an ETS binding site
Unlike full-length Elk3 protein, the predicted Elk3d isoform has a truncated ETS DNA-binding domain (Fig. 3) and it was uncertain whether this would be sufficient for specific interaction with DNA containing an ETS binding site. [35S]methionine-labelled Elk3d, full-length Elk3 and control proteins were each synthesized in rabbit reticulocyte lysates, resolved by SDS-PAGE and quantified by PhosphorImager (Fig. 5A). Equimolar amounts of the in vitro translated proteins were analyzed by mobility shift assay with the oligonucleotide probe PEA3* that contains a strong binding site for ETS proteins. Elk3d did not form a detectable DNA complex, in contrast to a number of ETS proteins with a range of affinities, such as full-length Elk3, the c-Ets1 DNA-binding domain (N70), full-length c-Ets1 and v-Ets (Fig. 5B; (Giovane et al., 1994; Wasylyk and Wasylyk, 1993)). Binding of Elk3d could not be detected in a wide range of conditions, including different times of incubation, with various non-specific inhibitors (poly dA-dT, poly dI-dC, salmon sperm DNA or no competitor) and with various probe sequences (data not shown). These results suggest that the 18 amino acids of the ETS domain that are deleted in Elk3d are critical for DNA-binding.
Fig. 5.
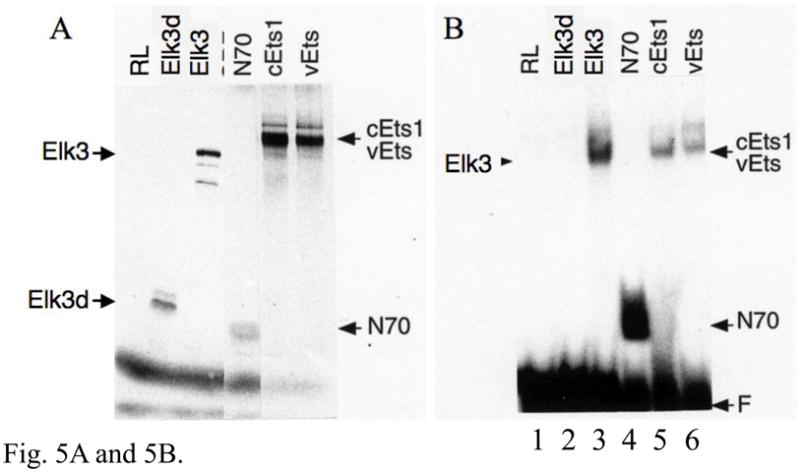
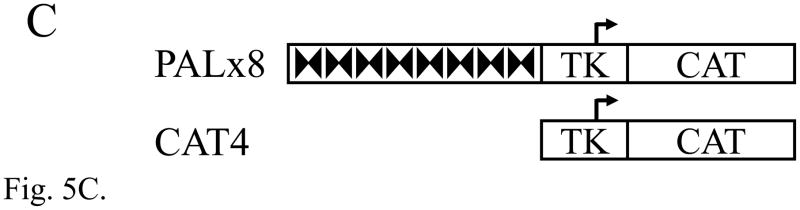
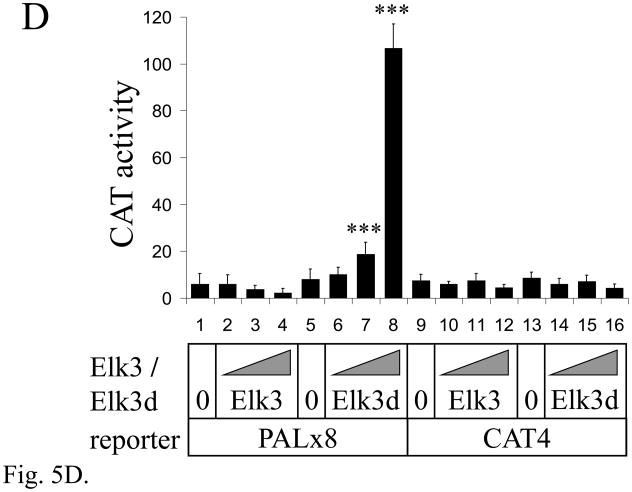
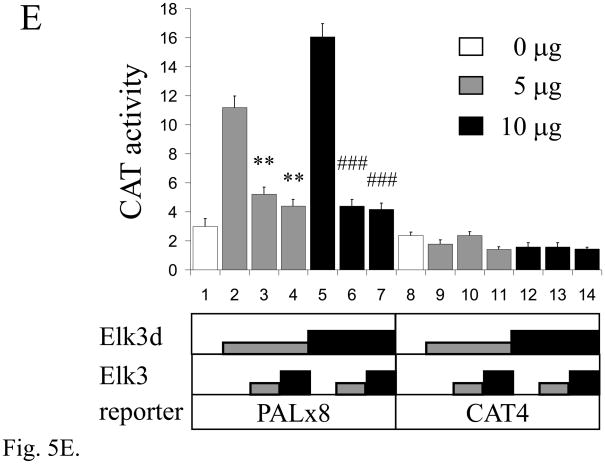
The Elk3d protein does not form complexes with DNA containing ETS binding sites, but can transactivate transcription through these sites. (A) ETS-family proteins labelled by in vitro synthesis in reticulocyte lysates with [35S]-methionine. Arrowheads indicate the proteins of expected size; RL, reticulocyte lysate incubated without added RNA; and N70, the c-Ets1 DNA-binding domain (Wasylyk and Wasylyk, 1993). (B) Mobility-shift assay of ETS-family proteins with [32P]-labelled PEA3* probe that contains a strong ETS binding site. Equimolar amounts of Elk3d (lane 2), full-length Elk3 (lane 3), c-Ets1 DNA-binding domain (N70, lane 4), full-length c-Ets1 (lane 5), v-Ets (lane 6) or an equal volume of mock-incubated reticulocyte lysate (lane 1) were analyzed, and note that Elk3d does not form a complex with PEA3* probe. Specific complexes or excess free probe (F) are indicated by arrowheads. (C) Schematic diagram of the transcriptional reporter PALx8-TK-CAT4 (PALx8), which contains eight tandem repeats of the stromelysin-1 palindromic ETS binding site upstream from a thymidine kinase (TK) promoter and CAT coding sequences of pBL-CAT4 (CAT4). (D) Elk3d activates transcription. The transfections contained the reporters PALx8 (bars 1–8) or CAT4 (bars 9–16) and the expression vectors pTL2-Net(Elk3) (1 μg, bars 2 and 10; 5 μg, bars 3 and 11; 10 μg, bars 4 and 12), pTL2-Elk3d (1 μg, bars 6 and 14; 5 μg, bars 7 and 15; 10 μg, bars 8 and 16) or control pSG5 (5 μg, bars 1, 5, 9 and 13). CAT activity is expressed in arbitrary units. Data are shown as means ± S.E.; *** P<0.001 t-test versus control (bars 1, 5). (E) Transcriptional activation by Elk3d is antagonized by Elk3. Transfections contained the reporters PALx8 (bars 1–7) or CAT4 (bars 8–14) and the expression vectors pTL2-Elk3d (5 μg, bars 2–4, 9–11; 10 μg, bars 5–7, 12–14), pTL2-Net(Elk3) (5 μg, bars 3, 6, 10, 13; 10 μg, bars 4, 7, 11, 14) or control pSG5 (bars 1,8). CAT activity is expressed in arbitrary units. Data are shown as means ± S.E.; ** P<0.005 t-test versus no Elk3 addition (bar 2); # # # P<0.001 t-test versus no Elk3 addition (bar 5).
Elk3d activates transcription through ETS binding sites, which is antagonized by Elk3
Full-length Elk3 protein has previously been shown to inhibit transcription under basal (low-serum) conditions (Giovane et al., 1994; Maira et al., 1996). By comparison to full-length Elk3 protein, the Elk3d isoform still includes most of the transcriptional activation domain, but lacks the transcriptional inhibitory domains NID and CID (Fig. 3). The transcriptional properties of Elk3d were investigated in transfection assays of NIH-3T3 cells using the PALx8 reporter that contains 8 tandem repeats of a palindromic ETS binding site (Fig. 5C; (Maira et al., 1996; Wasylyk et al., 1991)). This element is inactive in NIH-3T3 cells in the absence of introduced ETS proteins (Fig. 5D, bars 1,5 vs. 9,13). Surprisingly, Elk3d specifically activated the PALx8 reporter (Fig. 5D, bars 6–8) whereas full-length Elk3 specifically inhibited the reporter (Fig. 5D, bars 2–4). The Elk3d and Elk3 proteins were expressed at similar levels in NIH-3T3 cells, as detected by Western blotting (data not shown), and Elk3d did not bind to the ETS binding sites of the PALx8 reporter in contrast to full-length Elk3 (data not shown and (Giovane et al., 1994)). In addition, the transcriptional activation by Elk3d was inhibited by co-expression of full-length Elk3 (Fig. 5E, bars 2–7). These results suggest that Elk3d activates transcription through ETS binding sites by an indirect mechanism (see Discussion), and demonstrate that alternative splicing of Elk3 mRNA produces different transcriptional regulators that have opposing activities.
