Abstract
Various factors/pathways including hormonal regulation have been suggested to control HSV-1 latency and reactivation. Our computer analysis identified a DNA repeat containing thyroid hormone response elements (TRE) in the regulatory region of HSV-1 LAT. Thyroid hormone (T3) exerts its function via its receptor (TR), a transcriptional factor. Present study investigated the roles of TR and T3 on HSV-1 gene expression using cultured neuoroblastoma cell lines. We demonstrated that liganded TR activated LAT transcription but repressed ICP0 transcription in the presence of LAT TRE. The chromatin immunoprecipitation (ChIP) assays showed that TRs were recruited to LAT TREs independently of T3 and hyperacetylated H4 was associated with promoters that were transcriptionally active. In addition, ChIP results showed that a chromatin insulator protein CTCF was enriched at the LAT TREs in the presence of TR and T3. In addition, chromatin remodeling factor BRG1 complex is found to participate in the T3/TR-mediated LAT activation since overexpression of BRG1 enhanced the LAT transcription and the dominant negative mutant K785R abolished the activation. This is the first report revealing that TR exerted epigenetic regulation on HSV-1 ICP0 expression in neuronal cells and could have a role in the complex processes of HSV-1 latency/reactivation.
Keywords: HSV-1, thyroid hormone, chromatin, transcription, LAT, ICP0, latency
Introduction
Herpes Simples Virus Type-1 (HSV-1) causes diseases ranging from mild oral lesions to severe keratitis and lethal encephalitis. HSV-1 infection is composed of four phases: acute infection, establishment of latency, maintenance of latency, and reactivation from latency. During acute infection, HSV-1 targets epithelial cells where the virus undergoes active gene expression in a cascade fashion and generates virion progeny after replication. The expression cascade is characterized by a sequential order with genes grouped into three categories: immediate early (IE), early (E), and late (L). After the completion of replication and egress, acute infection may be followed by the establishment of latency in the innervating neurons of the peripheral nervous system. In contrast to the acute infection, viral transcriptions are largely void except in one region: latency-associated transcript (LAT) [1–4]. The latent virus can survive in the neurons throughout the life of the host and may reactivate in response to various stimuli after the reversal of viral gene silencing [3, 5–8]. A variety of mechanisms have been proposed to describe the establishment of viral latency and reactivation such as altered immune response [9–12], microRNAs-induced gene silencing [13], differential neuronal suppression [14–17], hormonal regulation [18–21], and repressive chromatin [22–28]. Our laboratory identified thyroid hormone responsive elements (TREs) in several HSV-1 regulatory regions and one of them is located in the LAT regulatory sequence, suggesting that thyroid hormone (T3) could have roles in the transcriptions of LAT and neighboring regions and therefore modulate the viral latency and reactivation.
TRE is the binding site of transcription factor thyroid hormone receptors (TRs). The regulatory activity of TR is dependent on the ligand thyroid hormone (TH). TH is produced by the addition of iodine to tyrosine in the thyroglobulin protein [29]. The primary hormone product is Thyroxine (T4) but it is converted to potent triiodothyronine (T3) in target tissues [30]. T3 interacts with TRs to exert its biological effects [31]. TRs belongs to the nuclear hormone receptors superfamily such as TRα and TRβ in vertebrates [32]. The TREs-TRs interaction is, Regulation of HSV-1 Gene Expression by Thyroid Hormone Receptor nonetheless, independent of T3, implicating that both unliganded as well as liganded TRs participated in the gene regulation [33].
The present study analyzes the interrelationship of T3, TRs, and chromatin during the HSV-1 gene regulation in neuronal cells. The gene regulation of LAT, and ICP0 was investigated in mouse neuroblastoma cells N2a and N2aTRβ, in which the TRβ is constitutively expressed. The effects of TR and T3 on histone modification and cofactor recruitment to the promoters were also assessed. Our results showed that TR can either activate or repress these genes dependent on the presence of T3 via recruitment of various co-activators or co-repressors for distinct histone modifications and chromatin remodeling.
Materials and Methods
Viruses, cell lines, and culture conditions
HSV-1 strain 17Syn+ was obtained from Dr. James Hill’s laboratory at Louisiana State University Health Science Center (New Orleans, LA). The N2a cell line was purchased from the American Type Culture Collection (ATCC). N2aTRβ cell is a gift from Dr. Robert Denver (University of Michigan, Ann Arbor, MI) and maintained in DMEM/F12 supplemented with 10% charcoal treated fetal bovine serum (FBS). All cells were grown in an incubator at 37° C with 5%CO2. T3 was purchased from Sigma (St. Louis, MO).
Plasmids
Plasmid pSG28 contains HSV-1 sequence from 106785 to 131534. It covers the LAT TRE and open reading frames such as ICP4, LAT, ICP0, etc. Plasmid pAA3 possesses the HSV-1 sequence from 122713 to 134792. It contains partial deletion of LAT and does not have LAT TRE. Plasmid phMGFP (Promega, Cat#: E6421) was used as transfection efficiency control. Plasmid pBRG1 containing the complete ORF of BRG1 and was a gift from Dr. Keji Zhao (NHLBI, NIH). The plasmid pK785R, another gift from Dr. Zhao, contains a mutation in the BRG1 ORF with amino acid change from K to R at position 785.
Transfection
Nucleofector II (Cat#: AAD-1001S) from Amaxa (Gaithersburg, MD) was used for high efficiency of transfection. The protocol was essentially described by the manufacturer. The experiments were performed using Kit V (Cat#: VCA-1003) and the protocol number was T-024.
Antibodies
The antibodies used for ChIP are listed in Table 1. The dilution was according to the manufacturer’s suggestions.
Table 1.
