Abstract
Genetically-modified mosquitoes that are unable to transmit pathogens offer opportunities for controlling vector-borne diseases such as malaria and dengue. Site-specific gene recombination technologies are advantageous in the development of these insects because anti-pathogen effector genes can be inserted at integration sites in the genome that cause the least alteration in mosquito fitness. Here we describe Anopheles stephensi transgenic lines containing φC31 attP “docking” sites linked to a fluorescent marker gene. Chromosomal insertion sites were determined and life-table parameters were assessed for transgenic mosquitoes of each line. No significant differences in fitness between the transgenic and non-transgenic mosquitoes were detected in this study. These transgenic lines are suitable for future site-specific integrations of anti-parasite transgenes into the attP sites.
Keywords: Life-table analyses, site-specific recombination, hybridizations in situ
Introduction
Transposon-mediated genetic transformation of anopheline mosquitoes (Catteruccia et al., 2000; Grossman et al., 2001; Terenius et al., 2007; Lombardo et al., 2009) allows testing hypotheses for malaria control through genetic manipulation of its vector (Terenius et al., 2008; Marshall & Taylor, 2009). Translation to the field of any strategies based on these hypotheses remains a challenge, and the potential negative impact of a transgene on mosquito survival and reproduction must be taken into consideration and minimized (Lambrechts et al., 2008; Marrelli et al., 2006).
Reduced fitness of laboratory-reared, genetically-engineered mosquitoes may result from factors both independent of and directly related to transgenesis. Inadequate mass-rearing conditions and introgression of novel alleles by crossing mosquito strains of different genetic backgrounds can impact fitness independent of the nature of the genetic modification (Menge et al., 2005; Marrelli et al., 2006). In addition, rearing of transgenic mosquitoes as homozygotes is associated with inbreeding depression, a condition that results in reduced fitness (Catteruccia et al., 2003; Irvin et al., 2004). Fitness loads also have been attributed to insertional mutagenesis and the expression of exogenous anti-pathogen effector molecules (Marrelli et al., 2006). However, measuring the contributions of each of these factors is challenging.
One approach for distinguishing between fitness costs associated with the effector molecule and those associated with insertional mutagenesis utilizes transgenic lines receptive to site-specific integration. Transgenes inserted into a “docking” site can mitigate variation in expression properties associated with random integration. The integrase and recognition sites of φC31 can be used to introduce multiple and alternative genes into the same chromosomal environment in mosquitoes (Nimmo et al., 2006). Here, we report the molecular characterization and fitness assessment of four Anopheles stephensi transgenic lines generated for use with the φC31 integrase system. These lines offer the advantage of allowing the stable and efficient integration of a transgene into a site whose location and impact on fitness are known.
Results and Discussion
Anopheles stephensi transformation and Southern hybridization analyses
A total of 990 embryos (G0) were injected with transformation plasmid pBac[3×P3-ECFPfa]attP (Fig. 1) and six transgenic lines (20, 30, 19A, 43, 44 and P11) were obtained. Lines 20, 30, 19A and 43 were selected for molecular characterization. Hybridization of a transgene-specific probe (ECFP) to genomic DNA digested with the restriction enzyme, BamHI, showed single integrations in lines 20, 30, and 43 and two integrations in line 19A (Fig. 1). Hybridizing fragment lengths of ∼16 kilobase-pairs (kb), 5 kb and 17 kb, were detected for lines 20, 30 and 43, respectively. Transgenic line 19A had hybridizing fragments of ∼3.5 kb and 15 kb. Genomic DNA from wild-type mosquitoes did not hybridize with the probe.
Figure 1.
