Abstract
We developed a novel brain atlas template to facilitate computational brain studies of Chinese subjects and populations using high quality magnetic resonance imaging (MRI) and well-validated image analysis techniques. To explore the ethnicity-based structural brain differences, we used the MRI scans of 35 Chinese male subjects (24.03±2.06yr) and compared them to an age-matched cohort of 35 Caucasian males (24.03±2.06yr). Global volumetric measures were used to identify significant group differences in the brain length, width, height and AC-PC line distance. Using the LONI BrainParser, 56 brain structures were automatically labeled and analyzed for all subjects. We identified significant ethnicity differences in brain structure volumes, suggesting that a population-specific brain atlas may be more appropriate for studies involving Chinese populations. To address this, we constructed a 3D Chinese brain atlas based on high resolution 3.0T MRI scans of 56 right-handed male Chinese volunteers (24.46±1.81yr). All Chinese brains were spatially normalized by using linear and nonlinear transformation via the “AIR Make Atlas” pipeline workflow within the LONI pipeline environment. This high-resolution Chinese brain atlas was compared to the ICBM152 template, which was constructed using Caucasian brains.
Keywords: Brain atlas, Chinese, MRI, 3.0 Tesla, Registration, Neuroimaging, Brain mapping
Introduction
Modern imaging technologies have profoundly advanced our understanding of human brain structure and function in health and disease (Demetriades, 2002; Giraud et al., 2001; Goldstein and Volkow, 2002; Kasai et al., 2002; Lee et al., 2001; Munte et al., 2002; Theodore and Gaillard, 2002). Human brains are highly variable between different individuals within a group, and between phenotypic groups (e.g., age, race). Therefore, spatial normalization is an important pre-processing step used to reduce inter-subject anatomical variability. A brain template, or an atlas, provides a standard anatomical reference for individual or population based assessment of brain structure and function (Ashburner and Friston, 1999; Collins et al., 1994; Evans AC, 1993; Lancaster et al., 1999; Mazziotta et al., 2001; Toga and Thompson, 2001).
A commonly used human brain coordinate system employed in different brain mapping studies is that of Talairach and Tournoux (Fox et al., 1985; Talairach J, 1988). The Talairach atlas, however, does not necessarily represent the in vivo anatomy of all living subjects since it was based on the postmortem sections of a 60-yr old French female. The uneven slice separations, ranging from 3 to 4mm, and the inconsistency of the orthogonal plane sections are also limitations of the Talairach atlas. The International Consortium for Brain Mapping (ICBM) created another standard brain template to address the limitations of the Talairach brain template by averaging a large group of MRI scans of 305 young normal subjects (239 males and 66 females, age: 23.4±4.1yr). These ICBM scans were first spatially normalized into the Talairach space by linear registrations (Collins et al., 1994; Evans AC, 1993; Toga and Thompson, 2001). One of the most popular brain atlases is the ICBM152 atlas,which is the average of 152 normal MRI scans aligned into a common space using a 9 parameter transformation (Ahsan et al., 2007; Chau and McIntosh, 2005; Shattuck et al., 2008; Yoon et al., 2009). The ICBM atlases are adopted by many groups, incorporated in several software packages and utilized in many volumetric studies of normal and abnormal brain anatomy and function. For example, the Statistical Parametric Mapping software (SPM, Institute of Neurology, University College of London, U.K.) promotes the ICBM atlas for diverse types of human brain mapping studies (Carmack et al., 2004; Friston KJ, 1995; Shen et al., 2005; Tzourio-Mazoyer et al., 2002).
Yet, the use of brain atlases in spatial normalization is typically limited to studies involving subject cohorts of similar phenotype (e.g., age, gender, race, disease conditions) to those used to construct the corresponding atlas (Altaye et al., 2008; Buckner et al., 2004; Jackson, 1992; Lee et al., 2005; Moriguchi et al., 2005; Smith et al., 2007; Thompson et al., 2000; Wilke et al., 2008; Zhang et al., 1998). Thus, new population-specific brain atlases are created and recommended for use with other cohorts substantially different from the populations used to generate existent atlases (Altaye et al., 2008; Lee et al., 2005; Moriguchi et al., 2005; Wilke et al., 2008). Genetics and environmental factors make the Oriental and Occidental populations dissimilar. Thus, using Caucasian atlases for Chinese populations leads to overall brain volume, shape and size differences (Chee et al., 2009; Kochunov et al., 2003). Additionally, differences in brain structures between these populations may underlie different brain functions. If a Caucasian template is used in functional studies involving Chinese subjects, such genotypic and phenotypic differences may cause inaccurate measurements, comparisons and interpretations of results.