The sources of antibodies
| Antibody | Species | Clonality | Source |
|---|---|---|---|
| TRβ1 | Rabbit | Polyclonal | Milipore/Upstate (Cat#: 06-539) |
| CTCF | Rabbit | Polyclonal | Milipore/Upstate (Cat#: 07-729) |
| BRG-1 | Rabbit | Polyclonal | Abcam (Cat#: ab4081) |
| Acetyl-histone H3 | Rabbit | Polyclonal | Milipore/Upstate (Cat#: 17-245) |
| Acetyl-histone H4 | Rabbit | Polyclonal | Milipore/Upstate (Cat#: 06-866) |
RT-PCR
Total RNA from cells was isolated by Trizol reagent (Invitrogen, Carlsbad CA). RT-PCRs were performed using Superscript One-Step RT-PCR (Invitrogen) with 0.2 μg of total RNA and correspondent primers. Their sequences are shown as follows: Actin: 5′-ATT CCT ATG TGG GCG ACG AG-3′ and 5′-TGG ATA GCA ACG TAC ATG GC-3′; LAT: 5′ CGG CGA CAT CCT CCC CCT AAG C 3′ and 5′ GAC AGA CGA ACG AAA CGT TCC G 3′; ICP0: 5′-TTC GGT CTC CGC CTG AGA GT-3′ and 5′-GAC CCT CCA GCC GCA TAC GA-3′. The RT-PCR reaction was carried out at 45°C for 20 min followed by 25 cycles of 94°C for 30 sec, 57°C for 30 sec, and 68°C for 30 sec. The RT-PCR products were analyzed using 2% agarose gel electrophoresis. The results were documented by Kodak Gel-Logic 100 imaging system. To increase the sensitivity of ICP0 assays, DIG-dNTP was included in the reaction for quantitative purpose. The gel was transferred to a nylon membrane, developed by CDP-star, and measured by Syngene G: BOX lightly cooled CCD camera and molecular biology software GeneTools® described previously [27].
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed using a DIG Gel shift Kit 2nd generation (Roche applied science, Indianapolis, IN) essentially described in the manufacturer’s protocol. Briefly, single-stranded oligonucleotides (oligo) for LAT TRE: 5′-GGG GAG AGG GGA GAG GGG GGG AGA GGG GAG AG -3′; Xenopus (positive control) TRE (xTRE): 5′-GAT CGC AGG TCA TTT CAG GAC AGC -3′ [34]; Mutant TRE: 5′-GGG GAG AGG GGA TTG GGG GGG ATT GGG GAG AG -3′ and their complementary oligos were synthesized (Invitrogen, San Diego, CA) and annealed (95°C for 10 min followed by slow cooling) to make double-stranded (ds) oligos. The cell extracts of N2a or N2aTRβ were isolated by first washing the cells twice with cold PBS followed by lysis with NP40 buffer (Invitrogen# FNN0021) containing protease inhibitor and PMSF. The ds oligos were terminally-labeled with non-radioactive DIG-11-ddUTP by terminal transferase and incubated with the cell extracts of N2a or N2aTRβ for 30 min at room temperature. For competition, 10× of unlabeled oligo were added to the labeled oligo for specificity analysis. The samples were electrophoresed on a 6% DNA Retardation Gel (Invitrogen) at 70 V for 1 h followed by electrophoretic transfer (Novex X Cell II Blot module) to positive-charged nylon membrane (Roche). The DIG labeled oligos were visualized by an enzymatic immunoassay using anti-Digoxigenin-alkaline phosphatase, Fab-fragments and chemiluminiscent substrate CSPD (Roche). The chemiluminiscent signal was captured using GeneGenome HR imaging system (Syngene, Frederick, MD).
ChIP
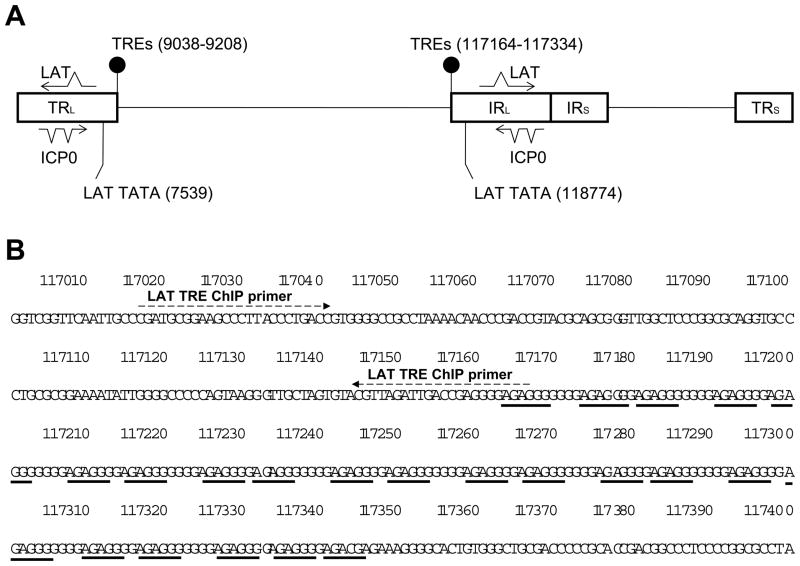
The protocol was described previously [27]. The sequences of PCR primers: ICP0 (promoter) ChIP: 5′-TAA TGG GGT TCT TTG GGG GAC ACC-3′ and 5′-TGC AAA TGC GAC CAG ACT GTC-3′; LAT ChIP: 5′-GCT ACG CCT TCG GGA ATG G′-3′ and 5′-AGA GGG GAG CCA GTT AGA TTG C-3′; ICP4 ORF: 5′-CGA CAC GGA TCC ACG ACC C-3′ and 5′-GAT CCC CCT CCC GCG CTT CGT CCG-3′. The location of LAT ChIP primers (117017–117165) is immediately upstream of LAT TRE (117161–117334) since LAT TRE is highly G-C rich and not suitable for PCR (Fig. 1B).
Fig. 1. HSV-1 LAT TREs.
1A: Two sets of positive TREs were identified in HSV-1 genome within LAT regulatory region. The first sets of TREs were found in the Terminal Repeat Long (TRL) and the second sets were located in the Internal Repeat Long (IRL) reiteration set 1.