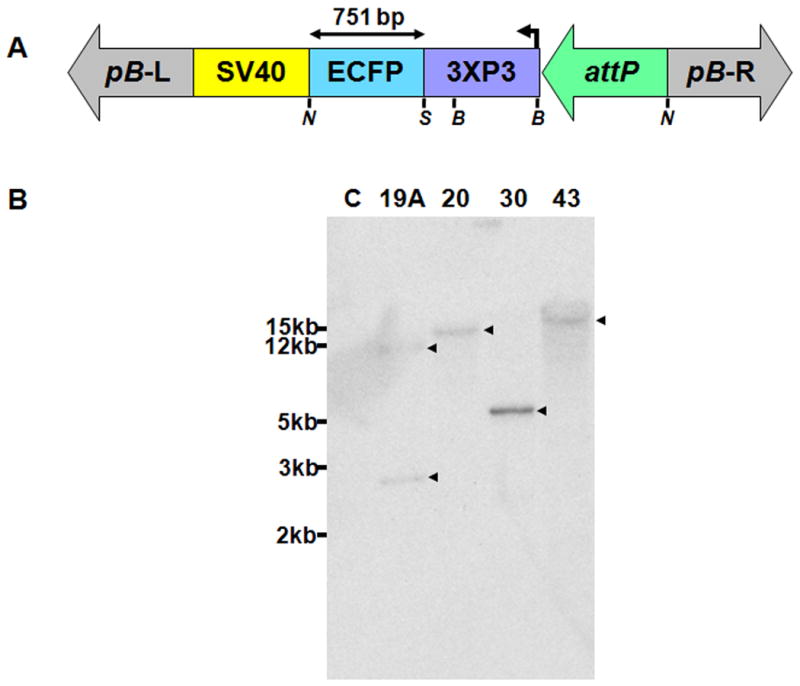
Transgene structure and Southern blot analyses of insertions in Anopheles stephensi. A) Schematic representation of the transgene construct. The construct contains a 3×P3-ECFP-SV40 polyA marker cassette, piggyBac left and right arms (pB-L and pB-R) and an attP docking site. Restriction enzymes, NotI (N) and SalI (S) generate a 751 bp ECFP probe. The BamHI (B) restriction endonuclease sites used in the Southern analyses are indicated. B) Southern blot detection of attP integration and copy number using a labeled ECFP probe from the transgene. Small arrowheads indicate positive signals corresponding to hybridizing DNA fragments. Relative sizes in kilobase-pairs (kb) of DNA markers are indicated on the right.
Molecular mapping of genome integration sites
Amplification (inverse-PCR) and sequencing of transgene-genome junctions and reconstructions of the wild-type integration sites show that the insertions occurred at TTAA sequences, the preferred site of piggyBac transposition, and at different locations within the genome (Fraser et al., 1995; Supplemental Figure 1). A single insertion site each was identified in lines 20 and 30, confirming the Southern blot results. Analyses of lines 19A and surprisingly, 43, resulted in the recovery and reconstruction of two insertion sites. Blast analyses of the reconstructed insertion site sequences of lines 20, 43 and 19A-A indicated that they do correspond to An. stephensi genomic sequences (Supplemental Figure 1) and independent amplifications of genomic DNA with primers designed to anneal to the DNA flanking the insertion sites further validated their contiguity within the mosquito genome (data not shown).
Physical mapping of the integration sites
Fluorescent hybridization in situ to polytene chromosomes derived from transgenic lines confirmed transgene copy number, with a single signal in lines 20 and 30, and two signals in line 19A (Fig. 2). Hybridization of the ECFP probe to a line 43 chromosomal preparation identified a single copy of the transgene in the X chromosome. It is likely that transgenic line 43 is a mixed population of individuals with one or two distinct insertions.
Figure 2.

Fluorescent in situ hybridization of polytene chromosomes of wild-type and transgenic Anopheles stephensi. Arrowheads indicate signals corresponding to insertion sites. A) A Cy3-labeled (red-orange signal) ECFP probe was hybridized to preparations of transgenic lines 19A, 30, and 43. Integrated attP transgenes were detected in the X chromosome of line 43, in an autosome of line 30, and in two chromosomal locations in line 19A. B) Hybridization of transgenic line 20 and wild-type chromosomes with two DNA probes: a Cy3-labeled ECFP probe (red signal and arrowhead), identifying the transgene, and a Cy5-labeled Ski probe (blue signal and arrowhead), identifying the 3R chromosomal arm. No binding of the ECFP probe was observed in wild-type samples.
Lines 20 and 30 were selected for further physical mapping analysis. Hybridizations with both a Cy3-labeled ECFP probe and a Cy5-labled chromosome marker probe were performed simultaneously to identify the chromosome to which the integrated transgene was linked. The Cy5-labeled marker probes, Vitellogenin, Ski, and PPO1, bind to unique locations in An. stephensi polytene chromosome arms 2R, 3R and 3L, respectively (Sharakhova et al., 2006; I. Sharakhov, personal communication). The ECFP signal found in line 20 samples was present near the Ski probe indicating that it integrated into 3R (Fig. 2). The ECFP signal found in line 30 samples was present in an autosome, but not linked to marker, consistent with the interpretation that it is located on the chromosomal arm 2L (Data not shown).