Multiple Pctaire2 mRNAs are expressed that are natural antisense transcripts to Elk3 mRNAs
We next focussed on the potential regulation of Elk3 at the mRNA level. The Elk3 gene is located next to but on the opposite DNA strand from the Pctk2 gene that encodes Pctaire2, a Cdc2-related serine/threonine protein kinase in which the amino acid sequence PCTAIRE corresponds to the conserved PSTAIRE motif of the Cdc2 catalytic domain ((Meyerson et al., 1992); genomic sequence NT_039500). Intriguingly, a bioinformatic study identified two Pctaire2 cDNA sequences from mouse retina with a different 3′-untranslated region (3′-UTR) sequence which overlaps the Elk3 transcription unit on the opposite DNA strand ((Katayama et al., 2005); BC031778; BC049904). To determine whether Elk3 and Pctaire2 mRNAs are co-expressed in tissues, and so could potentially form sense-antisense heteroduplex mRNA, primers were designed to amplify each of the two potential Pctaire2 mRNA isoforms. These are referred to here as isoform Pctaire2a and Pctaire2b, that respectively either do not include or do include Elk3-antisense sequence.
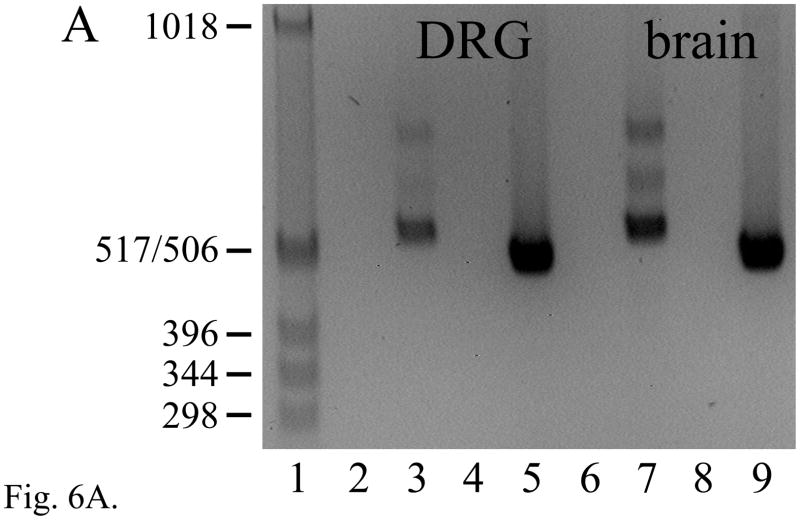
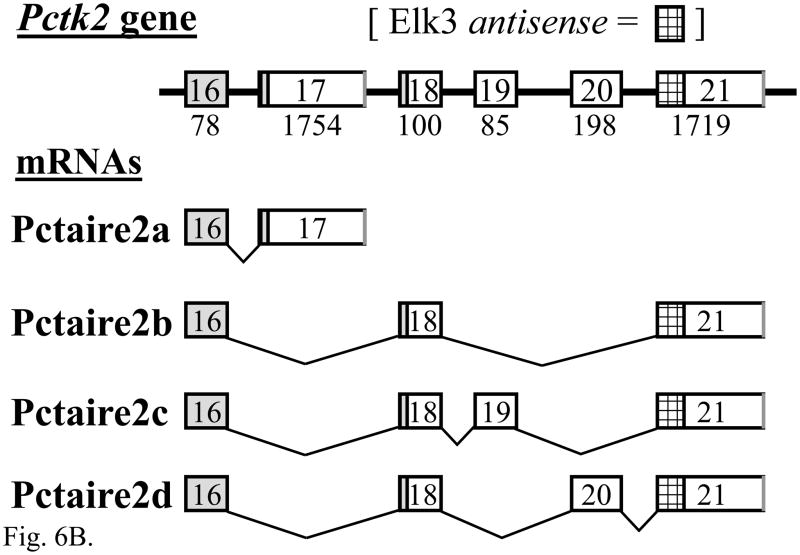
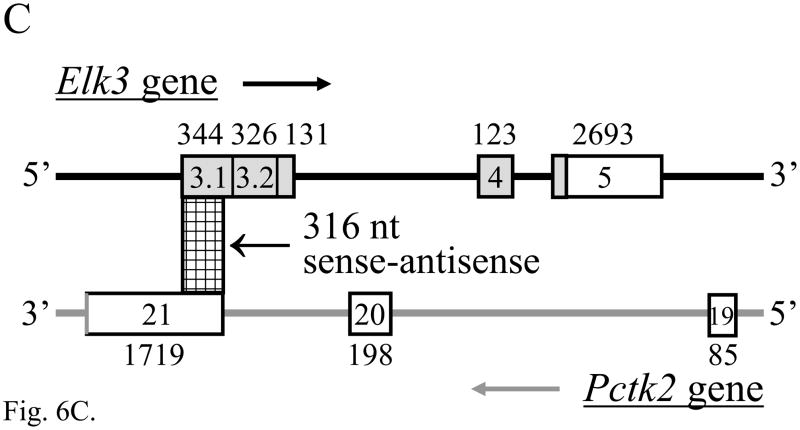
RT-PCR products of the expected size and sequence for both the Pctaire2a (501 bp) and Pctaire2b (547 bp) isoforms were expressed in adult mouse DRG (each n = 10) and brain (each n = 6) (Fig. 6A). The alternative splicing of Pctaire2 mRNA isoforms containing Elk3-antisense sequence proved to be complex, as two novel products Pctaire2c (632 bp) and Pctaire 2d (745 bp) were also detected in brain (each n = 6) and, more faintly, in DRG (each n = 10) (Fig. 6A, lanes 3 and 7). These result from the use of two newly recognized cassette exons, and each of the four isoforms result from alternative splicing using canonical GT and AG dinucleotides (Fig. 6B; Table S3). The Pctaire2b, Pctaire2c and Pctaire2d mRNA isoforms each include sequence antisense to 316 nucleotides of Elk3 mRNA transcribed from within the exon 3.1 portion of exon 3 (Fig. 6C; nt sequence alignment in Fig. S2), and therefore would be expected to affect expression of Elk3 (see Discussion).
Fig. 6.
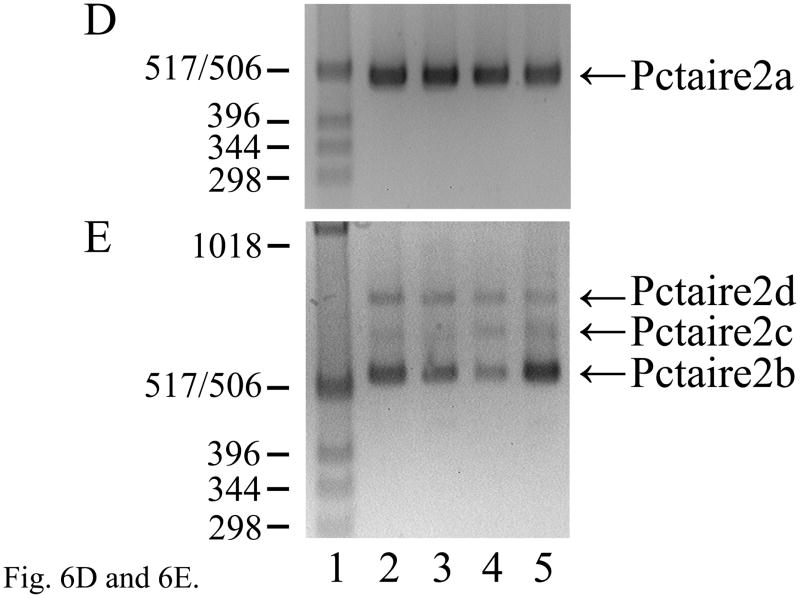
The expression of multiple Pctaire2 mRNA isoforms that are natural antisense transcripts to Elk3 mRNAs. (A) RT-PCR analysis showing expression of Pctaire2b (547 bp), Pctaire2c (632 bp) and Pctaire2d (745 bp) mRNA isoforms in adult DRG (lane 3) and brain (lane 7), together with the expression of Pctaire2a (501 bp) in DRG (lane 5) and brain (lane 9). Lane designations follow Fig. 1 legend, with marker fragment sizes of 1018 - 298 bp. (B) Schematic diagram of the Pctk2 gene (3′ end) and the transcribed Pctaire2 mRNA isoforms. Exon numbering to 17 follows the reference RNA sequence (NM_146239.2) with downstream exons designated here as 18 – 21, and coding regions are in gray. Note that the Pctaire2b, Pctaire2c and Pctaire2d mRNA isoforms each include sequence that is antisense to 316 nucleotides of Elk3 mRNA (cross-hatched). (C) Schematic overview of the Elk3 and Pctk2 gene organizations in the region of exon overlap (cross-hatched) responsible for potential Elk3/Pctaire2 sense-antisense mRNA hybrids. Arrows indicate the general direction of gene transcription, with exons numbered (Fig. 6B and (Giovane et al., 1997)) and sized in nt, and coding regions in gray. (D) and (E) Expression of Pctaire2a (D; 501 bp) and Pctaire2b/Pctaire2c/Pctaire2d (E; respectively, 547, 632 and 745 bp) mRNA isoforms detected by RT-PCR in adult brain (positive-control, lane 2), heart (lane 3), kidney (lane 4) and testis (lane 5), with marker fragment sizes indicated in bp (lane 1).
Each of the four different Pctaire2 mRNA isoforms are also expressed in mouse heart, kidney and testis (Fig. 6D and E, lanes 3–5), as confirmed for Pctaire2a and Pctaire2b by DNA sequencing. By comparison to the predicted Pctaire2a protein sequence, the other three mRNA isoforms encode a common protein in which the C-terminal 12 amino acids are replaced by the sequence ALS encoded by exon 18 (Fig. 6B). These results demonstrate that Elk3 and Pctaire2 mRNAs are co-expressed and subject to alternative mRNA splicing in multiple tissues, with the potential for sense-antisense heteroduplex formation between Elk3/Elk3b/Elk3c and Pctaire2b/Pctaire2c/Pctaire2d mRNA isoforms.