1B: The positive TRE sequence 5′-GGGAGA- 3′ was repeated 21 times in these regions separated by one or four nucleotides. These TREs are approximately 1.4 kb upstream of the LAT TATA box. The figure is based on HSV-1 complete genome sequence (Accession #: X14112). LAT TRE ChIP primers were designed adjacent to one end.
T3 removal assays
N2a and N2aTRβ cells were plated on a 6-well plate with 40–50% confluency with the addition of T3 (10 nM) into the media refreshed daily. On day 6, viral infection was performed for 1 h at moi of 1. The inoculum was then removed and the cells were washed twice with 1 ml of PBS followed by the addition of 1 ml of fresh media containing T3 for 1 day. The T3 incubation was stopped after 1 day by removing the medium completely and washing the cells twice with PBS. New medium was added in each well with or without T3. The media were collected 48 h postinfection (hpi) and subjected to RT-PCR and plaque assays. The paradigm was shown in Fig. 7A.
Fig. 7. Long-term T3 treatment prevented the ICP0 expression and the washout of T3 derepressed the ICP0 expression and increased virus release.
7A: Cells were pre-treated with T3 for five days and subsequently infected with 17Syn+-EGFP viruses at moi of 1 with T3. The washout was done a day after the infection and the assays were performed 48 hpi.
7B: Total RNA was isolated at 48 hpi and subjected to RT-PCR assays using ICP0 primers to study TR-mediated regulation.
7C: The media of infected cells were collected 48 hpi and subjected to plaque assays using CV1 cells to investigate the release of infectious viruses. The P value measured by a Student’s paired t-test with a two-tailed distribution was 0.0015, indicating a significant increase.
Results
Location of HSV-1 LAT TREs
We identified putative TREs close to the LAT regulatory region in HSV-1 genome (Fig. 1A). The first sets of TREs were found in the Terminal Repeat Long (TRL) component from 9038 to 9208, coexisting with the reiteration set-1 (RE-1) (Fig. 1A). The second sets were located from 117164 to 117334, overlapping with Internal Repeat Long (IRL) RE-1 (Fig. 1A). The positive TRE sequence 5′-GGGAGA- 3′ was repeated 21 times in these regions separated by one or four nucleotides (Fig. 1B). The TRE is 1.4 kb upstream of the LAT TATA box. We hypothesize that transcription factor TR binds to this TRE and regulates the transcription of LAT and neighboring gene ICP0.
Liganded TR mediated positive regulation on LAT but repressed ICP0 transcription
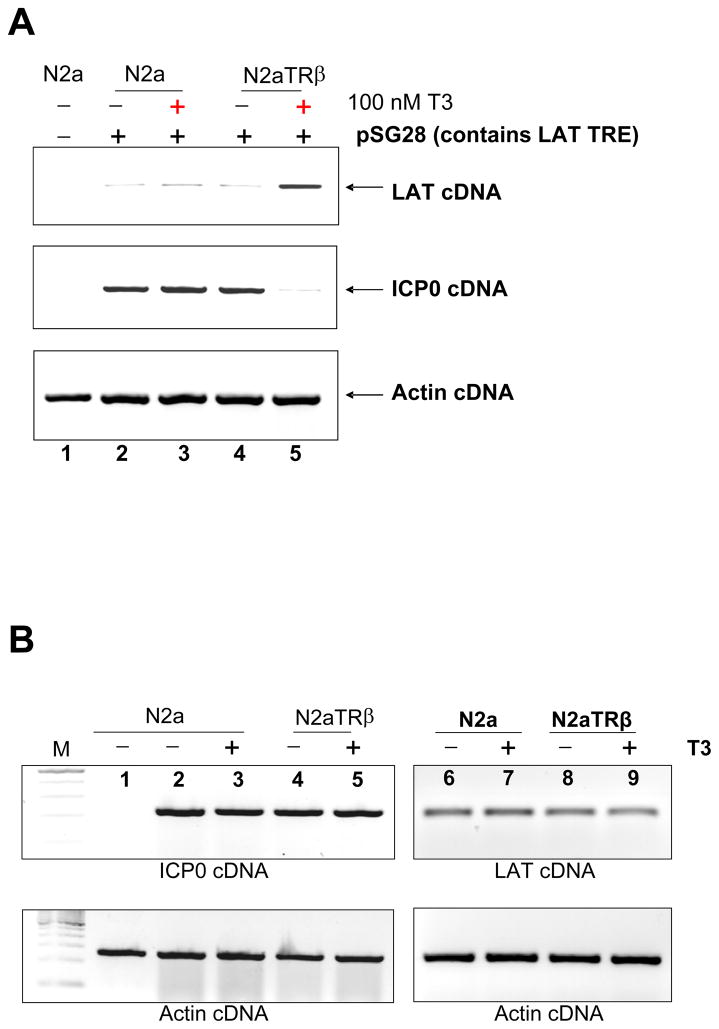
The regulatory effect of TR on LAT and ICP0 was examined by transfecting N2a or N2aTRβ cells with plasmid pSG28, which contains ICP0 and LAT including LAT TREs. RT-PCR assays showed that LAT transcription was up-regulated by TR and T3 (Fig. 2A). ICP0, however, was down-regulated by liganded TR (Fig. 2A). To address whether or not ICP0 was directly regulated by TR and T3, transfections were performed using plasmid pAA3, which contains ICP0 promoter but not LAT TREs. The results showed no regulation on ICP0 transcription (Fig. 2B, compare lane 4 and 5). Effect of TR on LAT was also investigated and the RTPCR assays revealed that liganded TR did not regulate LAT transcription in the absence of LAT TRE (Fig. 2B, compare lane 8 and 9). Transfection of phMGFP followed by fluorescent microscopy revealed similar numbers of cells emitting green fluorescence, suggesting that transfection efficiency were similar between these two cells (data not shown). Together, these observations indicated that LAT was positively regulated by TR and T3. ICP0 was repressed by liganded TR and this TR-mediated repression required LAT TREs.