Fitness assessments
Hemizygous mosquitoes from transgenic (T) lines 20, 30, 43 and 19A were compared to non-transgenic (NT) individuals for the effects of docking site insertions on their fitness (Table 1 and Fig. 3). Assaying hemizygous T mosquitoes circumvents the recessive effects of inbreeding depression (Catteruccia et al., 2003) and mimics what would happen during a potential field release of homozygotes as the first mating with natural mosquitoes will produce mosquitoes carrying a single copy of the transgene. The only variable between T and NT mosquitoes in our experiments was the presence of the transgene, allowing for the assessment of the effect of insertional mutagenesis with respect to the genomic locations where the transgenes integrated. The negative effects of transgenesis on fitness were not uniform for the traits and lines analyzed (Table 1). For example, line 20-derived NT mosquitoes exhibited significantly higher egg hatchability than T mosquitoes (F1,82= 4.29, P < 0.05). NT mosquitoes exhibited significantly higher fecundity (χ2 =9.18, d.f. = 1, P < 0.05) in line 30 comparisons. Line 43-derived NT mosquitoes exhibited significantly higher larval viability (χ2 = 4.32, d.f. = 1, P < 0.05), female wing span (F1,104= 5.96, P < 0.05) and shorter larval-to pupal developmental time (t = 9.04, d.f. = 17, P < 0.001). We observed that a substantial proportion of T females did not lay eggs after blood feeding in lines 30 and 19A (χ2 = 4.78, d.f. = 1, P < 0.05), significantly higher than the NT counterparts. In six instances, T mosquitoes performed better than their NT counterparts: fecundity (χ2 = 16.23, d.f. = 1, P < 0.001), male wing span (F1,98 = 12.52, P < 0.001) and larval-to pupal developmental time (t = 4.33, d.f. = 7, P < 0.01) in line 20, male and female wing span in line 30 (F1,98 = 12.06, P < 0.001 in males; F1,98 = 23.59, P < 0.001 in females), and larval viability in line 19A (χ2 = 4.13, d.f. = 1, P < 0.05).
Table 1.
Comparisons of values for life-table parameters of non-transgenic (NT) and transgenic (T) Anopheles stephensi.1
| Insertions2 | Single insertions | Double insertions | ||||||
|---|---|---|---|---|---|---|---|---|
| Line | 20 | 30 | 19A | 43 | ||||
| Genotype3 | NT | T | NT | T | NT | T | NT | T |
| Fecundity4 | 100.08±5.96 (39; 4-165) |
128±4.35*** (45; 33-171) |
141.32±10.62 (60; 3-254) |
93.16±6.26* (44; 6-171) |
119.07±5.00 (46; 31-177) |
110.60±5.66 (40, 25-175) |
169±10.06 (46, 10-238) |
174.26±10.00 (50, 22-256) |
| Percentage of females that laid no eggs5 | 0 (0) | 6.67 (0) | 0 (0) | 30.16 (0) | 21.25 (21.25) | 31.25* (18.75) | 0 (0) | 0 (0) |
| Egg hatchability6 | 0.60±0.03 (0-0.95) |
0.49±0.04* (0-0.92) |
0.53±0.04 (0-0.97) |
0.54±0.04 (0-0.97) |
0.58±0.03 (0-1) |
0.66±0.03 (0-0.97) |
0.60±0.06 (0-0.99) |
0.66±0.05 (0-0.99) |
| Larval-to-Pupal development in days7 | 6.85±0.10 (7) | 6.36±0.04** (8) | 7.15±0.25 (8) | 6.63±0.16 (14) | 8.34±0.26 (8) | 8.35±0.17 (11) | 7.51±0.09 (10) | 8.75±0.11*** (10) |
| Larval viability7 | 0.88±0.02 (10) | 0.89±0.06 (10) | 0.91±0.03 (8) | 0.93±0.01 (14) | 0.81±0.06 (8) | 0.91±0.04* (11) | 0.85±0.07 (10) | 0.72±0.04* (10) |
| Female longevity in days | 27.36±0.64 | 30.20±0.53** | 29.14±0.49 | 26.05±0.67*** | 21.61±0.51 | 20.67±0.46 | 23.85±0.54 | 20.66±0.59*** |
| Male longevity in days | 29.13±0.80 | 26.79±0.58*** | 26.84±0.60 | 25.32±0.69 | 24.53±0.58 | 21.69±0.67 | 24.66±0.60 | 18.54±0.60*** |
| Female wing span (mm)7 | 3.09±0.03 (50) | 3.19±0.03 (50) | 3.09±0.03 (50) | 3.30±0.03*** (50) | 3.20±0.01 (52) | 3.19±0.01 (52) | 3.24±0.03 (53) | 3.14±0.02* (53) |
| Male wing span (mm)7 | 2.86±0.03 (50) | 3.00±0.03 (50)*** | 2.84±0.04 (50) | 3.00±0.02*** (50) | 2.94±0.02 (51) | 2.92±0.02 (53) | 2.96±0.02 (53) | 2.89±0.03 (53) |
P < 0.05,
P < 0.01, and
P < 0.001.