In this study, we demonstrated significant differences in brain structures between Chinese and Caucasian populations. We also developed a novel brain atlas using high quality T1-weighted 3.0T structural magnetic resonance (MR) images constructed using the scans of 56 normal Chinese males. This Chinese brain atlas facilitates a more robust and accurate studies of brain anatomy and activation in the Chinese population.
Materials and Methods
Subjects
Sixty-three normal righted-handed Chinese young male volunteers, without a history of any neurological, psychiatric or significant medical illness, were recruited from the local community through the research center for sectional and imaging anatomy at Shandong University School of Medicine or from bulletin advertisements. All participants were examined by two neurosurgeons to exclude prior neurological diseases. The Edinburgh handedness inventory (Oldfield, 1971) was used to assess handedness. The subjects ranged in age from 20 to 30yr (mean age=24.49±1.76yr). The local ethics committee at Shandong University School of Medicine approved the study, and all participants provided written informed consents before entering the study. Thirty-five normal right-handed Caucasian young male subjects (24.03±2.06yr) were randomly selected from the ICBM database (http://ida.loni.ucla.edu). The subjects’ age distribution is presented in Table 1.
Table 1.
Age distribution of the subjects used in this study
| Groups |
|||||
|---|---|---|---|---|---|
| Chinese | Caucasian | ||||
| sex | Age (yr) |
Number of subjects |
sex | Age (yr) |
Number of subjects |
| male | 21 | 1 | male | 21 | 1 |
| male | 22 | 8 | male | 22 | 8 |
| male | 23 | 11 | male | 23 | 8 |
| male | 24 | 12 | male | 24 | 8 |
| male | 25 | 13 | male | 25 | 3 |
| male | 26 | 10 | male | 26 | 0 |
| male | 27 | 5 | male | 27 | 4 |
| male | 28 | 2 | male | 28 | 2 |
| male | 29 | 1 | male | 29 | 1 |
Mean±S.D.(yr) of Chinese group= 24.49±1.76
Mean±S.D.(yr) of Caucasian group= 24.03±2.06
Data acquisition and preparation
The 35 Caucasian subjects selected from the ICBM database were scanned with a standardized MRI protocol described in previous publications (Mazziotta et al., 2001). High resolution structural brain MRI scans were acquired at the ICBM site using a 1.5 Tesla GE SIGNA MRI scanner. All scans were collected according to the standard ICBM MRI protocol. For each subject, three-dimensional (3D) T1-weighted MRI images were acquired using a sagittal 3D spoiled gradient echo (SPGR) sequence. The typical 1.5T acquisition parameters were echo time (TE) of 4.0 ms, repetition time (TR) of 24 ms, flip angle of 35°, and 24 cm of filed of view. The acquisition matrix was 256×256×124 in the x-, y-, and z-dimensions yielding a voxel size of 0.98×0.98×1.20 mm3.
For the 63 Chinese volunteers, high-resolution T1-weighted 3D SPGR MR images were acquired on a 3.0 Tesla GE SIGNA scanner (GE Medical Systems, Milwaukee, USA). The imaging parameters were as follows: 1.40mm axial slices, TE=2.88 ms, TR=6.68 ms, inversion time=450 ms, flip angle=25° and field of view=24×24cm. The acquisition matrix was 512×512×248 in the x-, y-, and z-dimensions yielding a voxel size of 0.47×0.47×0.70 mm3.
All data was acquired in DICOM format and then imported into ANALYZE format for analysis using the import function of LONI Image Data Archive (IDA) (Van Horn et al., 2009).
Image preprocessing
Skull and other non-brain tissues of every individual brain were removed using the Brain Extraction Tool (BET) programs distributed as a part of the FSL package (Smith, 2002), an automated software program that identifies the brain region in the MRI images by using the CSF layer between pia and arachnoid matter to guide its processing. Errors in the automated segmentation were manually corrected using the BrainSuite software package (Shattuck and Leahy, 2002), which provides users with the ability to display simultaneous views of three orthogonal planes through the MRI volumes.