Fig. 2. TR/T3 mediated regulation on HSV-1 LAT and ICP0.
2A: N2a and N2aTRβ cells were transfected with plasmid containing intact LAT TRE, LAT, and ICP0 to analyze the TR-mediated regulatory effect. Total RNA was purified after 48 h and subjected to RT-PCR assays using primers against LAT and ICP0. Actin primers were used as control. It showed that liganded TR up-regulated LAT but down-regulated ICP0 (compare lane 4 and 5).
2B: Transfection of N2a and N2aTRβ cells with plasmids without LAT TREs but contains complete ICP0 gene. Total RNA was purified followed by RT-PCR assays using primers against LAT and ICP0. Actin primers were used as control. Results showed that neither ICP0 nor LAT was regulated in the absence of LAT TRE.
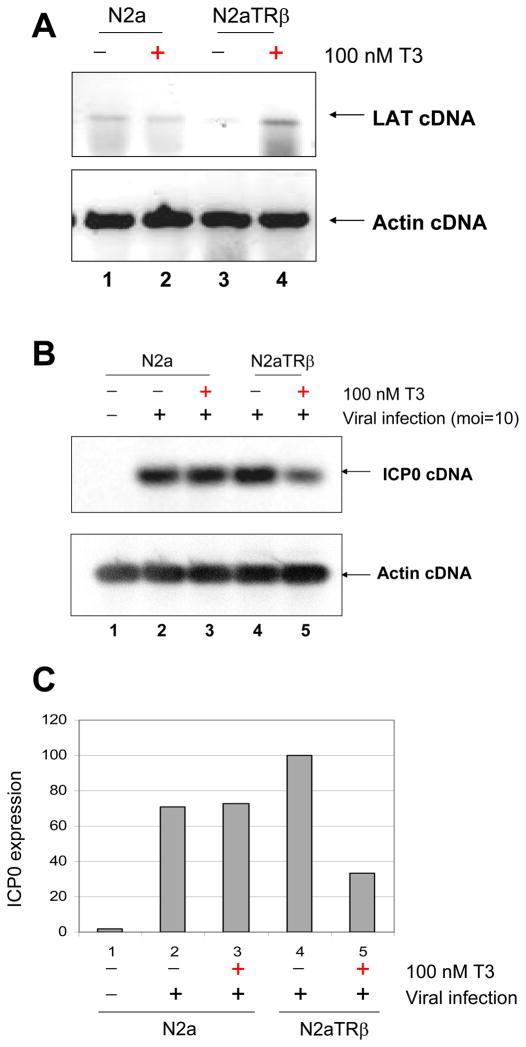
Viral infections were instigated to confirm the observation from the transfection experiments. Strain 17Syn+ was used at moi=10 to infect N2a and N2aTRβ cells with or without 100 nM T3 followed by RNA isolation and RT-PCR assays. The results showed no TR-mediated regulatory effect on LAT at 8 hpi, probably due to high moi and IE transactivation (data not shown). After the addition of 50μg/ml of CHX during infection to inhibit IE protein synthesis, LAT transcription was up-regulated by liganded TR at 8 hpi (Fig. 3A). TR and T3, at the same condition, caused a 60% reduction on ICP0 transcription measured by Syngene’s molecular biology software GeneTools® (Fig. 3B and C). These results demonstrated that liganded TR produced an up-regulation on LAT and down-regulated ICP0 during infection.
Fig. 3. Regulation of LAT and ICP0 by TR and T3 during infection.
3A: N2a and N2aTRβ cells were infected with wild-type virus 17Syn+ at moi=10 with the addition of 50 μg/ml cycloheximide to prevent the α transactivation. The total RNA was purified at 8 hpi and subjected to RT-PCR by primers against LAT. Actin primers were used as controls.
3B: The RNA samples from 3A were used for RT-PCR assays to assess ICP0 transcription. DIG-dNTP was included in the reaction for quantitative purpose.
3C: The chemiluminescence signal of 3B was captured by GENEgnome® camera from Syngene (Fredrick, MD). The quantitative analyses were measured by Syngene’s molecular biology software GeneTools®.
TRβ was recruited to LAT TRE and CTCF was enriched to the same region in the presence of T3
Direct interaction of TR to TRE was addressed by EMSA. Labeled oligos were incubated with cell extract of N2a and no shifted bands were observed (Fig. 4A, lane 1 to 8). In N2aTRβ, labeled LAT TRE oligo generated shifted band in the presence of extract (Fig. 4A, lane 12) and this band was effectively abolished by unlabeled oligo competition (Fig. 4A, lane 13). This protein-oligo shift pattern was confirmed by positive control xTRE (Fig. 4A, lane 14, 15, 16). Mutant LAT TRE (from GGGAGA to GGGATT) failed to produce similar band shift (Fig. 4A, lane 9 and 10), indicating the importance of the context. Collectively, these results demonstrated the in vitro interaction of TR/LAT TRE.
Fig. 4. TR bound to LAT TRE and facilitated CTCF enrichment to LAT RE-1 in the presence of T3.
4A: EMSA using extract from N2a (Lane 1 to 8) and N2aTRβ (Lane 9–16). Labeled oligos of LAT TRE, mutant TRE, and positive xenopus TRE (xTRE) were used for analyses. Free labeled oligo and labeled oligo-TR complex were marked. Noted that extract of N2aTRβ, not N2a, produced a shifted band while using labeled LAT TRE oligo (Lane 12) and positive control (Lane 15). Mutant oligo failed to generate the pattern (Lane 10).
4B: Cells were transfected by pSG28 and treated with or without T3 for 48 h. The cells were then subjected to ChIP using Abs against TRβ or CTCF for immuno-precipitation. The recruitment of TRβ or CTCF was analyzed by PCR. IgG controls showed no signal, indicating the specificity of the assays (data not shown).