The mean number ± standard error (S.E) is provided for each parameter.
“Insertion” refers to the number of independent insertions of the transgenes into each line.
“Genotype” refers to the control (NT) and transgenic mosquitoes that carry no or one or two hemizygous transgenes (T).
Mean number of eggs per female. The numbers in parentheses are the total number of mosquitoes examined; range of fecundity. Females that did not lay eggs were excluded from the calculation.
The numbers in parentheses are percentage of females that died during the experiment.
Average fraction of deposited eggs that hatched. The numbers in parentheses indicate the range of egg hatchability.
Numbers of replicates or samples for each experiment are in parentheses.
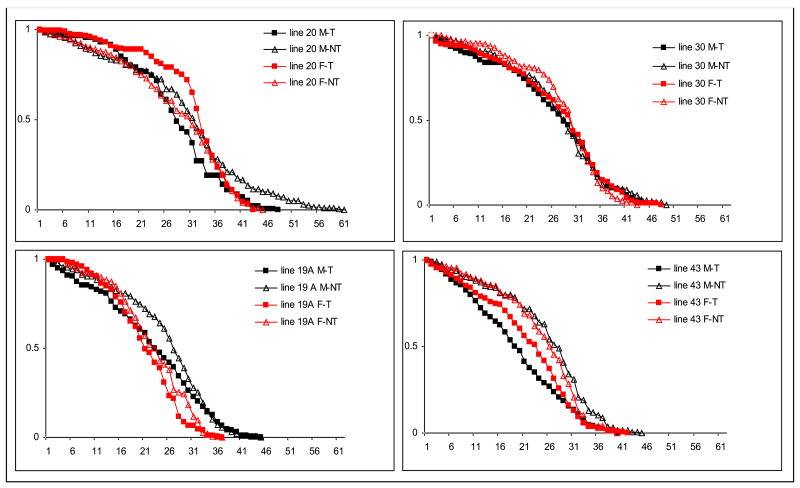
Figure 3.
Adult survivorship curve of NT and T Anopheles stephensi females (F) and males (M). The Y-axis indicates the proportion of the total population remaining at a given day, and the numbers in the X-axis correspond to days post adult eclosion.
Transgenes generally exhibited negative effects on adult mosquito survivorship (Fig. 3). Specifically, the average longevity of NT male mosquitoes in line 20 was 29.1 days, 2.3 days longer than T male mosquitoes (χ2 = 8.71, d.f. = 1, P < 0.01; Table 1). Similar phenomena were observed in female mosquitoes of line 30 (29.1 days in NT vs. 26.1 days in T; P < 0.001), female mosquitoes (23.9 days in NT vs. 20.7 days in T; P < 0.001) and male mosquitoes (24.5 days in NT vs. 18.5 days in T; P < 0.001) of line 43. The only exception was females of line 20 in which T mosquitoes lived significantly longer than NT (30.2 days in T vs. 27.4 days in NT; P < 0.01). The mean longevity between T and NT mosquitoes in line 19A was similar.