Delineation of global brain features
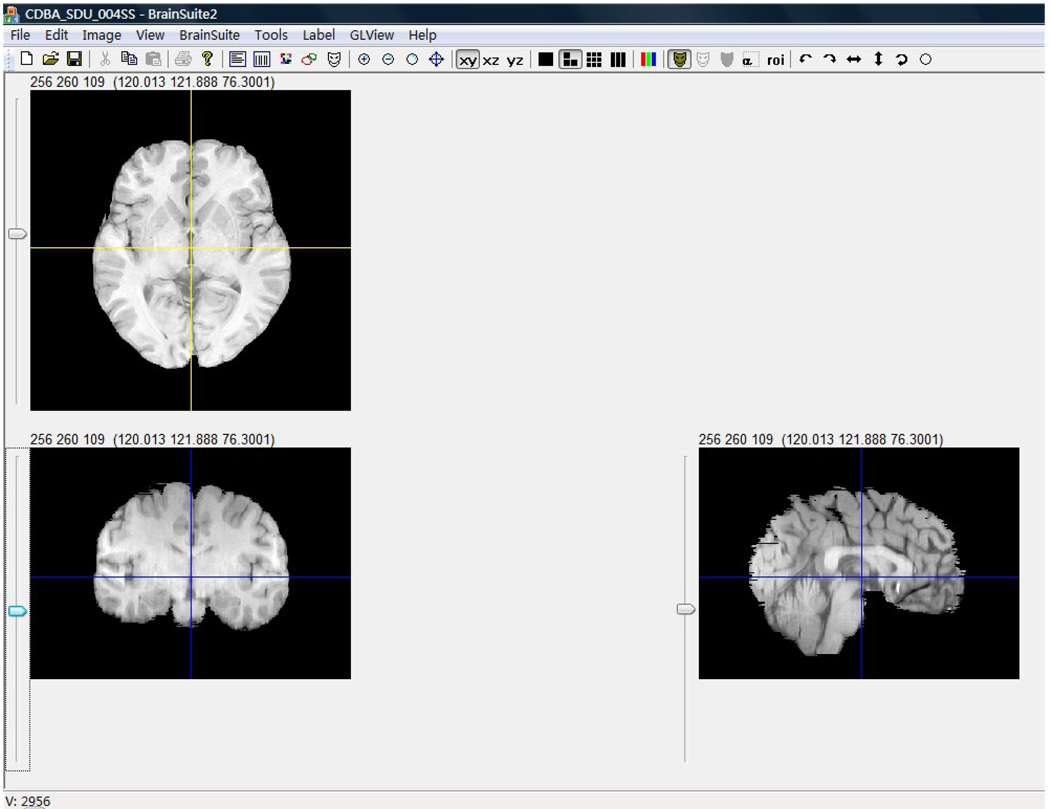
Although the global brain shape and size do not provide detailed morphometry, they are useful as a baseline for comparing different brains. The length, width, height of whole brain and the AC-PC line length are important measurements of brain shape and size. For the 35 Chinese brains and their 35 Caucasian age- and gender-matched counterparts, we computed the global morphometrics using the BrainSuite software package (Shattuck and Leahy, 2002), which provides an accurate value of voxel size and has the ability to display simultaneous views of three orthogonal planes aiding the user in determining the boundaries of different brain structures. A protocol was developed to measure four specific values (Figure 1). Using the transverse plane through the anterior and posterior commissures, we determined the following four measures: (1) the length of AC-PC line was estimated as the distance from the center of the anterior commissure to the center of the posterior commissure; (2) the length of the whole brain was the distance from the anterior pole to the posterior pole on the transverse plane cross the AC-PC line; (3) the width of the whole brain was determined as the distance of the line running through the middle point of the AC-PC line from the left pole to the right pole on the transverse plane; (4) the brain height was the distance from the superior pole to the inferior pole on the coronal plane.
Fig.1. The measurements of brain length, width, height and AC-PC line distance using BrainSuite.
The transverse plane was through the AC-PC line, the coronal plane was through the middle point of AC-PC line and vertical to the transverse plane, the sagittal plane was the median sagittal plane. Using the methods mentioned above and the value label, the four parameters were measured directly.