4C: The binding specificity of TR and CTCF to LAT TRE was further analyzed by ChIP using primers against other regions (ICP4 ORF and ICP0 promoter, respectively). Lane 1 to 6: ChIP-TRβ using primers against ICP4 ORF; Lane 7 to 10: ChIP-CTCF using primers against ICP0 promoter.
The anti-TRβ antibody was used for ChIP to confirm the in vivo binding of receptor to the LAT TREs. The transfection and ChIP assays showed that TRβ was recruited to the LAT TRE regardless the ligand status (Fig. 4B). In addition, this region was previously described as a chromatin insulator-like element [22]. The potential for TRβ to modulate the CTCF interaction was addressed by ChIP using anti-CTCF Ab. The results demonstrated that liganded TRβ increased the interaction of CTCF to the element (Fig. 4B). To address the specificity of interaction, ChIP-TRβ and ChIP-CTCF were performed using primers away from the target regions (primers against ICP4 ORF and ICP0 promoter, respectively). These negative controls produced no signal (Fig. 4C), demonstrating that TRs interacted with LAT TREs and the liganded TRβ could facilitate the recruitment of CTCF to the HSV-1 chromatin insulator.
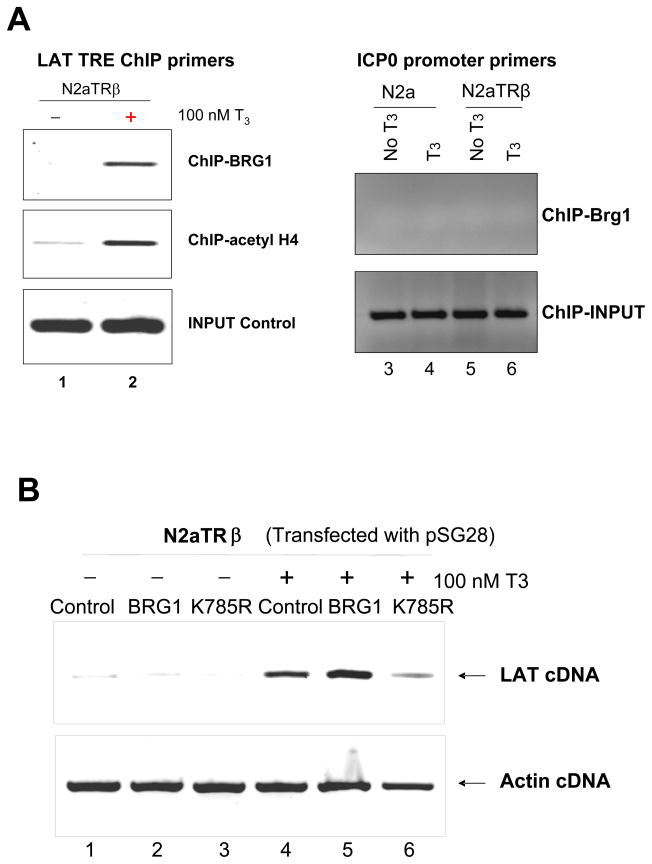
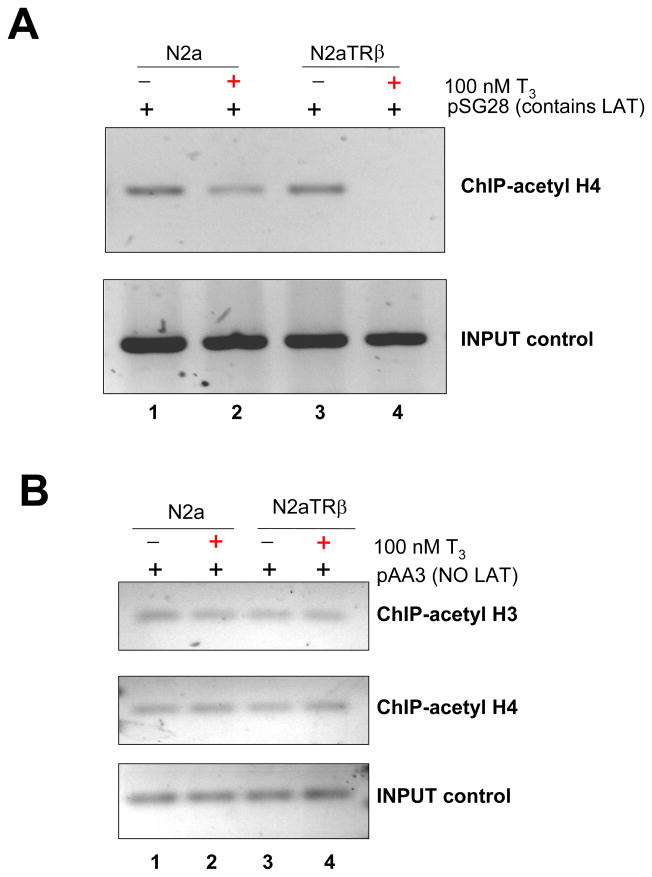
Participation of histone modification and chromatin remodeling in the gene regulation of LAT
TR modulated histone modification and recruitment of chromatin remodeling complex at the TRE-containing promoters for gene regulation [35–39]. It is likely that liganded TR exerted LAT regulation through these mechanisms. This hypothesis was investigated by ChIP using corresponding antibodies. The LAT promoter was found to be hyperacetylated in the presence of TR and T3 (Fig. 5A, compare 1 and 2). In addition, chromatin remodeling complex BRG1 was enriched at the LAT promoter by liganded TR (Fig. 5A, compare lane 1 and 2) but not at the ICP0 promoter (Fig. 5A, lane 3 to 6), suggesting that BRG1 complex may participate in the T3/TR-mediated regulation. To confirm the roles of BRG1 in HSV-1 gene expression, BRG1 over-expression vector pBRG1 and its dominant negative mutant pK785R were transfected with pSG28 in N2aTRβ cells. The RT-PCR assays showed that the overexpression of BRG1 had little effect on LAT in the absence of TR but further increased the TR-mediated LAT transcription in the presence of T3 (Fig. 5B, compare lane 1–5). The dominant negative mutant K785R, on the contrary, abolished the activation (Fig. 5B, lane 6). These results demonstrated that histone modification and chromatin remodeling both contribute to the T3/TR-mediated LAT regulation.