Multiple Mfsd4 mRNAs are expressed that are natural antisense transcripts to Elk4 mRNAs
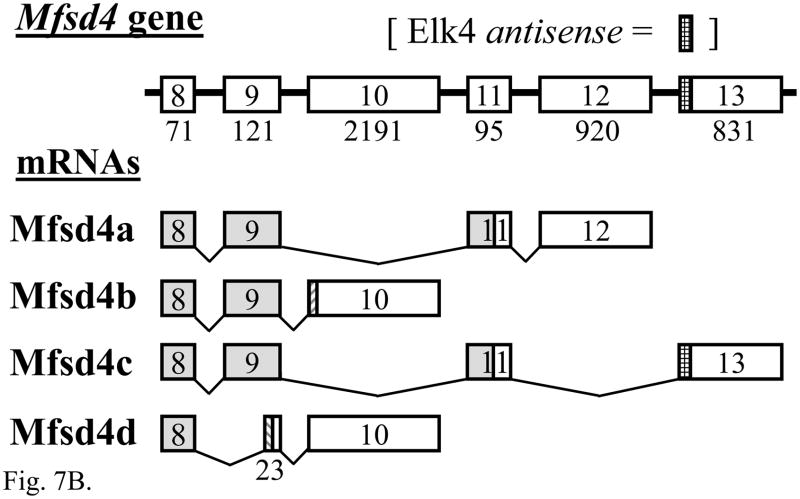
Searches of a bioinformatic sense-antisense database (SADB) did not reveal a natural antisense transcript to Elk1 mRNA, whereas the Elk4 gene is overlapped by the Mfsd4 (Major facilitator superfamily domain containing 4) transcription unit on the opposite DNA strand. Little information is available concerning Mfsd4, but there are two alternatively spliced reference RNA sequences (isoforms ‘a’ and ‘b’) plus a single database cDNA sequence from mouse normal colon with a different 3′-UTR (BC058790) that we will refer to as isoform ‘c’. It is part of this different 3′-UTR that includes sequence antisense to Elk4 mRNA.
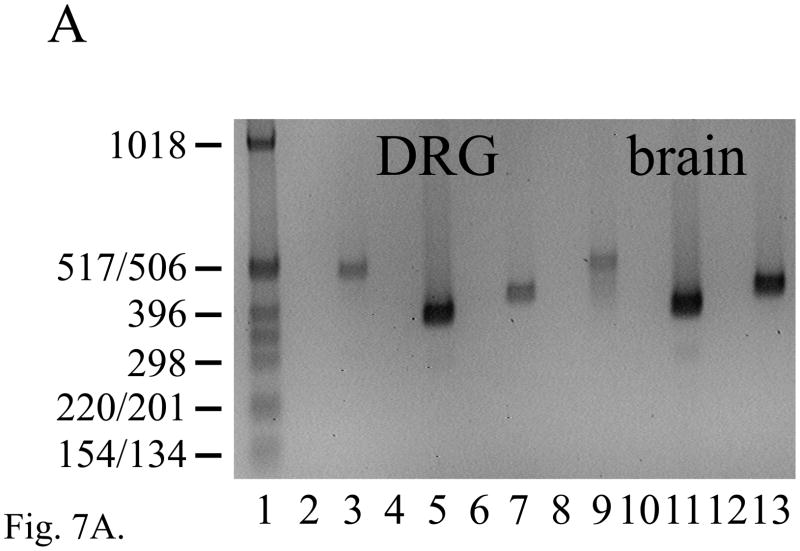
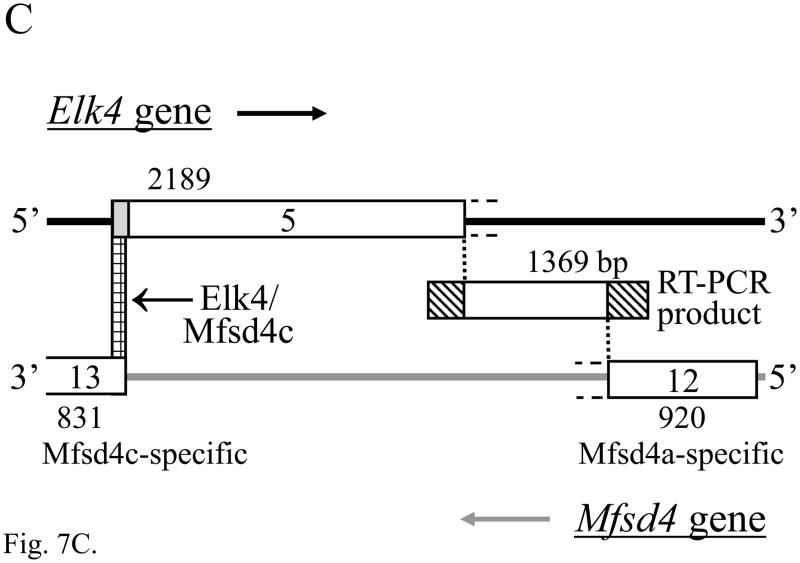
RT-PCR products of the expected sizes and sequences for mRNA isoforms Mfsd4a, Mfsd4b and Mfsd4c were each expressed in adult mouse DRG (Fig. 7A, lanes 3, 5 and 7; each n = 10) and brain (Fig. 7A, lanes 9, 11 and 13; each n = 6). A barely detected product of 288 bp in both DRG and brain (Fig. 7A, lanes 5 and 11) that results from the skipping of most of exon 9 (Fig. 7B) is here designated as Mfsd4d, and each of the Mfsd4 isoforms result from alternative splicing using canonical GT and AG dinucleotides (Table S4). The expression of both Elk4 and Mfsd4c mRNAs in DRG and brain predicts the potential for Elk4/Mfsd4c sense-antisense interactions, between 84 nucleotides of Elk4 mRNA coding sequence and the Mfsd4c mRNA 3′-UTR (Fig. 7C; nt sequence alignment in Fig. S3). The Mfsd4c isoform is also expressed in heart and kidney, though in testis a smaller, unrelated product was amplified (data not shown).
Fig. 7.
The expression of multiple Mfsd4 mRNA isoforms that are natural antisense transcript to Elk4 mRNAs. (A) RT-PCR analysis showing the expression of Mfsd4a (511 bp), Mfsd4b (386 bp) and Mfsd4c (432 bp) mRNA isoforms in adult DRG (lanes 3, 5 and 7, respectively) and brain (lanes 9, 11 and 13, respectively). The novel Mfsd4d isoform (288 bp) was barely detected in DRG and brain (lanes 5 and 11, respectively). Lane designations follow Fig. 1 legend, with marker fragment sizes indicated in bp. (B) Schematic diagram of the Mfsd4 gene (3′ end) and the transcribed mRNA isoforms. Exon numbering to 12 follows the reference RNA sequences (NM_172510, NM_001114662) with a different cDNA 3′-UTR sequence (BC058790) designated as exon 13. Coding regions are in gray, with those resulting from frameshifts relative to Mfsd4a shown in diagonal bars. Note that the Mfsd4c mRNA isoform includes sequence that is antisense to 84 nt of Elk4 mRNA (cross-hatched). (C) Schematic overview of the Elk4 and Mfsd4 gene organizations in the region of exon overlaps. Details are as Fig. 6C, with the Elk4 terminal exon 5 of 2132 nt (NM_007923) incorporating a cDNA-derived extension of 57 nt (AA472317), and indicated Elk4/Mfsd4c exon overlap (cross-hatched) and incomplete 3′-end exon sequences (dashed extensions). The relative position of a 1369 bp RT-PCR product is shown, including sequences overlapping either Elk4 exon 5 (221 nt) or Mfsd4 exon 12 (252 nt; diagonal bars).
Other Mfsd4 isoforms could also be involved in sense-antisense interactions with the currently uncloned portion of the Elk4 mRNA 3′-UTR (Fig. 7C), as only 3.37 of the > 9.5 kilobase (kb) Elk4 transcript had been cloned (NM_007923; (Giovane et al., 1994)). Using RT-PCR we cloned a 1369 bp product from DRG and heart that overlaps the available Elk4 and Mfsd4a mRNA sequences by > 220 nt (Figs. 7C and S4; data not shown). Therefore, Elk4 and Mfsd4 mRNAs are co-expressed in multiple tissues, resulting in the potential for sense-antisense interactions between the Mfsd4a or Mfsd4c mRNA isoforms and different sites common to each of the four Elk4 transcripts (Figs. 7C and 1E).
The Mfsd4a and Mfsd4c mRNA isoforms encode the same predicted protein with a Major Facilitator Superfamily domain (MFS; (Finn et al., 2008)) and twelve predicted transmembrane segments ((Moller et al., 2001; Siintola et al., 2007); Fig. S5). MFS proteins transport small solutes by using chemiosmotic ion gradients (Pao et al., 1998), and although the substrate-specificity of Mfsd4 is not yet known, it is most closely related to sodium-dependent glucose transporter 1 (NP_001106856; rat NaGLT1 (Horiba et al., 2003)).
The effect of peripheral nerve injury on ELK and Pctaire2 mRNA expression
In preliminary experiments using oligonucleotide microarrays we had noted an upregulation of Elk3 mRNA by 2.66- or 1.56-fold in mouse lumbar DRG seven days after peripheral nerve transection (axotomy) of the sciatic nerve. The sequences of both oligonucleotide clusters on the microarrays were antisense to the Elk3 mRNA 3′-UTR, and therefore could detect each of the four isoforms. In contrast, the Elk1 hybridization signals were low and Elk4 was not present on the microarrays (Affymetrix mouse 430 2.0 array).