In summary, while all transgenic mosquito lines showed significant differences with respect to non-transgenic mosquitoes for some individual life-table parameters, none were uniformly less fit than wild-type controls. These results suggest that there is no unequivocal impact of the insertion of the piggyBac-based construct at the specific genomic insertion sites in these lines. Additional research to (1) evaluate the stability of the transgene over the generations; (2) introgress the transgene to an homozygous state and compare the effect on fitness of hemizygotes versus homozygotes transgenic mosquitoes; (3) pursue site-specific integration of an effector molecule and evaluate the impact of its expression; and (4) assess the mating competitiveness of transgenic mosquitoes with respect to wild type out-bred ones are still necessary. This will greatly contribute to the goal of producing genetically modified mosquitoes as fit as their wild type competitors, and potentially benefit malaria control programs.
Experimental Procedures
Mosquito husbandry
A colony of Anopheles stephensi (gift of M. Jacobs-Lorena, John Hopkins University) maintained in our insectary for >5 years was used in the experiments. The mosquitoes are kept at 27°C with 77% humidity and 12 hr day/night, 30 min dusk/dawn lighting cycle. Adult females are allowed to feed to repletion on mice and larvae are maintained on a diet of powdered fish food (Tetramin). All adult mosquitoes have continuous access to water and raisins.
Docking-site plasmid
The docking-site transgene construct, pBac[3×P3-ECFPfa]attP, was donated kindly by Paul Eggleston (Keele University, UK) (Nimmo et al., 2006).
Microinjection and Southern hybridizations
Microinjection procedures, selection and establishment of transformed lines were carried out as described previously (Catteruccia et al., 2000). Anopheles stephensi embryos were microinjected with the docking-site donor plasmid pBac[3×P3-ECFPfa]attP (0.5 μg/μl) and piggyBac helper plasmid (0.3 μg/μl) (Handler and Harrell 1999). Each surviving adult male was crossed to 15 wild-type females from the parental strain to produce single-founder families. Adult females were pooled in groups of 6-10 mosquitoes and mated with an equal number of wild-type males. Transgenic progeny expressing ECFP in the eyes were identified during larval stages and used to establish transgenic lines. Insertions of the piggyBac-based elements into the genomic DNA were detected analyzing genomic DNA from 12 adult female mosquitoes and standard Southern blotting and hybridization techniques (Sambrook et al., 1989).
Insertion site characterization by gene amplification
Gene amplification (inverse-PCR [iPCR]) was carried out to assemble and verify the transgene integration sites using genomic DNA from transgenic An. stephensi and primers complementary to the DNA adjacent to the transposon inverted terminal repeats (Supplemental Figure 1) (Nimmo et al., 2006). Primer sets piggyBac3for and piggyBac3rev, and piggyBac5for and piggyBac5rev (Nimmo et. al., 2006) were used to identify the 3′- and the 5′-end primary sequence of the integration sites, respectively; while primers based on the sequence of each identified contig (Supplemental Figure 1) were used for the amplification of non-transgenic mosquito genomic DNA. Amplification conditions were 1 min at 94°C followed by six cycles of 30s at 94°C, 45s at 65°C (-2°C/each cycle), and 1 min at 72°C; followed by 30 cycles of 30s at 94°C, 45s at 55°C and 1 min at 72°C. Amplification conditions for the analyses of the transposon-genome junctions from non-transgenic mosquito genomic DNA, were 1 min at 94°C followed by 30 cycles of 30s at 94°C, 45s at 58-62°C, and 1 min at 72 °C. DNA sequences of flanking regions were used to reconstruct the wild-type insertion site for each line, and were analyzed using the Blast service at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the TEfdatabase (http://tefam.biochem.vt.edu/tefam/resource.php), VectorBase (http://agambiae.vectorbase.org/Tools?BLAST/) and Full-Malaria/Parasites and Full-Arthropods website (http://fullmal.hgc.jp/).