Fig. 5. Participation of chromatin remodeling and histone acetylation in LAT transcription by liganded TR.
5A: ChIP was performed using anti-BRG1 and anti-acetyl histone H4 Ab followed by PCR using specific primers. IgG controls showed no signal, indicating the specificity of the assays (data not shown). Lane 1 and 2: LAT TRE ChIP primers; Lane 3 to 6: ICP0 promoter primers.
5B: Cells were cotransefected with pSG28 and with or without expression vectors of BRG1 or K785R specified in the figure. After 48 h, the transfected cells were subjected to RNA isolation and RT-PCR assays using primers recognizing LAT transcript. Actin primers were used as controls.
Histone H4 hypoacetylation was enriched at the ICP0 promoter by liganded TR in the presence of LAT TRE
The remodeling complex of BRG1 appeared to participate in the LAT regulation but it did not exert a regulatory effect on ICP0 since overexpression of BRG1 and K786R exhibited no impact on ICP0 expression (data not shown). Nonetheless, the ICP0 promoter was hypoacetylated in the presence of liganded TR since the ChIP assays showed that the histone H4 acetylation was reduced at the ICP0 promoter in the presence of TR and T3 (Fig. 6A). This TR/T3-mediated hypoacetylation required the presence of LAT TREs, evidenced by the fact that lack of LAT TRE did not show hypoacetylation at the ICP0 promoter (Fig. 6B). Collectively, these results demonstrate that liganded TR induced hypoacetylation at the ICP0 promoter and it is likely the results of chromatin insulation mediated via LAT TREs.
Fig. 6. T3/TR-mediated ICP0 promoter histone modification required the presence of LAT TRE.
6A: Cells were transfected with pSG28 (containing LAT TRE) and incubated with or without T3 for 48 h followed by ChIP with anti-acetyl histone H4 Ab. The IP chromatin solution was purified and the resulting DNA was amplified by primers recognizing ICP0 promoter.
6B: Parallel experiments to 5A using plasmid pAA3 (containing no LAT TRE) were done with the anti-acetyl histone H4 Ab as well as acetyl histone H3 Ab for IP. The histone modification was analyzed by the same ICP0 primers.
T3 washout de repressed ICP0 and increased infectious virus release
To investigate the effect of T3 on TR-mediated regulation during HSV-1 replication and release of infectious virus, N2a and N2aTRβ cells were pre-treated with T3 for 5 days followed by infection at moi of 1 with or without T3 washout (Fig. 7A). RT-PCR assays showed that the expression of ICP0 was de-repressed upon washout of T3 at 48 hpi in N2aTRβ but not N2a cells (Fig. 7B). The plaque assays indicated that the T3 washout increased the virus release 60-fold more than the cells being treated with T3 in N2aTRβ (Fig. 7C). There was no change in N2a cells upon T3 removal (Fig. 7C). These results suggested that subtraction of hormone reversed the ICP0 repression and this viral protein may play an active role during viral replication in resting cells and enhanced the viral gene expression and virus release.
Discussion
This study focused on the roles of TR and its ligand T3 in the regulation of HSV-1 gene expression in neuronal cells. Our data demonstrated that transcription factor TR utilized its ligand to exert differential regulatory effect on HSV-1 genes LAT and ICP0 in neuronal cells. The results, for the first time, showed that liganded TR bound LAT TRE and activated LAT promoter via histone modification as well as BRG1-mediated chromatin remodeling. In addition, liganded TR mediated down-regulation on ICP0, at least in part, by histone hypoacetylation and LAT TRE was required. Current model using T3 -treated N2aTRβ cell was particularly useful during initial in vitro studies since N2aTRβ cells were differentiated after T3 incubation, mimicking the sensory neurons in vivo. Investigations using animal models are underway and the preliminary data indicated that T3 treatment decreased and delayed the virus release during TG explant assays, supporting the hypothesis that T3 participated in the HSV-1 latency and reactivation (data not shown).
BRG1 complexes disrupts histone-DNA interactions and have been implicated in the regulation of transcriptional activation [40]. Involvement of BRG1 chromatin remodeling complex in the gene regulation via nuclear hormone receptors was identified by modifying the nucleosomal structure to allow transcriptional activation or repression [41]. In vitro assays with several nuclear hormone receptors, including the estrogen receptor and the glucocorticoid receptor, have shown that BRG1 can modify the pattern of nucleosomal positioning and aid local disruption within preassembled nucleosomal display [42]. Our results indicated that BRG1 was recruited to the LAT promoter by liganded TR and its dominant negative mutant eliminated the activation, confirming the specificity. BRG1 contributed to HSV-1 IE gene expression through VP16-HCF-Oct1 complex [43]. Intact VP16 complexes may not be able to enter the nucleus of sensory neurons but BRG1, on the other hand, is present in neurons. It is likely that BRG1 is recruited to HSV-1 regulatory sequences via T3 and TR to regulate LAT transcription. More experiments are necessary to analyze the enrichment of chromatin remodeling complex at the HSV-1 genome in neuron.
The functions of LAT and its role on ICP0 during HSV latency and reactivation are elusive and the specific roles in gene regulation are vague. In neuronal cells, LAT was shown to reduce viral gene expression and replication during productive infection [44]. In vivo, LAT mutant virus enhanced gene expression in sensory neurons during lytic and latent infection [45]. A more recent report showed that LAT augments transcriptions of several lytic genes during the latent stage in rabbits, possibly by chromatin modifications [46]. Chromatin insulator motifs were identified within the LAT regulatory regions [22, 24]. Our results indicated that TR was bound to the LAT TREs and facilitate the enrichment of chromatin insulator binding protein CTCF to the region only in the presence of liganded TR. Under the same condition the ICP0 promoter was hypoacetylated by liganded TR. TREs and CTCF motifs were often adjacent and possessed overlapping functions [47–49]. In addition, ICP22 intron contains reiterative repeats similar in position and composition to the reiterative elements but different in its actual sequences. It is likely that the liganded TRs facilitate the binding of CTCF to the elements or directly recruit the CTCF to the sites therefore induce heterochromatin by forming a boundary elements (between LAT TRE/RE-1 and ICP22 intron) to regulate the gene expression. Mutant viruses with specific deletions are underway to investigate the T3/TR-mediated effects of euchromatin/heterochromatin on viral gene expression and silencing in animal models.