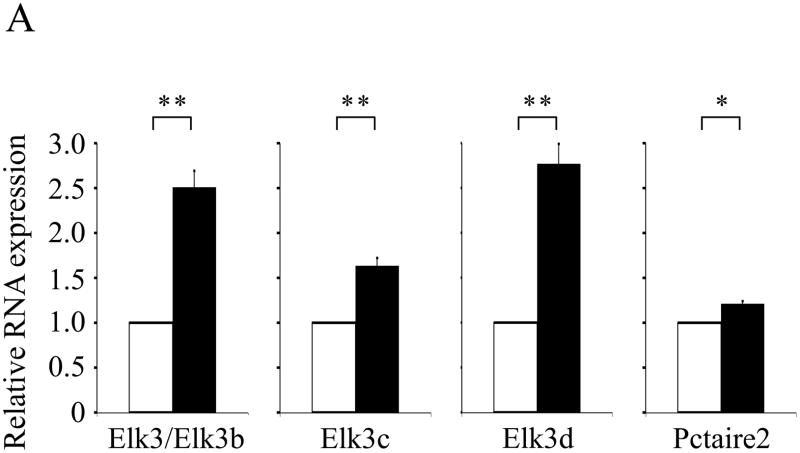
In order to validate and extend these results, real-time quantitative RT-PCR assays were designed to discriminate specifically between the Elk3/Elk3b, Elk3c and Elk3d mRNA isoforms (see Experimental methods). Expression levels were normalized to GAPDH (glyceraldehyde 3-phosphate dehydrogenase) mRNA, which is unchanged following peripheral nerve injury ((Kerr et al., 2008) and references therein). Seven days after axotomy, the expression of Elk3/Elk3b mRNAs increased by 2.51 - fold (P<0.005), Elk3c mRNA increased by 1.63 - fold (P<0.005) and Elk3d mRNA increased by 2.77 - fold (P<0.005) in injured compared to uninjured lumbar L4 and L5 DRG (Fig. 8). In the Elk3/Elk3b and Elk3d assays, the threshold cycle values (Ct) at which fluorescence exceeded the threshold limit were respectively 27.55 and 31.84 in uninjured DRG, whereas the less highly expressed Elk3c mRNA (Fig. 1C; (Giovane et al., 1997)) had a Ct value of 35.24 which approaches the sensitivity limits of the detection system (Applied Biosystems Application Note, 127AP05-03 available at http://www.appliedbiosystems.com).
Fig. 8.
Induction of Elk3 and Pctaire2 mRNA isoforms in DRG following peripheral nerve injury. In quantitative RT-PCR assays the expression of Elk3/Elk3b mRNAs increased to 2.514 ± 0.178 of control (n = 4; P=0.0034), Elk3c mRNA increased to 1.634 ± 0.081 of control (n = 4; P=0.0044), Elk3d mRNA increased to 2.767 ± 0.226 of control (n = 4; P=0.0043) and Pctaire2b/Pctaire2c/Pctaire2d mRNAs that include sequence antisense to Elk3 increased to 1.212 ± 0.031 of control (n = 4; P=0.0063) in pooled ipsilateral (axotomized) lumbar L4 and L5 DRG compared with contralateral (unaxotomized) controls seven days after axotomy. Data are shown as means ± S.E., in which expression after axotomy (filled boxes) was compared with contralateral controls of 1.00 relative units (unfilled boxes), with statistical significance of differences judged by one sample t-test (** P<0.005; * P<0.01).
Three additional quantitative RT-PCR assays were designed to detect (i) all three Pctaire2 mRNA isoforms that include sequence antisense to Elk3 mRNA (Pctaire2b/Pctaire2c/Pctaire2d); (ii) both the full-length Elk1 and Elk1b mRNA isoforms; and (iii) all four Elk4 mRNA isoforms. There was a small but significant 21 % increase in Pctaire2b/Pctaire2c/Pctaire2d mRNA expression after axotomy (P<0.01; Fig. 8), whereas the expressions of Elk1 and Elk4 mRNA isoforms were unchanged compared to uninjured DRG (respectively, 1.032 ± 0.037 and 1.082 ± 0.036 of control; each n = 4, P>0.05). In summary, the Elk3/Elk3b, Elk3c and Elk3d mRNAs increased by an average of 2.31-fold after axotomy and there was a small increase in Pctaire2b/Pctaire2c/Pctaire2d mRNAs, indicating that they are part of the in vivo response to peripheral nerve injury in DRG, unlike either Elk1 or Elk4 mRNAs.
Discussion
This study demonstrates that adult mouse DRG and other tested tissues express the previously described Elk1, Elk3/Elk3b/Elk3c and Elk4 mRNAs, together with the novel alternatively spliced isoforms Elk1b, Elk3d and Elk4c/Elk4d/Elk4e. The novel isoforms could not have been detected in previous RT-PCR analyses, due to the relative positions of primers used (Galang et al., 2004; Giovane et al., 1997; Hollenhorst et al., 2004; Kobberup et al., 2007; Sahin et al., 2009), and are not conserved between different ELK factors except for the in-frame skipping of the whole of exon 3 in Elk3d and Elk4d mRNAs (Fig. 1B and E). Among the five novel isoforms, only the Elk1b transcript is a potential substrate for nonsense-mediated mRNA decay (NMD), due to a frameshift and resulting premature stop codon > 50–55 nt upstream from an exon-exon junction, though NMD may be exploited as a form of post-transcriptional regulation (McGlincy and Smith, 2008). Currently, no rat or human sequences corresponding to the novel isoforms are present in mRNA databases, though the full extent of alternative splicing is far from being fully characterized, as shown by the recently isolated ETS-family isoforms of Gabp-α (FN545817) and ETS1 (Laitem et al., 2009).
Alternative splicing of ELK mRNAs results in multiple predicted proteins with different domain structures (Fig. 3; (Giovane et al., 1997)). COS-7 cells transfected with Elk3 resulted in localization predominantly to the nucleus, whereas Elk3d was predominantly cytoplasmic (Fig. 4). By comparison to Elk3, the Elk3d isoform includes the nuclear export sequence located within the ETS domain, but lacks the two characterized nuclear localization signals and the domain J that is necessary for nuclear export in response to stress signals (Ducret et al., 1999; Ducret et al., 2000). As Elk3d protein has no unique epitope, it will be of interest in the future to characterize which classes of DRG neurons express Elk3 or Elk3d mRNAs by single-cell RT-PCR.
The ETS DNA-binding domain is composed of three α-helices and a four-stranded antiparallel β-sheet, arranged in the order α1-β1-β2-α2-α3-β3-β4, which form a winged helix-turn-helix (wHTH) topology. All amino acids that make base-specific contacts occur within the α3 DNA-recognition helix, while backbone contacts to opposite DNA strands are made by the loop between the β3 and β4 strands (the ‘wing’) and by the α2 helix and proceeding loop (the ‘turn’ of HTH) (Mo et al., 1998; Mo et al., 2000). Similar secondary structures are predicted for five examples of the ETS-family using the programme PSIPRED (Elk1, Elk3, Elk4, Erg and Ets1; (Bryson et al., 2005)). In contrast, Elk3d lacks predicted structures corresponding to β3 and β4 strands, and this lack of a β3-loop-β4 winged segment is most likely responsible for Elk3d not binding to the oligonucleotide probe PEA3* (Fig. 5B). Of the other novel isoforms, Elk1b and Elk4d lack predicted structures corresponding to a β4 strand (Bryson et al., 2005), whereas Elk4c and Elk4e have intact ETS DNA-binding domains (Fig. 3). A truncation mutant of Elk4 equivalent to isoform Elk4c has been shown to bind specifically to ETS binding sites (Treisman et al., 1992), but the lack of a transactivation domain suggests that Elk4c may act as dominant negative inhibitor of activation to potentially interfere with multiple ETS-family factors (Giovane et al., 1997; Hever et al., 2003; Laitem et al., 2009).
Elk3, Elk3b and Elk3c have previously been shown to inhibit transcription from the PALx8 reporter when tested in NIH-3T3 cells under basal (low-serum) conditions (Giovane et al., 1994; Giovane et al., 1997; Maira et al., 1996). In contrast, human ELK1 activated transcription (Giovane et al., 1994; Maira et al., 1996) and ELK4 was essentially inactive (Maira et al., 1996). Here we show that despite not binding directly to the PALx8 reporter, Elk3d activated transcription which was inhibited by co-expression of Elk3 (Fig. 5D and E). The cytoplasmic localization of Elk3d argues for the activation resulting from signalling downstream of cytosolic Elk3d protein-protein interactions, with the inhibition of Elk3d-mediated transactivation by Elk3 being reminiscent of its inhibition of Ets1-mediated transactivation (Giovane et al., 1994). However, we cannot exclude the possibility that Elk3d is being recruited indirectly to the ETS binding sites by endogenous ETS proteins expressed in NIH-3T3 cells (Hever et al., 2003; Maira et al., 1996; O’Leary et al., 2005; Rodrigo et al., 1999). ELK1 proteins are known to interact via their transactivation domain (Gille et al., 1996), with amino acids 375–399 being sufficient for this interaction (Yang et al., 1999), and the corresponding Elk3 amino acids 351–389 are present in the Elk3d isoform (Fig. 3).