Delineation of brain structures
For all 35 Chinese and 35 Caucasian brain MRI scans, 56 brain structures were automatically obtained (these include 50 cortical structures, 4 sub-cortical areas, the brainstem, and the cerebellum). This was achieved using the LONI BrainParser software (Tu et al., 2008) – a machine learning-based approach, which relies on a pre-trained models of common structures of interest. The BrainParser and LONI pipeline environment (Dinov et al., 2009), which contains the LONI BrainParser workflow, are available for downloading and include all necessary pre- and post-processing steps (http://www.loni.ucla.edu/Software/). The data processing workflow included the skull-stripping of all MRI brain volumes and reorientation (Woods et al., 1998a; Woods et al., 1998b) to ensure that all images shared the same orientation. The FLIRT program was used to align all the subjects to a reference brain atlas LPBA40 (Shattuck et al., 2008) using a 12-parameter transformation. Then all the individual brain volumes were registered and deformed to the reference template LPBA40 to facilitate the segmentation of 56 brain structures. We used “ITK” registration, which included the 3D Multi-modality B-spine Deformable Registration module, followed by the auto BrainParsing module, Inverse Deformation Field module and Deformation Field Warping module. Subsequently, all labeled volumes were warped back to their original (native) spaces, again using FLIRT. All the results were inspected by two trained neuroanatomists (Tang and Sun) and the residual errors were manually corrected using the BrainSuite software package (Shattuck and Leahy, 2002) using a special protocol developed for this study. Finally, the volume of every structure was measured for all 70 subjects. The complete results table included 56 contiguously labeled structures in the delineation space for all 35 Chinese and 35 Caucasian subjects.
Template construction
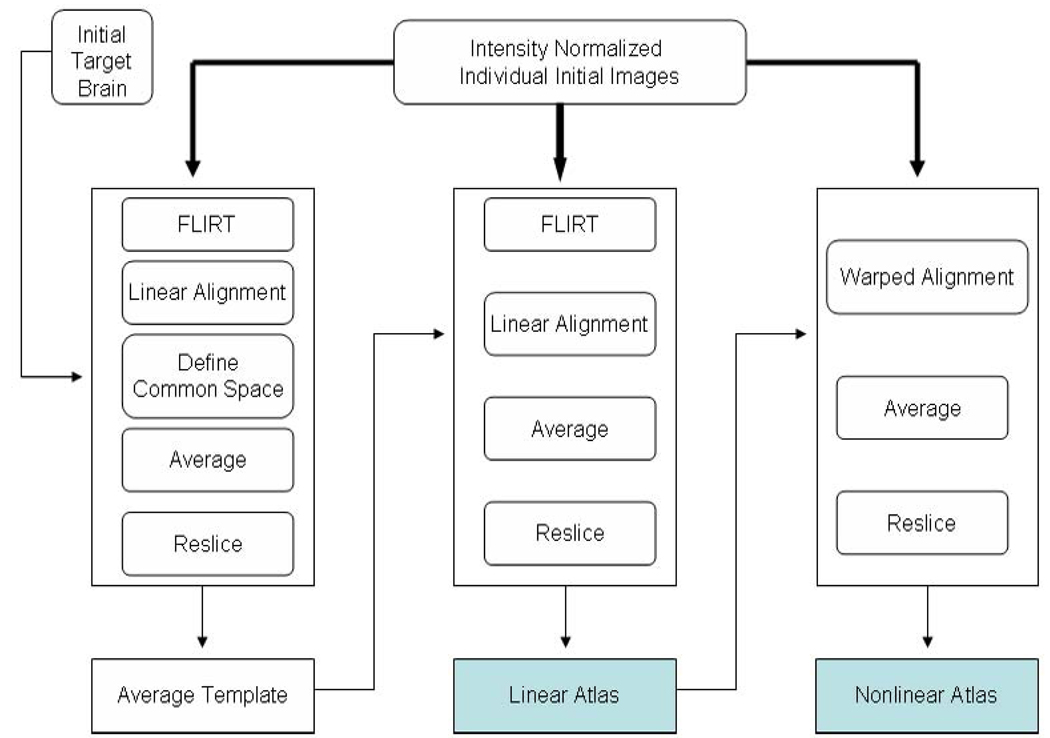
Using a modified LONI pipeline (Rex et al., 2003; Dinov et al., 2009) “AIR Make Atlas”, we constructed the Chinese brain atlas (Chinese_56) composed of high resolution 3D structure MRI data from 56 normal Chinese subjects. The Automated Image Registration Program (AIR, version 5.2.5) was used for the linear and nonlinear registrations (Woods et al., 1998a; Woods et al., 1998b). One of the brain volumes was randomly selected as an intermediate target brain and all the 56 brains (including the target brain) were linearly aligned to this target brain using a 12-parameter transformation. Then we calculated an intensity average brain template with a common position using the Define Common and Soft Mean modules. Taking the intensity average brain template as the new registration target and repeating the above steps we obtained a linearly aligned brain template by averaging the intensities of the resliced volumes across the 56 Chinese subjects. All the 56 brains were rigidly registered to the first target brain using a 6-parameter transformation to get 56 new brains with the same spatial coordinate and scale to the average brain template. The next step involved the non-linear warping of these 56 new brains (based on fifth order polynomial transformation) to the linear average brain template using the “align wrap” module. Non-brain tissue was removed by the automatic BET program and subsequent manual corrections were applied as necessary to get the 56 skull-stripped brain volumes. These were used to generate a new skull–stripped nonlinear brain atlas (Figure 2).