Our results further indicated that TR and T3 repressed HSV-1 TK expression and modulate viral replication/release in neuronal cells (unpublished data). TK is required to provide dNTP for viral replication in resting cells such as neurons and viral replication is required for efficient α and β expressions in neurons during reactivation [50]. TK was further suggested to initiate α transcription and subsequent replication during reactivation [51] and the mutant virus lacking TK exhibited greatly reduced α and β expression during reactivation, suggesting the importance of TK [52]. Based on these results, T3 and TR may exert additional regulation to control HSV-1 replication and gene expression during latency/reactivation.
Together, our studies suggested that thyroid hormone and its receptor are able to recruit different cofactors to control critical transcription of various HSV-1 genes in neuronal cells and may have implication to control latency and reactivation. More experiments especially mutant viruses in cell culture and animal models are necessary to identify key mechanisms involved in HSV-1 latency and viral reactivation through the TR/T3 - mediated regulations.
Acknowledgments
This project is supported by NIH/NCRR P20RR016456. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCRR/NIH. We thank Dr. Keji Zhao from NHLBI/NIH for BRG1 expression vectors. We are grateful for the 17Syn+-EGFP virus from Dr. James Hill of LSUHSC in New Orleans, LA. Our gratitude also goes to Dr. Robert Denver (Ann Arbor, MI) for the N2aTRβ cell line.
Abbreviations
- HSV-1
Herpes Simplex Virus Type 1
- IE gene(s)
Immediate-Early gene(s)
- ICP0
The infected cell protein No. 0
- HDACs
Histone Deacetylase Complexes
- LAT
Latency-associated transcript
- ChIP
Chromatin Immunoprecipitation
- CTCF
CCCTC-binding factor
- BRG1
brahma-related gene 1
- hpi
hours post infection
References
- 1.Goldenberg D, et al. The abundant latency-associated transcripts of herpes simplex virus type 1 are bound to polyribosomes in cultured neuronal cells and during latent infection in mouse trigeminal ganglia. J Virol. 1997;71(4):2897–904. doi: 10.1128/jvi.71.4.2897-2904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Javier RT, et al. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology. 1988;166(1):254–7. doi: 10.1016/0042-6822(88)90169-9. [DOI] [PubMed] [Google Scholar]
- 3.Jones C. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin Microbiol Rev. 2003;16(1):79–95. doi: 10.1128/CMR.16.1.79-95.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu TT, et al. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70(9):5962–7. doi: 10.1128/jvi.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook ML, Thompson RL, Stevens JG. A herpes simplex virus mutant is temperature sensitive for reactivation from the latent state: evidence for selective restriction in neuronal cells. Virology. 1986;155(1):293–6. doi: 10.1016/0042-6822(86)90192-3. [DOI] [PubMed] [Google Scholar]
- 6.Ecob-Prince M, Hassan K. Reactivation of latent herpes simplex virus from explanted dorsal root ganglia. J Gen Virol. 1994;75(Pt 8):2017–28. doi: 10.1099/0022-1317-75-8-2017. [DOI] [PubMed] [Google Scholar]
- 7.Kriesel JD, et al. Neuronal reactivation of herpes simplex virus may involve interleukin-6. J Neurovirol. 1997;3(6):441–8. doi: 10.3109/13550289709031190. [DOI] [PubMed] [Google Scholar]
- 8.Sawtell NM, Thompson RL. Comparison of herpes simplex virus reactivation in ganglia in vivo and in explants demonstrates quantitative and qualitative differences. J Virol. 2004;78(14):7784–94. doi: 10.1128/JVI.78.14.7784-7794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koelle DM, Corey L. Herpes Simplex: Insights on Pathogenesis and Possible Vaccines. Annu Rev Med. 2008;59:381–395. doi: 10.1146/annurev.med.59.061606.095540. [DOI] [PubMed] [Google Scholar]
- 10.Vider-Shalit T, et al. Phase-dependent immune evasion of herpesviruses. J Virol. 2007;81(17):9536–45. doi: 10.1128/JVI.02636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bystricka M, Russ G. Immunity in latent Herpes simplex virus infection. Acta Virol. 2005;49(3):159–67. [PubMed] [Google Scholar]
- 12.Morrison LA, Knipe DM. Mechanisms of immunization with a replication-defective mutant of herpes simplex virus 1. Virology. 1996;220(2):402–13. doi: 10.1006/viro.1996.0328. [DOI] [PubMed] [Google Scholar]
- 13.Cui C, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80(11):5499–508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Block T, et al. Long term herpes simplex virus type 1 infection of nerve growth factor-treated PC12 cells. J Gen Virol. 1994;75(Pt 9):2481–7. doi: 10.1099/0022-1317-75-9-2481. [DOI] [PubMed] [Google Scholar]
- 15.Moxley MJ, et al. Herpes simplex virus type 1 infection prevents detachment of nerve growth factor-differentiated PC12 cells in culture. J Gen Virol. 2002;83(Pt 7):1591–600. doi: 10.1099/0022-1317-83-7-1591. [DOI] [PubMed] [Google Scholar]
- 16.Su YH, et al. The HSV 1 genome in quiescently infected NGF differentiated PC12 cells can not be stimulated by HSV superinfection. J Neurovirol. 2000;6(4):341–9. doi: 10.3109/13550280009030760. [DOI] [PubMed] [Google Scholar]
- 17.Su YH, et al. Stability and circularization of herpes simplex virus type 1 genomes in quiescently infected PC12 cultures. J Gen Virol. 2002;83(Pt 12):2943–50. doi: 10.1099/0022-1317-83-12-2943. [DOI] [PubMed] [Google Scholar]
- 18.Garza HH, Jr, Hill JM. Effect of a beta-adrenergic antagonist, propranolol, on induced HSV-1 ocular recurrence in latently infected rabbits. Curr Eye Res. 1997;16(5):453–8. doi: 10.1076/ceyr.16.5.453.7051. [DOI] [PubMed] [Google Scholar]
- 19.Hardwicke MA, Schaffer PA. Differential effects of nerve growth factor and dexamethasone on herpes simplex virus type 1 oriL- and oriS-dependent DNA replication in PC12 cells. J Virol. 1997;71(5):3580–7. doi: 10.1128/jvi.71.5.3580-3587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquart M, et al. Ocular reactivation phenotype of HSV-1 strain F(MP)E, a corticosteroid-sensitive strain. Curr Eye Res. 2003;26(3–4):205–9. doi: 10.1076/ceyr.26.3.205.14890. [DOI] [PubMed] [Google Scholar]