The truncated transactivation domain of Elk3d retains four of the five C-terminal Ser/Thr-Pro motifs of Elk3 that are phosphorylated by MAP kinases (Thr337, Ser359, Ser365 and Ser398; (Buchwalter et al., 2004)), the conserved Phe354Trp355 critical for transactivation (Price et al., 1995) and the FXFP motif (Fig. 3). Among vertebrate ETS proteins, only the ELK subfamily include a conserved FXFP motif that is a docking site for activated but not unactivated MAP kinases of the ERK subfamily (Jacobs et al., 1999; Lee et al., 2004b; Zhang et al., 2008). The FXFP motif of ELK1 is critical for the phosphorylation by ERK2 of the nearby Ser383/Ser389 (Fantz et al., 2001; Jacobs et al., 1999; Zhang et al., 2008) which are required for transactivation (Price et al., 1995), and phosphorylation of the corresponding Ser359/Ser365 of Elk3 (Ducret et al., 2000) and Ser381/Ser387 of ELK4 (Price et al., 1995) have been shown to be particularly important for efficient transactivation. ERK substrates can be localized to the nucleus or cytoplasm (Yoon and Seger, 2006), and Elk3d protein is a putative substrate for ERK via the FXFP motif, which can function independently of the domain D docking site for MAP kinases (Fantz et al., 2001; Jacobs et al., 1999; Zhang et al., 2008) that is not present in Elk3d (Fig. 3). The FXFP and four C-terminal Ser/Thr-Pro motifs are also retained in the novel Elk4d and Elk4e proteins (Fig. 3; NP_031949).
The overlapping gene pairs Elk3/Pctk2 and Elk4/Mfsd4 are each co-expressed in the tested tissues, resulting in potential Elk3/Pctaire2 and Elk4/Mfsd4 sense-antisense mRNA pairs (Figs. 6 and 7). [Note that the distinction between which pair member is the sense or antisense transcript is arbitrary when both encode proteins (Beiter et al., 2009).] It will be of interest to determine whether these sense-antisense (S/AS) pairs are conserved in human, especially as a very short (focal) region of genome amplification in human chromosome 12q23.1 that only contains the genes ELK3 and PCTK2 occurs at a significantly high frequency among 3131 cancer specimens ((Beroukhim et al., 2010); http://www.broadinstitute.org/tumorscape). A large proportion of S/AS pairs are co-expressed (Ge et al., 2008; Katayama et al., 2005; Sun et al., 2005) and bioinformatic studies have found many examples of exon-overlapping S/AS pairs conserved between mouse and human (Numata et al., 2007; Numata et al., 2009; Okada et al., 2008). This suggests that gene expression can be regulated by the formation of S/AS perfectly complementary double-stranded RNA (dsRNA; (Numata et al., 2007; Sun et al., 2005; Werner et al., 2009)), the latter being detectable by RNase protection assays (Munroe and Zhu, 2006). S/AS pairing can result in RNA masking of regulatory elements within either transcript, so inhibiting interaction with trans-acting factors (Beiter et al., 2009; Munroe and Zhu, 2006), and there is evidence of the dsRNA region being processed into discrete, strand-specific small RNAs of 17–30, 60 or 80 nucleotides (Carlile et al., 2008; Okada et al., 2008; Watanabe et al., 2008). Such endogenous small interfering RNAs (endo-siRNAs) have been implicated in RNA interference (Carlile et al., 2008; Watanabe et al., 2008; Werner et al., 2009). Endo-siRNAs from mouse oocytes have been deep-sequenced, and when mapped on the genome have localized to a number of clusters (‘small RNA clusters’), 17 of which were predicted to result from S/AS pairing (Watanabe et al., 2008). Here we show that an additional small RNA cluster includes at least 11 endo-siRNAs corresponding to Elk4 mRNA exon 5 sequence, to which the Mfsd4a and Mfsd4c antisense mRNAs are complementary (Table S5; Fig. 7C; (Watanabe et al., 2008)).
There is no evidence from sense-antisense databases (SADB, NATsDB) of Elk1 mRNAs overlapping another gene. On the mouse X chromosome the nearest gene located in an antisense orientation is Timp1 (tissue inhibitor of metalloproteinase 1), which is a nested gene located wholly within DNA complementary to intron 5 of the host gene Syn1 (synapsin I; (Derry and Barnard, 1992; Gibson et al., 2005)), and exon-overlapping S/AS mRNA pairs are significantly under-represented on the mouse and human X chromosomes (Katayama et al., 2005; Werner et al., 2009).
Evidence for the differential induction or repression of individual ELK subfamily members has been scarce. At the protein level, Elk3 has been shown to be downregulated by hypoxia, unlike either Elk1 or Elk4, with mRNA levels being essentially unchanged (Gross et al., 2007). Here we detected differential regulation at the mRNA level, with the induction of Elk3 mRNA isoforms (Fig. 8), but not Elk1 or Elk4, in adult mouse DRG seven days after peripheral axotomy. Following such injury the altered gene expression within the cell bodies of the DRG is thought to promote neuronal survival and axonal regeneration (Navarro et al., 2007), though which transcription factors co-ordinate this response is not well understood. c-JUN and ATF3 are known to be upregulated and promote axon regeneration (c-Jun) and neurite outgrowth (ATF3; (Navarro et al., 2007)), whereas STAT3 is phosphorylated and activated (Lee et al., 2004a; Navarro et al., 2007) and increases motoneuron survival (Schweizer et al., 2002). In addition, using oligonucleotide arrays the DRG mRNAs with a > 2-fold change after sciatic nerve axotomy included the transcription factors Sox11 (Additional File 2 of (Costigan et al., 2002); AA030427 and W97408 of (Bonilla et al., 2002); (Tanabe et al., 2003)) and CREM (Costigan et al., 2002), and the putative transcriptional modulator ANKRD1/CARP (U50736 of (Costigan et al., 2002); (Stam et al., 2007)), while the transcription factor Ddit3/Gadd153 was identified by serial analysis of gene expression (SAGE; (Mechaly et al., 2006)). The inductions of mouse Sox11 and Ddit3/Gadd153 mRNAs have been validated (Mechaly et al., 2006; Tanabe et al., 2003) and RNAi-mediated knockdown of Sox11 decreases neurite growth (Jankowski et al., 2006). It will now be of interest to extend our findings by characterizing whether the Elk3 isoforms that are expressed in DRG can differentially affect neuronal survival and/or regeneration after nerve injury, especially given the potential for their interaction with Erk1/2 which is activated in DRG cell bodies seven days after axotomy (Obata et al., 2003).
Experimental methods
Animals, surgery and tissue collection
All animals were fed standard chow and water ad libitum, and animal care procedures were carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines. All reasonable efforts were made to minimize animal suffering and to use the minimum number of animals necessary to perform statistically valid analyses. Lumbar DRG and other tissues from adult (10–12 week old) male 129/OlaHsd mice (Bristol University colony) were frozen on dry ice and stored at −80 °C. For studies on peripheral nerve transection (axotomy), the right sciatic nerve of 10–14 week old male 129/OlaHsd mice were transected at the mid-thigh level (Kerr et al., 2004), prior to killing 7 days later by cervical dislocation to obtain ipsilateral (axotomized) and contralateral (control) lumbar L4 and L5 DRG pools each from eleven animals.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA isolation, DNase treatment and re-extraction, and RT reactions were as published (Kerr et al., 2004). Partial-length cDNAs were each amplified by RT-PCR using 5 μl RT reaction (100 ng of total RNA equivalent), HPLC-purified primers (sequences detailed below) and hot start DNA polymerase that included a proofreading enzyme (FastStart Taq DNA polymerase, Roche) under the following conditions: 94 °C for 5 min, and 40 cycles of 94 °C, 30 sec; 66 °C, 45 sec; 72 °C, 45 sec; with a final 72 °C for 10 min (Kerr et al., 2004; Kerr et al., 2008), except for extensions of 1 min 20 sec in the case of expected products of > 1 kb. Products were excised from 3 % agarose gels, purified and TA-cloned into pCRII-TOPO (Invitrogen) (Kerr et al., 2007), with DNA sequencing performed by Geneservice Ltd., Oxford University.
The Elk1 primers F2 (5′-GCTTCTGAGAGAACAAGGTAATGGC-3′) and R2 (5′-CTCAACTCTTCGGATTTCTGGTTTG-3′) correspond, respectively, to nt 234–258 and 902-878 and of the reference RNA sequence and span exons 2–4 (NM_007922). The Elk3 primers F6 (5′-CCCTGTGGAAGAAGTCAGGACTG-3′), R10 (5′-GTGGGGAACTGGAAAA-GTGTGTTCG-3′), F2 (5′-GCACTTGCTGCTGGACCAGAAAC-3′) and R9 (5′-TGGGGCGACCGGACTAAGACT-3′) correspond, respectively, to nt 932–954, 1549-1525, 446–468 and 1499-1479 of the reference RNA sequence (NM_013508), except for the underlined nucleotides corresponding to the reverse complement of the T1490G and C1481T substitutions found in all Elk3 sequenced clones (Table S1). The Elk3-F6/R10 and Elk3-F2/R9 primer pairs span, respectively, exons 3.1 - 4/5 and exons 2–4 ((NM_013508; (Giovane et al., 1997)), and primer Elk3-F6 avoids Pctaire2 antisense sequence (see text). The Elk4 primers F2 (5′-CAGCTCCTGCAGGAGCCTCAGA-3′), F3 (5′-GAAGAAACTATCCAAGCTTTGGAGAC-3′) and R3 (5′-AGGGGCTCGGAGTCAGCAAGATG-3′) correspond, respectively, to nt 382–403, 994–1019 and 1450-1428 of the reference RNA sequence (NM_007923). The Elk4-F2/R3 and Elk4-F3/R3 primer pairs span, respectively, exons 2–4 and 3–4 (NM_007923).