Fig.2. The construction protocol for the Chinese Brain template (Chinese_56).
The thick lines indicate the flow of the 56 images while narrow lines represent the flow of a single image. The entire protocol included three steps. First, the averaged template was constructed to be used as an Average Minimum Deformation Target (MDT). Second, each individual image was linearly aligned to the MDT to get a linear atlas. Third, non-linear fifth order polynomial warping was used to co-register all datasets.
Comparison between Chinese brain template (Chinese_56) and ICBM152
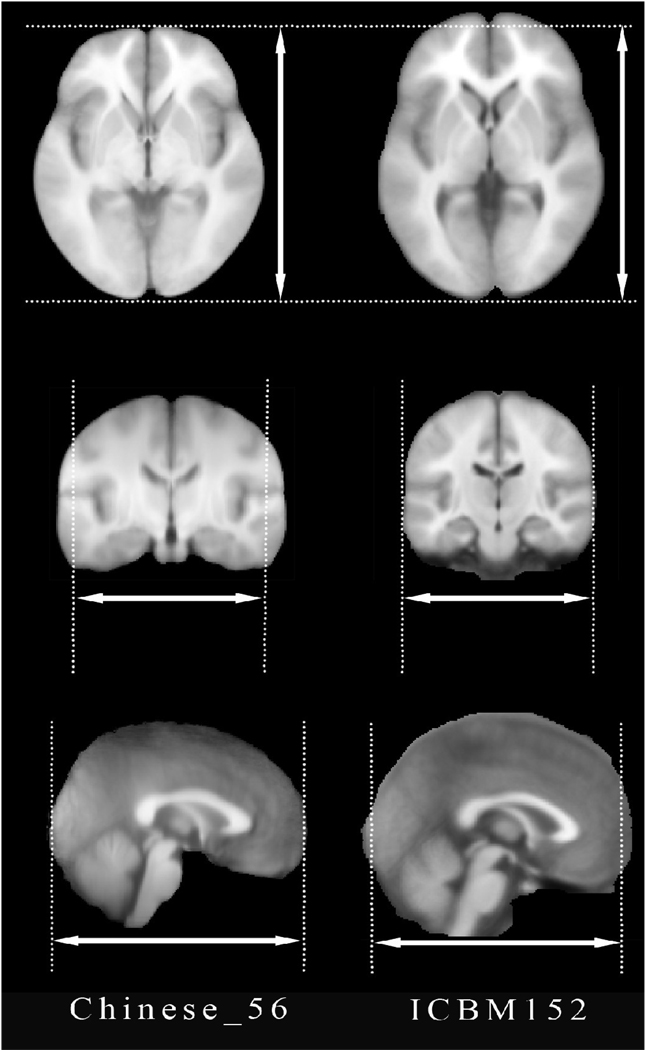
Both, the Chinese brain template (Chinese_56) and the ICBM152 were co-registered by aligning the AC-PC vector. Then the Chinese_56 template was linearly registered to the ICBM152 using a 6-parameter transformation to preserve its original characteristics in size and volume (Figure 3). Subsequently, the global features (length, width, height, AC-PC distance and ratios) of these two templates were measured.
Fig.3. Differences between the Chinese brain template (Chinese_56) (left column) and the ICBM152 atlas (right column).
Rigid body transformation (6 parameters) was applied to align two different spatial coordinates. The ethnic differences between the two populations are clearly visible–the Chinese_56 template (constructed using Chinese population) is relatively shorter but wider compared to the widely-used ICBM152 brain atlas (based on Caucasian brains).
Comparison of Image Registrations using Different Brain Atlases
In order to test the accuracy of registrations to different brain atlases, we aligned 7 new and distinct Chinese brain MRI volumes to the Chinese brain template (Chinese_56) and separately to the ICBM152 template. Both protocols used 6-parameter and 12-parameter transformations separately, and we performed the quantitative assessments of the global brain morphometry.