Fluorescent hybridization in situ
DNA probes complementary to either the ECFP gene or the line-specific, wild-type reconstructed insertion site DNA were labeled with Cy3-AP3-dUTP (GE Healthcare) using the Random Primers DNA Labeling System (Invitrogen). Chromosome marker probes, Ski, PPO1, and Vitellogenin (Accession nos. AY578814, AY559300 and DQ442990, respectively) were amplified from An. stephensi genomic DNA and labeled with Cy5-AP3-dUTP (GE Healthcare). Polytene chromosomes from both transgenic and wild-type half-gravid females were prepared (Sharakhova et al. 2006) and hybridized overnight at 37°C (Trask 1991). The preparations were placed in 0.2 × SSC (0.03 M sodium chloride, 0.003 M sodium citrate) for at least 20 minutes before counterstaining with YOYO-1 (Invitrogen) and mounting with DABCO (Sigma-Aldrich). Either a Zeiss Axioskop or a Zeiss LSM 710 laser scanning microscope was used to archive images of the samples.
Production of hemizygous mosquitoes
Transgenic lines were maintained routinely as intercrossed colonies following efforts to establish homozygosity for each transgene. Hemizygous individuals for fitness assessment were generated by outcrossing 200 transgenic females with 20 wild-type males and 20 transgenic males with 200 wild-type females. Progeny for each experiment were derived either from the male or female outcrosses. Sibling mosquitoes not expressing the ECFP marker gene in the eyes were used as controls and are referred to as non-transgenic (NT) and individuals positive for ECFP are referred to as transgenic (T). This crossing scheme results in T mosquitoes with one or two copies per genome of the transgene, with the latter comprising copies from independent insertion events.
Fitness Assessments
Fitness was assessed in hemizygous T mosquitoes and their NT siblings using the following parameters: fecundity (number of eggs laid per female), fertility as a function of egg hatchability (number of larvae/number of eggs), larval-to-pupal developmental time, larval viability (proportion of larvae surviving to pupal stage), pupal sex-ratio, adult wing-span and adult longevity. Insectary procedures were standardized to ensure that all mosquitoes were treated similarly and to maximize confidence that differences in fitness trait measurements do not result from manipulations and/or environmental conditions alone.
Larval development rate and survivorship
Fifty T and NT first-instar larvae from the outcrosses were reared separately in bowls containing 400 ml of distilled water. Ten replicates were set up for each treatment. Larvae were fed to repletion once a day and the number of surviving larvae counted daily.
Fecundity
Approximately 200 T females (2-5 day old) were mated with 20 NT males of the same age. Similarly, 200 NT females (2-5 day old) were mated with NT males. The T and NT females were offered a single blood meal and after two days, fed females were put individually into glass vials lined with damp filter paper as oviposition sites. The females were maintained on a 1M sucrose solution and egg deposition was monitored at 72 hours post blood meal. The number of eggs laid by each female after the single blood meal was counted and recorded as an index of fecundity, excluding females that laid no eggs. These analyses were carried out with single blood meals because experiments with multiple blood meals are compromised by the significant lethality in females after feeding. The numbers of larvae hatching from the laid eggs were recorded as a measure of fertility.
Adult lifespan
Fifty 1-day old T and NT mosquitoes each in five replicates were placed in separate cages and maintained on raisins and wet cotton balls. Dead mosquitoes were removed and recorded until all died.
Wing span
Fifty 3-5-day old T and NT mosquitoes per sex were selected randomly and their wing lengths measured from the axial incision to the apical margin, not including the fringe of scales.
Data Analyses
Data analysis was conducted using the JMP statistical software (SAS Institute, Cary, NC). The Wilcoxon/Kruskal-Wallis non-parametric test (Sokal & Rohlf, 1994) was applied to compare NT and T mosquitoes from each line for fecundity and larval viability because the data did not conform to normal distribution after various transformations. Analysis of variance was used to assess egg hatchability, male and female wing span. Unpaired t-test with unequal variance was used to compare larval-to-pupal developmental time. Adult survivorship was analyzed using the Kaplan-Meier survival analysis (Sokal and Rohlf, 1994), and average longevity was calculated for both sexes of T and NT mosquitoes.
Supplementary Material
Primary nucleotide sequence of the reconstructed genome locus of integration for the transgenic lines 20, 30, 19A and 43. The right and left integration sites were identified by gene amplification (iPCR) and were linked to form a contig verified subsequently by amplification of genomic DNA from non-transgenic mosquitoes. The TTAA integration site is bolded. The primers used for genome junction validation are underlined. Coordinates are provided of the alignment comparisons for each contig with the results of whole-genome sequencing available at http://tefam.biochem.vt.edu/tefam/resource.php.