- 21.Noisakran S, et al. Role of the hypothalamic pituitary adrenal axis and IL-6 in stress-induced reactivation of latent herpes simplex virus type 1. J Immunol. 1998;160(11):5441–7. [PubMed] [Google Scholar]
- 22.Amelio AL, McAnany PK, Bloom DC. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol. 2006;80(5):2358–68. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedadala GR, Pinnoji RC, Hsia SC. Early growth response gene 1 (Egr-1) regulates HSV-1 ICP4 and ICP22 gene expression. Cell Research. 2007;17(5):1–10. doi: 10.1038/cr.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, et al. A CTCF-dependent chromatin boundary element exists between the LAT and ICP0 promoters in the HSV-1 genome. J Virol. 2007 doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubat NJ, et al. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J Virol. 2004;78(22):12508–18. doi: 10.1128/JVI.78.22.12508-12518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubat NJ, et al. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. Journal of Virology. 2004;78(3):1139–49. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinnoji RC, et al. Repressor element-1 silencing transcription factor/neuronal restrictive silencer factor (REST/NRSF) can regulate HSV-1 immediate-early transcription via histone modification. Virol J. 2007;4:56. doi: 10.1186/1743-422X-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6(3):211–21. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 29.Deme D, Pommier J, Nunez J. Kinetics of thyroglobulin iodination and of hormone synthesis catalysed by thyroid peroxidase. Role of iodide in the coupling reaction. Eur J Biochem. 1976;70(2):435–40. doi: 10.1111/j.1432-1033.1976.tb11034.x. [DOI] [PubMed] [Google Scholar]
- 30.Papavasiliou SS, et al. Thyroid hormonelike actions of 3,3′,5′-L-triiodothyronine nad 3,3′-diiodothyronine. J Clin Invest. 1977;60(6):1230–9. doi: 10.1172/JCI108882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14(2):184–93. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 32.Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–86. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 33.Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81(3):1097–142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 34.Hsia SC, Wang H, Shi YB. Involvement of chromatin and histone acetylation in the regulation of HIV-LTR by thyroid hormone receptor. Cell Res. 2001;11(1):8–16. doi: 10.1038/sj.cr.7290061. [DOI] [PubMed] [Google Scholar]
- 35.Hsia SC, Shi YB. Chromatin disruption and histone acetylation in regulation of the human immunodeficiency virus type 1 long terminal repeat by thyroid hormone receptor. Mol Cell Biol. 2002;22(12):4043–52. doi: 10.1128/MCB.22.12.4043-4052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsia SC, et al. Role of chromatin disruption and histone acetylation in thyroid hormone receptor action: implications in the regulation of HIV-1 LTR. Histol Histopathol. 2003;18(1):323–31. doi: 10.14670/HH-18.323. [DOI] [PubMed] [Google Scholar]
- 37.Huang ZQ, et al. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. Embo J. 2003;22(9):2146–55. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee KC, et al. Transcriptional activation by thyroid hormone receptor-beta involves chromatin remodeling, histone acetylation, and synergistic stimulation by p300 and steroid receptor coactivators. Mol Endocrinol. 2003;17(5):908–22. doi: 10.1210/me.2002-0308. [DOI] [PubMed] [Google Scholar]
- 39.Wallberg AE, et al. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12(5):1137–49. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 40.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–89. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 41.Yoshinaga SK, et al. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Zhang B, Faller DV. BRG1/BRM and prohibitin are required for growth suppression by estrogen antagonists. Embo J. 2004;23(11):2293–303. doi: 10.1038/sj.emboj.7600231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. Journal of Virology. 2004;78(18):9689–96. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mador N, et al. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate-early gene mRNA levels in a neuronal cell line. J Virol. 1998;72(6):5067–75. doi: 10.1128/jvi.72.6.5067-5075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garber DA, Schaffer PA, Knipe DM. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J Virol. 1997;71(8):5885–93. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giordani NV, et al. During Hsv-1 Infection of Rabbits, the Ability to Express the Lat Increases Latent-Phase Transcription of Lytic Genes. J Virol. 2008 doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Awad TA, et al. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274(38):27092–8. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- 48.Burke LJ, et al. The thyroid hormone receptor and the insulator protein CTCF: two different factors with overlapping functions. J Steroid Biochem Mol Biol. 2002;83(1–5):49–57. doi: 10.1016/s0960-0760(02)00256-x. [DOI] [PubMed] [Google Scholar]
- 49.Lutz M, et al. Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. Embo J. 2003;22(7):1579–87. doi: 10.1093/emboj/cdg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichol PF, et al. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. Journal of Virology. 1996;70(8):5476–86. doi: 10.1128/jvi.70.8.5476-5486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tal-Singer R, et al. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995;69(7):4471–83. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosz-Vnenchak M, et al. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J Virol. 1993;67(9):5383–93. doi: 10.1128/jvi.67.9.5383-5393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]