Results
Differences of global brain features between Chinese and Caucasian brains
We measured the shape and size of each individual brain in the two groups and found differences in global brain features between the Chinese and Caucasian cohorts. The mean values of length, width, height and AC-PC line distance of Chinese brains were 160.99mm, 142.64mm, 110.72mm and 26.28mm, respectively; while the corresponding mean values of Caucasian brains were 171.68mm, 127.48mm, 106.31mm and 28.13mm, respectively. The ratios of width/length, height/length and height/width of Chinese brains were 0.89, 0.69 and 0.78, respectively; whereas the corresponding ratios of Caucasian brains were 0.74, 0.62 and 0.83. Thus, the Caucasian brains were generally longer and the Chinese brains were generally rounder in shape. A 2-sample t-test (2-tailed) statistical analysis of these data showed that the differences of brain shape and size between Chinese and Caucasian were significant, p<0.01 (Table 2).
Table 2.
The comparisons of brain shape and size between Chinese and Caucasian subjects
| Measurement Variable |
Chinese (Mean±S.D.) |
Caucasian (Mean±S.D.) |
P |
|---|---|---|---|
| AC-PC (mm) | 26.28±1.13 | 28.13±1.42 | 4.28E-07* |
| Length (mm) | 160.99±7.30 | 171.68±9.71 | 5.46E-06* |
| Width (mm) | 142.64±5.08 | 127.48±5.04 | 4.81E-14* |
| Height (mm) | 110.72±4.54 | 106.31±6.07 | 1.68E-3* |
| W/L | 0.89±0.05 | 0.74±0.05 | 3.36E-15* |
| H/L | 0.69±0.03 | 0.62±0.03 | 6.13E-11* |
| H/W | 0.78±0.04 | 0.83±0.05 | 2.75E-07* |
P < 0.01
Differences of brain structures between Chinese and Caucasian Brains
After analyzing the volumes of all the 56 structures in the two groups, we found that in some regions, the Chinese and Caucasian brain were significantly different in volume (p<0.01) (Table 3). These differences included the left middle orbitofrontal gyrus, left gyrus rectus, left precuneus, left middle temporal gyrus, left parahippocampal gyrus, left cingulated gyrus, left lateral orbitofrontal gyrus, left superior parietal gyrus, left middle occipital gyrus, left inferior temporal gyrus, left insular cortex, left insular cortex, left putamen, right superior frontal gyrus, right precentral gyrus, right lateral orbitofrontal gyrus, right gyrus rectus, right postcentral gyrus, right precuneus, right superior occipital gyrus, right middle occipital gyrus, right superior temporal gyrus, right middle temporal gyrus,right inferior temporal gyrus, right parahippocampal gyrus, right insular cortex, right caudate and right putamen.
Table3.
The volume comparisons of 56 brain structures between Chinese and Caucasian brains
| Structure of Interest | Structure volume (mm3, mean±S.D.) |
P-value | |
|---|---|---|---|
| Chinese | Caucasian | ||
| L superior frontal gyrus | 57644.48±4989.81 | 57064.27±7201.00 | 0.67 |
| R superior frontal gyrus | 50888.59±7025.47 | 56250.75±6357.12 | 1.15E-03** |
| L middle frontal gyrus | 56006.27±5145.71 | 53958.89±7167.56 | 0.17 |
| R middle frontal gyrus | 58514.13±6040.09 | 57379.84±5870.66 | 0.42 |