Acknowledgments
We are grateful to Lynn Olson, who assisted in preparing the manuscript, and Goufa Zhou, who helped with the statistical analyses. DA, MB and GY were supported in part by NIH grants (D43 TW01505), MB, AI, NJ, OM and AAJ by NIH grant R01 AI29746.
References
- Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, Crisanti A. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405:959–062. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- Catteruccia F, Godfray HC, Crisanti A. Impact of genetic manipulation on the fitness of Anopheles stephensi mosquitoes. Science. 2003;299:1225–1227. doi: 10.1126/science.1081453. [DOI] [PubMed] [Google Scholar]
- Fraser MJ, Cary L, Boonvisudhi K, Wang HG. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology. 1995;211:397–407. doi: 10.1006/viro.1995.1422. [DOI] [PubMed] [Google Scholar]
- Handler AM, Harrell RA. Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- Irvin N, Hoddle MS, O'Brochta DA, Carey B, Atkinson PW. Assessing fitness costs for transgenic Aedes aegypti expressing the GFP marker and transposase genes. Proc Natl Acad Sci USA. 2004;101:891–896. doi: 10.1073/pnas.0305511101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L Koella JC, Boëte C. Can transgenic mosquitoes afford the fitness cost? Trends Parasitol. 2008;24:4–7. doi: 10.1016/j.pt.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Lycett GJ, Lanfrancotti A, Coluzzi M, Arca B. Analysis of apyrase 5′ region validates improved Anopheles gambiae transformation technique. BMC Res Notes. 2009;2:24. doi: 10.1186/1756-0500-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrelli MT, Moreira CK, Kelly D, Alphey L, Jacobs-Lorena M. Mosquito transgenesis: what is the fitness cost? Trends Parasitol. 2006;22:197–202. doi: 10.1016/j.pt.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Marshall JM, Taylor CE. Malaria control with transgenic mosquitoes. PLoS Med. 2009;6:e20. doi: 10.1371/journal.pmed.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menge DM, Guda T, Zhong D, Pai A, Zhou G, Beier JC, Gouagna L, Yan G. Fitness consequences of Anopheles gambiae population hybridization. Malar J. 2005;4:44. doi: 10.1186/1475-2875-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo DD, Alphey L, Meredith JM, Eggleston P. High efficiency site-specific genetic engineering of the mosquito genome. Insect Mol Biol. 2006;15:129–136. doi: 10.1111/j.1365-2583.2006.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. A Laboratory manual. 2nd. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning; p. 31. [Google Scholar]
- Sharakhova MV, Xia A, McAlister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1603/0022-2585(2006)43[861:ascpft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The Principles and Practices of Statitics in Biological Research. Third. W.H. Freemand and Company; San Francisco: 1994. [Google Scholar]
- Terenius O, Juhn J, James AA. Injection of An. stephensi embryos to generate malaria-resistanct mosquitoes. J Vis Exp. 2007;5:216. doi: 10.3791/216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius O, Marinotti O, Sieglaff D, James AA. Molecular genetic manipulation of vector mosquitoes. Cell Host Microbe. 2008;4:417–423. doi: 10.1016/j.chom.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ. DNA sequence localization in metaphase and interphase cells by fluorescence in situ hybridization. Methods Cell Biol. 1991;35:3–35. doi: 10.1016/s0091-679x(08)60567-1. [DOI] [PubMed] [Google Scholar]
- Wakaguri H, Suzuki Y, Katayama T, Kawashima S, Kibukawa E, Hiranuka K, Sasaki M, Sugano S, Watanabe J. Full-malaria/parasites and full-arthropods: databases of full-length cDNAs of parasites and arthropods, update 2009. Nucleic Acids Res. 2009;37:D520–D525. doi: 10.1093/nar/gkn856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary nucleotide sequence of the reconstructed genome locus of integration for the transgenic lines 20, 30, 19A and 43. The right and left integration sites were identified by gene amplification (iPCR) and were linked to form a contig verified subsequently by amplification of genomic DNA from non-transgenic mosquitoes. The TTAA integration site is bolded. The primers used for genome junction validation are underlined. Coordinates are provided of the alignment comparisons for each contig with the results of whole-genome sequencing available at http://tefam.biochem.vt.edu/tefam/resource.php.