Primers were designed to amplify Pctaire2 mRNA isoforms either not including (reference RNA sequence NM_146239.2; ‘Pctaire2a’) or including Elk3-antisense sequence (BC031778; BC049904; ‘Pctaire2b’). The Pctaire2 common forward primer F2 (5′-TGCAAAAGGACCCGGGTTTTCGAA-3′) corresponds to nt 1844–1867 of NM_146239.2 and nt 1391–1414 of BC031778. The isoform-specific reverse primers R2 (5′-CCTCCTTCCATGTCACTGGCTCA-3′) and R3 (5′-TGAGGAGAAACAGATGCGTT-CACAT-3′) correspond, respectively, to nt 1937-1915 of BC031778 and nt 2344-2320 of NM_146239.2. All sequenced Pctaire2a clones included the substitutions T1875C and A2174C compared to NM_146239.2, and Pctaire2b clones included the substitutions T1422C, A1540G and A1675G, plus the A-rich simple-sequence repeat C(A)4C(A)4C(A)8C(A)6C(A)8 rather than C(A)4C(A)6C(A)4C(A)6C(A)10 of BC031778. (Note that the Pctaire2b substitutions A1540G and A1675G correspond, respectively, to the reverse complements of the Elk3 substitutions T929C and T794C detailed in Table S1.)
Primers were designed to amplify the three different Mfsd4 mRNA isoforms: ‘a’ (NM_172510.4), ‘b’ (NM_001114662) and the cDNA sequence referred to here as ‘c’ (BC058790). The Mfsd4 common forward primer F2 (5′-CTCCAGTATAAAG/GTTGTG-CGACCA-3′, where ‘/’ represents the exon 7/8 junction) corresponds to nt 1443–1467 of both NM_172510.4 and NM_001114662, and to nt 1292–1316 of BC058790. The Mfsd4 isoform-specific reverse primers R3 (5′-TTCTCTTCTGAGAGTGGATCTGTAG-3′), R4 (5′-TGCT-CGATTTGTCCAGGGAAGACT-3′) and R1 (5′-AGGAGAGGTTAGGACTGGGTGCT-3′) correspond, respectively, to nt 1953-1929 of NM_172510.4, nt 1827-1804 of NM_001114662 and nt 1722-1700 of BC058790. Primer R1 avoids Elk4 antisense sequence (see text).
The primers designed to extend the known sequence of the Elk4 mRNA 3′-UTR were Ufor1 (5′-TGAGTTAAAGGAGAAAAGCCATTGGT-3′), Urev1 (5′-CACTGCTCTGAG-CCACGGTGCA-3′) and Urev2 (5′-GTTTATAGCCAGGTTGGGTGACAGAA-3′) that correspond, respectively, to nt 3508–3533 of the reference RNA sequence (NM_007923) and to the reverse complement of two downstream genomic DNA sequences (NT_078297).
Western blot analysis
Frozen tissue from adult male 129/OlaHsd mice (9 week old, Bristol University colony) was ground and homogenized (Kerr et al., 2007) in ice-cold radio-immunoprecipitation assay (RIPA) buffer including protease inhibitor cocktail (Sigma-Aldrich). Lysates were clarified by centrifugation at 10,000 × g for 2 × 10 min at 4 °C, the supernatant protein concentration was determined (Kerr et al., 2007), and aliquots of 100 μg whole cell lysate were separated by discontinuous SDS-polyacrylamide gel electrophoresis (SDS-PAGE; (Hobson et al., 2006)) together with 12,000–225,000 Da full-range rainbow markers (Amersham Biosciences). The resolved proteins were transferred to nitrocellulose membranes (Kerr et al., 2007), blocked, incubated overnight at 4 °C in 1:2000 affinity-purified Net#1996 antiserum, and immunoreactivity detected (Hobson et al., 2006). The rabbit anti-mouse Net#1996 antiserum was raised against the C-terminal 25 amino acids of Elk3/Net ((Gross et al., 2007); as was PAb375 (Giovane et al., 1994; Maira et al., 1996), see below), and was purified through three steps: caprylic acid, ammonium sulphate precipitation and immunoaffinity column chromatography against the peptide (Harlow and Lane, 1988).
Recombinant plasmids
pTL2-Elk3d was constructed by inserting a XmaI-HindIII PCR fragment of Elk3d into the corresponding sites of the expression vector pTL2 (pSG5-derived), followed by insertion into the XhoI and EcoRI sites of the XhoI-EcoRI fragment from pTL2-Net (Giovane et al., 1994) that corresponds to the 3′ end untranslated region of Elk3/Net. Sequences encoding the c-Ets1 DNA-binding domain (N70), c-Ets1 and v-Ets were as published (Wasylyk and Wasylyk, 1993).
Immunocytochemistry
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % foetal calf serum (FCS) (Gross et al., 2007). Cells were plated with 1 ml medium in plastic Leighton tubes and transfected 4 h later by the BBS calcium phosphate method (Wasylyk et al., 2005) with 1 μg expression vector (pTL2-Net(Elk3) or pTL2-Elk3d) in 100 μl of BBS/Ca per tube. 14 h later the cells were washed and incubated for a further 24 h, washed twice with PBS, fixed with methanol/acetone (2/1) for 30 min at −20 °C, and air-dried. A piece of the coverslip was saturated with 3 % bovine serum albumin (BSA) in PBS for 40 min at 37 °C, washed twice with PBS, incubated with affinity-purified rabbit antiserum PAb375 ((Giovane et al., 1994); diluted 1:1000 with PBS containing 0.3 % BSA) for 2 h at 37 °C, washed 3 times with PBS for 10 min, incubated with secondary antibody (CY3-Fcγ, Jackson ImmunoResearch Labs; diluted 1:250 in PBS containing 0.3 % BSA) for 1 h at 37 °C, washed 3 times for 10 min with PBS containing 0.02 % Tween 20, incubated for 2 min in Hoeschst dye (2 μg/ml in PBS) and washed briefly with PBS. The coverslip piece was placed on a glass slide, covered with Vectashield mounting medium and a coverslip, and examined by fluorescence microscopy.
In vitro transcription and translation, and electrophoretic mobility shift assays
Linearized pTL2 expression vectors were purified and transcribed with T7 RNA polymerase in the presence of m7GpppG for 1 h at 37 °C. RNA was analyzed by agarose gel electrophoresis, with optimized amounts of purified RNA used for protein synthesis in rabbit reticulocyte lysates (Promega) with [35S]-methionine that was then analyzed by SDS-PAGE (10 % polyacrylamide) and fluorography (Amplify, GE Healthcare) (Giovane et al., 1994). Proteins were quantified using a Phosphor-Imager (Fuji), taking into account the number of methionines.
Equimolar amounts of proteins (1–5 μl, adjusted to a constant volume with mock extracts incubated without added RNA) and an excess of PEA3* probe in 20 μl of buffer [20 mM Hepes (pH 7.9), 20 % glycerol, 0.1 mM EDTA, 2.5 mM DTT, 1 μg/20μl of poly[d(I-C)], 50 mM KCl] were incubated for 30 min at 25 °C. The samples were loaded on pre-run (20 min at 15 mA (75 V)) 4 % polyacrylamide (acrylamide:bis, 29:1) gels in 0.25 x Tris-borate/EDTA (TBE) and run for 60 min at 30 mA (150V) with recirculating buffer at 20 °C, similar to (Giovane et al., 1994). The PEA3* oligonucleotide 5′-TCGAGC-CGGAAGTGACGTCGA-3′ ((Giovane et al., 1994; Wasylyk and Wasylyk, 1993); ETS core consensus sequence underlined) and its complementary oligonucleotide were each 5′-labelled with T4 polynucleotide kinase and [α-32P]ATP, annealed overnight, and double-stranded blunt-ended probes were purified on native 10 % polyacrylamide gels (Maira et al., 1996).
Cell culture and transfection of reporters
NIH-3T3 cells were maintained in DMEM supplemented with 10 % FCS. Cells were transfected (Maira et al., 1996) with 20 μg per 100 mm dish of plasmid DNA containing expression vectors (pTL2-Net(Elk3) or pTL2-Elk3d; detailed above), 2.5 μg of reporter (PALx8-TK-CAT4 or control pBL-CAT4; (Maira et al., 1996; Wasylyk et al., 1991)), 3 μg of pSG5-lacZ (control for transfection efficiency) and completed with pSG5 (control for expression vectors). 30 % confluent cells were incubated with precipitated DNA for 24 h, washed three times with DMEM and then incubated with DMEM plus 0.05 % FCS for 24 h, scraped, freeze-thawed three times in solution A, and centrifuged (Giovane et al., 1994; Giovane et al., 1997). β-Galactosidase activity was measured first to correct for transfection efficiency, and chloramphenicol acetyltransferase (CAT) assays were as published (Giovane et al., 1994). For co-expression of Elk3 and Elk3d, the amounts of expression vectors used are indicated in the relevant Figure. Experiments were repeated at least three times.
Real-time quantitative RT-PCR assays
Relative mRNA expression levels were derived by the comparative threshold cycle (Ct) method, as previously described together with the GAPDH primer and probe set (Kerr et al., 2007). Primer and probe set design used default parameters of Primer Express software, and the probes detailed below had a 5′ fluorescent reporter dye FAM (6-carboxy-fluorescein) and the 3′ quencher dye TAMRA (6-carboxy-tetramethyl-rhodamine) (Applied Biosystems).