| L inferior frontal gyrus | 25583.79±2636.89 | 24342.97±3242.68 | 0.09 |
| R inferior frontal gyrus | 23727.10±1735.86 | 23338.74±2838.07 | 0.44 |
| L precentral gyrus | 24666.88±2606.66 | 25092.05±3050.72 | 0.52 |
| R precentral gyrus | 21763.39±5537.82 | 27866.06±3312.28 | 1.93E-06** |
| L middle orbitofrontal gyrus | 7566.84±804.30 | 6928.57±1002.36 | 8.10E-03** |
| R middle orbitofrontal gyrus | 8521.66±1028.10 | 7861.66±1032.50 | 1.04E-2* |
| L lateral orbitofrontal gyrus | 4266.28±574.53 | 6033.33±873.46 | 2.13E-10** |
| R lateral orbitofrontal gyrus | 4239.32±579.68 | 4974.21±799.83 | 5.71E-05** |
| L gyrus rectus | 3017.42±504.78 | 2129.76±510.19 | 5.96E-08** |
| R gyrus rectus | 2471.57±480.46 | 1702.01±418.11 | 2.85E-08** |
| L postcentral gyrus | 20284.75±2347.15 | 21049.22±2111.04 | 0.17 |
| R postcentral gyrus | 16600.85±3763.45 | 19677.61±2190.97 | 2.82E-04** |
| L superior parietal gyrus | 23390.42±2603.26 | 21074.49±2154.87 | 2.79E-04** |
| R superior parietal gyrus | 21975.47±2675.48 | 23564.16±2198.74 | 1.46E-02* |
| L supramarginal gyrus | 11521.73±1545.67 | 11811.24±1126.79 | 0.36 |
| R supramarginal gyrus | 12469.25±1464.76 | 12710.69±1434.01 | 0.52 |
| L angular gyrus | 15203.77±1632.96 | 15037.26±1443.83 | 0.64 |
| R angular gyrus | 18447.18±1916.54 | 19477.31±1817.84 | 2.70E-02* |
| L precuneus | 12137.93±1522.83 | 13205.68±1607.81 | 6.97E-03** |
| R precuneus | 10995.08±1248.35 | 11848.78±1327.95 | 9.61E-03** |
| L superior occipital gyrus | 6311.36±704.28 | 6200.30±686.15 | 0.44 |
| R superior occipital gyrus | 5211.84±660.73 | 6485.93±648.84 | 2.98E-09** |
| L middle occipital gyrus | 15073.21±1311.90 | 18661.44±1934.18 | 6.76E-10** |
| R middle occipital gyrus | 18403.01±1873.96 | 19654.59±2021.89 | 1.02E-02* |
| L inferior occipital gyrus | 9142.70±1231.22 | 9660.32±1762.57 | 0.17 |
| R inferior occipital gyrus | 11419.87±1530.46 | 10663.02±2170.79 | 0.08 |
| L cuneus | 5244.20±867.77 | 5496.47±910.06 | 0.25 |
| R cuneus | 6223.08±969.28 | 6065.47±888.31 | 0.48 |
| L superior temporal gyrus | 28594.71±2693.58 | 26973.74±3191.58 | 3.66E-02* |
| R superior temporal gyrus | 29649.06±3231.65 | 26432.93±2826.63 | 1.25E-04** |
| L middle temporal gyrus | 25696.55±2867.01 | 22483.18±2299.08 | 1.64E-05** |
| R middle temporal gyrus | 26577.45±2215.72 | 23933.98±2511.38 | 8.54E-05** |
| L inferior temporal gyrus | 23633.17±2576.37 | 21178.43±2721.99 | 3.25E-04** |
| R inferior temporal gyrus | 24720.58±2749.27 | 20895.54±2221.88 | 1.41E-06** |
| L parahippocampal gyrus | 6424.13±653.62 | 5453.03±757.55 | 2.46E-06** |
| R parahippocampal gyrus | 5073.07±495.95 | 5694.38±610.20 | 5.80E-05** |
| L lingual gyrus | 15748.01±1779.30 | 13572.10±2145.05 | 2.51E-04** |
| R lingual gyrus | 17354.04±1783.37 | 16356.78±2205.38 | 3.95E-02* |
| L fusiform gyrus | 15200.09±1707.50 | 14488.17±2004.46 | 0.16 |
| R fusiform gyrus | 11942.85±1142.30 | 12436.13±1484.28 | 0.15 |
| L insular cortex | 8502.61±839.56 | 9233.53±1184.69 | 4.08E-03** |
| R insular cortex | 7318.65±607.24 | 8493.78±1128.77 | 1.20E-06** |