Elk1 primers and probe to detect both the full-length Elk1 and Elk1b mRNA isoforms were for products that crossed intron 4 of 0.5 kb (NM_007922; NT_039700): forward primer 5′-TGCTCCCCACACATACCTTGA-3′, reverse primer 5′-GAAGGAGAGCTTGGCTG-GACTAC-3′ and non-extendable Taqman probe 5′-CCCGGTGCTGCTGACACCCA-3′ correspond, respectively, to nt 1265–1285, 1383-1361 and 1287–1306 of the Elk1 reference RNA sequence (NM_007922). The assay was optimized using plasmid containing cDNA insert corresponding to nt 234–1383 of NM_007922 (data not shown).
Assays to discriminate between Elk3 mRNA isoforms used a common reverse primer 5′-TGTGTTCGGCCCTTGCA-3′ and Taqman probe 5′-ACTGTTTCTGGCCTCGAG-TCCGCTG-3′ that correspond, respectively, to nt 1532-1516 and 1430–1454 of the Elk3 reference RNA sequence (NM_013508). The isoform-specific forward primers to detect the Elk3/Elk3b (Elk3full-F42; 5′-CCTTCTTCACCGCACAG/ACA-3′, where ‘/’ represents the exon 3.3 to 4 junction), Elk3c (Elk3c-F20; 5′-TCAGGACTGTGATCAG/ACACCAA-3′, where ‘/’ represents the exon 3.1 to 4 junction) or Elk3d (Elk3d-F37; 5′-CTGAGATACTATTACGACAAG/ACACCAA-3′, where ‘/’ represents the exon 2 to 4 junction) transcripts correspond, respectively, to nt 1402–1421, (946–961 + 1419–1425) and (597–617 + 1419–1425) of NM_013508. The isoform specificity of each assay was confirmed using plasmids containing cDNA inserts of full-length Elk3 transcribed from exons 2–5 and the corresponding Elk3b, Elk3c and Elk3d sequences amplified from adult heart using the primers Elk3 F2 (see above) and R2 (nt 1570-1548 of NM_013508) (data not shown).
Elk4 primers and probe to detect all four mRNA isoforms were for products that crossed intron 4 of 4.09 kb (NT_078297): forward primer 5′-CCAGACTGCAAGGTGC-TAATACG-3′, reverse primer 5′-TCCAGGCCTGACAGAGTGAAC-3′ and Taqman probe 5′-CAG/TTTCCCTCTGTACTCAACAGTCATGGCC-3′ (where ‘/’ represents the exon 4/5 junction) correspond, respectively, to nt 1508–1530, 1589-1569 and 1537–1567 of the Elk4 reference RNA sequence (NM_007923). The assay was optimized using plasmid containing cDNA insert corresponding to nt 1189–1590 of NM_007923 (data not shown).
Pctaire2 primers and probe were targeted to exon 16 and 18 sequences in order to detect mRNA isoforms ‘b’–‘d’, each of which also include Elk3-antisense sequence in the downstream exon 21 (Fig. 6B), and were for products that crossed intron16/exon17/intron17 of 2.64 kb (NT_039500): forward primer 5′-GACCCGGGTTTTCGAAACTC-3′, reverse primer 5′-AGGTTCAGTCTTTCCAAGTTTTAAGTCT-3′ and Taqman probe 5′-AG/CCTTATCCTGATCCTGGAAACTGCAGA-3′ (where ‘/’ represents the exon 16/18 junction) correspond, respectively, to nt 1399–1418, 1494-1467 and 1434–1462 of the Pctaire2 cDNA sequence BC031778. The specificity of the assay for Pctaire2 mRNA isoforms ‘b’–‘d’, but not isoform ‘a’, was confirmed using plasmids containing the corresponding cDNA inserts (data not shown).
Each of the assays amplified a single product of the expected size from DRG, as detected by gel electrophoresis on 4 % agarose gels (data not shown).
DNA and protein sequence analyses
Transcript, genomic and protein sequences were analyzed using BLAST programmes (http://www.ncbi.nlm.nih.gov/), and the sense-antisense databases SADB ((Katayama et al., 2005); http://fantom31p.gsc.riken.jp/s_as/) and NATsDB (http://natsdb.cbi.pku.edu.cn/) were used. Protein molecular weights were calculated using the Protein Information Resource ((Wu et al., 2003); http://pir.georgetown.edu); ETS family protein secondary structures were predicted using PSIPRED version 2.6 ((Bryson et al., 2005); http://bioinf.cs.ucl.ac.uk/psipred/); Mfsd4 sequences were analyzed using the Pfam protein families database release 23.0 ((Finn et al., 2008); http://pfam.sanger.ac.uk) with membrane-spanning regions predicted using the TMHMM version 2.0 programme ((Moller et al., 2001); www.cbs.dtu.dk/services/TMHMM/TMHMM2.0b.guide.html).
Supplementary Material
Acknowledgments
We thank Dr. Linda Hunt (Institute of Child Health, University of Bristol) for advice on statistics. The Wynick lab acknowledges financial support from the Medical Research Council and National Institute on Aging (AG10668), and the Wasylyk lab from the CNRS, INSERM, ARC and Ligue Contre le Cancer.
Footnotes
The nucleotide sequences reported in this paper were submitted to the EMBL and GenBank™ databases with Accession Numbers FN434115 – FN434128.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- Bacon A, Kerr NC, Holmes FE, Gaston K, Wynick D. Characterization of an enhancer region of the galanin gene that directs expression to the dorsal root ganglion and confers responsiveness to axotomy. J Neurosci. 2007;27:6573–6580. doi: 10.1523/JNEUROSCI.1596-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: a critical look in both directions. Cell Mol Life Sci. 2009;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Carlile M, Nalbant P, Preston-Fayers K, McHaffie GS, Werner A. Processing of naturally occurring sense/antisense transcripts of the vertebrate Slc34a gene into short RNAs. Physiol Genomics. 2008;34:95–100. doi: 10.1152/physiolgenomics.00004.2008. [DOI] [PubMed] [Google Scholar]
- Cesari F, Brecht S, Vintersten K, Vuong LG, Hofmann M, Klingel K, Schnorr JJ, Arsenian S, Schild H, Herdegen T, Wiebel FF, Nordheim A. Mice deficient for the ets transcription factor elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol Cell Biol. 2004;24:294–305. doi: 10.1128/MCB.24.1.294-305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Derry JM, Barnard PJ. Physical linkage of the A-raf-1, properdin, synapsin I, and TIMP genes on the human and mouse X chromosomes. Genomics. 1992;12:632–638. doi: 10.1016/0888-7543(92)90286-2. [DOI] [PubMed] [Google Scholar]
- Ducret C, Maira SM, Dierich A, Wasylyk B. The net repressor is regulated by nuclear export in response to anisomycin, UV, and heat shock. Mol Cell Biol. 1999;19:7076–7087. doi: 10.1128/mcb.19.10.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret C, Maira SM, Lutz Y, Wasylyk B. The ternary complex factor Net contains two distinct elements that mediate different responses to MAP kinase signalling cascades. Oncogene. 2000;19:5063–5072. doi: 10.1038/sj.onc.1203892. [DOI] [PubMed] [Google Scholar]
- Fantz DA, Jacobs D, Glossip D, Kornfeld K. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J Biol Chem. 2001;276:27256–27265. doi: 10.1074/jbc.M102512200. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- Ge X, Rubinstein WS, Jung YC, Wu Q. Genome-wide analysis of antisense transcription with Affymetrix exon array. BMC Genomics. 2008;9:27. doi: 10.1186/1471-2164-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Thomson NH, Abrams WR, Kirkham J. Nested genes: biological implications and use of AFM for analysis. Gene. 2005;350:15–23. doi: 10.1016/j.gene.2004.12.045. [DOI] [PubMed] [Google Scholar]
- Gille H, Kortenjann M, Strahl T, Shaw PE. Phosphorylation-dependent formation of a quaternary complex at the c-fos SRE. Mol Cell Biol. 1996;16:1094–1102. doi: 10.1128/mcb.16.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Pintzas A, Maira SM, Sobieszczuk P, Wasylyk B. Net, a new ets transcription factor that is activated by Ras. Genes Dev. 1994;8:1502–1513. doi: 10.1101/gad.8.13.1502. [DOI] [PubMed] [Google Scholar]
- Giovane A, Sobieszczuk P, Ayadi A, Maira SM, Wasylyk B. Net-b, a Ras-insensitive factor that forms ternary complexes with serum response factor on the serum response element of the fos promoter. Mol Cell Biol. 1997;17:5667–5678. doi: 10.1128/mcb.17.10.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Buchwalter G, Dubois-Pot H, Cler E, Zheng H, Wasylyk B. The ternary complex factor net is downregulated by hypoxia and regulates hypoxia-responsive genes. Mol Cell Biol. 2007;27:4133–4141. doi: 10.1128/MCB.01867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1988. [Google Scholar]
- Hever A, Oshima RG, Hauser CA. Ets2 is not required for Ras or Neu/ErbB-2 mediated cellular transformation in vitro. Exp Cell Res. 2003;290:132–143. doi: 10.1016/s0014-4827(03)00315-x. [DOI] [PubMed] [Google Scholar]
- Hobson SA, Holmes FE, Kerr NC, Pope RJ, Wynick D. Mice deficient for galanin receptor 2 have decreased neurite outgrowth from adult sensory neurons and impaired pain-like behaviour. J Neurochem. 2006;99:1000–1010. doi: 10.1111/j.1471-4159.2006.04143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiba N, Masuda S, Takeuchi A, Takeuchi D, Okuda M, Inui K. Cloning and characterization of a novel Na+-dependent glucose transporter (NaGLT1) in rat kidney. J Biol Chem. 2003;278:14669–14676. doi: 10.1074/jbc.M212240200. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- Jankowski MP, Cornuet PK, McIlwrath S, Koerber HR, Albers KM. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143:501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kerr NC, Gao Z, Holmes FE, Hobson SA, Hancox JC, Wynick D, James AF. The sodium channel Na(v)1.5a is the predominant isoform expressed in adult mouse dorsal root ganglia and exhibits distinct inactivation properties from the full-length Na(v)1.5 channel. Mol Cell Neurosci. 2007;35:283–291. doi: 10.1016/j.mcn.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr NC, Holmes FE, Wynick D. Novel isoforms of the sodium channels Nav1.8 and Nav1.5 are produced by a conserved mechanism in mouse and rat. J Biol Chem. 2004;279:24826–24833. doi: 10.1074/jbc.M401281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr NC, Holmes FE, Wynick D. Novel mRNA isoforms of the sodium channels Na(v)1.2, Na(v)1.3 and Na(v)1.7 encode predicted two-domain, truncated proteins. Neuroscience. 2008;155:797–808. doi: 10.1016/j.neuroscience.2008.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobberup S, Nyeng P, Juhl K, Hutton J, Jensen J. ETS-family genes in pancreatic development. Dev Dyn. 2007;236:3100–3110. doi: 10.1002/dvdy.21292. [DOI] [PubMed] [Google Scholar]
- Laitem C, Leprivier G, Choul-Li S, Begue A, Monte D, Larsimont D, Dumont P, Duterque-Coquillaud M, Aumercier M. Ets-1 p27: a novel Ets-1 isoform with dominant-negative effects on the transcriptional properties and the subcellular localization of Ets-1 p51. Oncogene. 2009;28:2087–2099. doi: 10.1038/onc.2009.72. [DOI] [PubMed] [Google Scholar]
- Lee N, Neitzel KL, Devlin BK, MacLennan AJ. STAT3 phosphorylation in injured axons before sensory and motor neuron nuclei: potential role for STAT3 as a retrograde signaling transcription factor. J Comp Neurol. 2004a;474:535–545. doi: 10.1002/cne.20140. [DOI] [PubMed] [Google Scholar]
- Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004b;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- Lopez M, Oettgen P, Akbarali Y, Dendorfer U, Libermann TA. ERP, a new member of the ets transcription factor/oncoprotein family: cloning, characterization, and differential expression during B-lymphocyte development. Mol Cell Biol. 1994;14:3292–3309. doi: 10.1128/mcb.14.5.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- Maira SM, Wurtz JM, Wasylyk B. Net (ERP/SAP2) one of the Ras-inducible TCFs, has a novel inhibitory domain with resemblance to the helix-loop-helix motif. EMBO J. 1996;15:5849–5865. [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Mechaly I, Bourane S, Piquemal D, Al-Jumaily M, Venteo S, Puech S, Scamps F, Valmier J, Carroll P. Gene profiling during development and after a peripheral nerve traumatism reveals genes specifically induced by injury in dorsal root ganglia. Mol Cell Neurosci. 2006;32:217–229. doi: 10.1016/j.mcn.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Enders GH, Wu CL, Su LK, Gorka C, Nelson C, Harlow E, Tsai LH. A family of human cdc2-related protein kinases. EMBO J. 1992;11:2909–2917. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Vaessen B, Johnston K, Marmorstein R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol Cell. 1998;2:201–212. doi: 10.1016/s1097-2765(00)80130-6. [DOI] [PubMed] [Google Scholar]
- Mo Y, Vaessen B, Johnston K, Marmorstein R. Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA. Nat Struct Biol. 2000;7:292–297. doi: 10.1038/74055. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol Life Sci. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro X, Vivo M, Valero-Cabre A. Neural plasticity after peripheral nerve injury and regeneration. Prog Neurobiol. 2007;82:163–201. doi: 10.1016/j.pneurobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Numata K, Okada Y, Saito R, Kiyosawa H, Kanai A, Tomita M. Comparative analysis of cis-encoded antisense RNAs in eukaryotes. Gene. 2007;392:134–141. doi: 10.1016/j.gene.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Numata K, Osada Y, Okada Y, Saito R, Hiraiwa N, Nakaoka H, Yamamoto N, Watanabe K, Okubo K, Kohama C, Kanai A, Abe K, Kiyosawa H. Identification of novel endogenous antisense transcripts by DNA microarray analysis targeting complementary strand of annotated genes. BMC Genomics. 2009;10:392. doi: 10.1186/1471-2164-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DA, Koleski D, Kola I, Hertzog PJ, Ristevski S. Identification and expression analysis of alternative transcripts of the mouse GA-binding protein (Gabp) subunits alpha and beta1. Gene. 2005;344:79–92. doi: 10.1016/j.gene.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Obata K, Yamanaka H, Dai Y, Tachibana T, Fukuoka T, Tokunaga A, Yoshikawa H, Noguchi K. Differential activation of extracellular signal-regulated protein kinase in primary afferent neurons regulates brain-derived neurotrophic factor expression after peripheral inflammation and nerve injury. J Neurosci. 2003;23:4117–4126. doi: 10.1523/JNEUROSCI.23-10-04117.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, Okubo K, Ikeda R, Saito R, Kanai A, Abe K, Tomita M, Kiyosawa H. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum Mol Genet. 2008;17:1631–1640. doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- Pao SS, I, Paulsen T, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MA, Rogers AE, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VN, Huebner K, Isobe M, ar-Rushdi A, Croce CM, Reddy ES. elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science. 1989;244:66–70. doi: 10.1126/science.2539641. [DOI] [PubMed] [Google Scholar]
- Rao VN, Reddy ES. Delta elk-1, a variant of elk-1, fails to interact with the serum response factor and binds to DNA with modulated specificity. Cancer Res. 1993;53:215–220. [PubMed] [Google Scholar]
- Rodrigo I, Cato AC, Cano A. Regulation of E-cadherin gene expression during tumor progression: the role of a new Ets-binding site and the E-pal element. Exp Cell Res. 1999;248:358–371. doi: 10.1006/excr.1999.4438. [DOI] [PubMed] [Google Scholar]
- Sahin A, Vercamer C, Kaminski A, Fuchs T, Florin A, Hahne JC, Mattot V, Pourtier-Manzanedo A, Pietsch T, Fafeur V, Wernert N. Dominant-negative inhibition of Ets 1 suppresses tumor growth, invasion and migration in rat C6 glioma cells and reveals differentially expressed Ets 1 target genes. Int J Oncol. 2009;34:377–389. [PubMed] [Google Scholar]
- Schweizer U, Gunnersen J, Karch C, Wiese S, Holtmann B, Takeda K, Akira S, Sendtner M. Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J Cell Biol. 2002;156:287–297. doi: 10.1083/jcb.200107009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The ETS-domain transcription factors Elk-1 and SAP-1 exhibit differential DNA binding specificities. Nucleic Acids Res. 1995;23:4698–4706. doi: 10.1093/nar/23.22.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siintola E, Topcu M, Aula N, Lohi H, Minassian BA, Paterson AD, Liu XQ, Wilson C, Lahtinen U, Anttonen AK, Lehesjoki AE. The novel neuronal ceroid lipofuscinosis gene MFSD8 encodes a putative lysosomal transporter. Am J Hum Genet. 2007;81:136–146. doi: 10.1086/518902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam FJ, MacGillavry HD, Armstrong NJ, de Gunst MC, Zhang Y, van Kesteren RE, Smit AB, Verhaagen J. Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur J Neurosci. 2007;25:3629–3637. doi: 10.1111/j.1460-9568.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- Stinson J, Inoue T, Yates P, Clancy A, Norton JD, Sharrocks AD. Regulation of TCF ETS-domain transcription factors by helix-loop-helix motifs. Nucleic Acids Res. 2003;31:4717–4728. doi: 10.1093/nar/gkg689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Hurst LD, Carmichael GG, Chen J. Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 2005;33:5533–5543. doi: 10.1093/nar/gki852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Bonilla I, Winkles JA, Strittmatter SM. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23:9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Criqui-Filipe P, Wasylyk B. Sumoylation of the net inhibitory domain (NID) is stimulated by PIAS1 and has a negative effect on the transcriptional activity of Net. Oncogene. 2005;24:820–828. doi: 10.1038/sj.onc.1208226. [DOI] [PubMed] [Google Scholar]
- Wasylyk C, Gutman A, Nicholson R, Wasylyk B. The c-Ets oncoprotein activates the stromelysin promoter through the same elements as several non-nuclear oncoproteins. EMBO J. 1991;10:1127–1134. doi: 10.1002/j.1460-2075.1991.tb08053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Wasylyk B. Oncogenic conversion of Ets affects redox regulation in-vivo and in-vitro. Nucleic Acids Res. 1993;21:523–529. doi: 10.1093/nar/21.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC. The Protein Information Resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Bumpass DC, Perkins ND, Sharrocks AD. The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol Cell Biol. 2002;22:5036–5046. doi: 10.1128/MCB.22.14.5036-5046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Shore P, Willingham N, Lakey JH, Sharrocks AD. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 1999;18:5666–5674. doi: 10.1093/emboj/18.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Li L, Papadopoulou N, Hodgson G, Evans E, Galbraith M, Dear M, Vougier S, Saxton J, Shaw PE. Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1. Nucleic Acids Res. 2008;36:2594–2607. doi: 10.1093/nar/gkn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.