| L cingulate gyrus | 14802.37±1684.33 | 13156.91±1707.91 | 1.46E-04** |
| R cingulate gyrus | 14318.17±1220.41 | 14115.34±1782.29 | 0.53 |
| L caudate | 1755.38±310.98 | 2139.05±393.75 | 1.07E-05** |
| R caudate | 1932.99±405.03 | 2602.73±494.21 | 1.49E-08** |
| L putamen | 4419.54±573.83 | 3010.55±639.22 | 5.71E-11** |
| R putamen | 3691.07±436.20 | 2580.09±327.13 | 4.49E-13** |
| L hippocampus | 3641.91±447.45 | 3557.26±446.24 | 0.38 |
| R hippocampus | 3539.88±400.84 | 3574.67±368.71 | 0.69 |
| cerebellum | 151769.23±9589.18 | 146131.74±13991.38 | 0.11 |
| brainstem | 31162.27±1721.06 | 29251.38±2818.32 | 3.12E-03** |
Note : R : right L : left
P<0.01 ;
P<0.05
An Average Chinese Brain Atlas
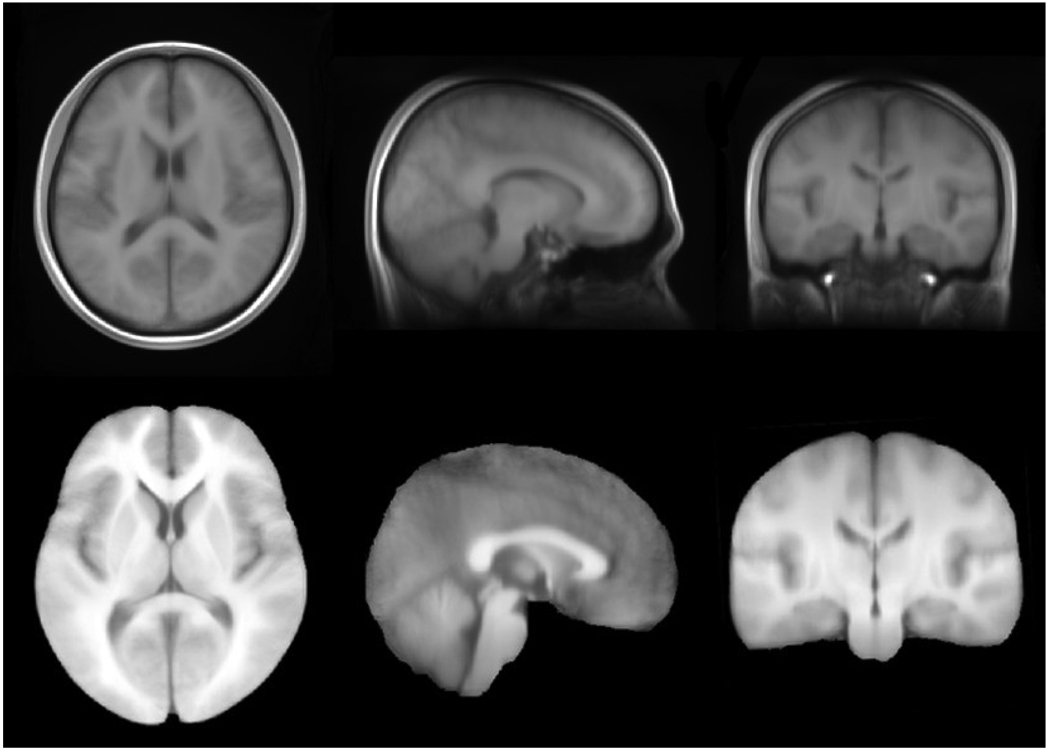
We constructed an average brain template composed of high quality brain MRI data from 56 Chinese young subjects (Figure 4). In order to compare the Chinese brain template (Chinese_56) to the ICBM152 template directly, the Chinese_56 atlas was rigidly aligned in ICBM152 space using a 6-parameter transformation to preserve its original characteristics in size and volume (Figure 3). Then the sizes of the bounding boxes and total volume were measured. As a result, the Chinese_56 is relatively shorter but wider than the ICBM152, which is based on a Caucasian population. Therefore, our results confirmed previously reported differences between Occidental and Oriental brains (Kim et al., 2005; Zilles et al., 2001). Note that the width/length ratio of the Chinese_56 is closer to 1 than that of the ICBM152-i.e., the Chinese brain template is closer to a cube compared to the ICBM template. We also measured the global features of the two brain templates and found some significant differences between them (Table 4).
Fig.4. Stereotactic representation of the Chinese brain template (Chinese_56) which is constructed using high quality brain MRI scans of 56 young Chinese adults.
Both the whole brain template (top panel) and skull-stripped brain template (lower panel) were generated automatically using a modified version of the LONI pipeline “AIR Make Atlas”. The resolution of the Chinese_56 atlas is 0.47×0.47×0.47 mm3, which is much higher than ICBM152 with the resolution of 1×1×1 mm3.
Table 4.
Comparisons between the Chinese and the ICBM brain templates
| Measurement | Chinese brain atlas (Chinese_56) | ICBM152 Atlas |
|---|---|---|
| AC-PC(mm) | 26.25 | 28.00 |
| Length (mm) | 168.77 | 177.00 |
| Width (mm) | 144.39 | 136.00 |
| Height (mm) | 110.64 | 124.00 |
| Width/Length | 0.86 | 0.77 |
| Height/Length | 0.66 | 0.70 |
| Height/Width | 0.77 | 0.91 |
The observed differences between the global Chinese_56 and ICBM152 atlases may be explained by dissimilarities of genetics and environmental exposures. The Chinese_56 atlas is relatively shorter but wider than the ICBM brain template, the width/length ratio of the Chinese brain template is closer to 1 than that of the ICBM152, the Chinese brain template is closer to a cubic square.
The Chinese_56 Atlas as an Image Registration Target
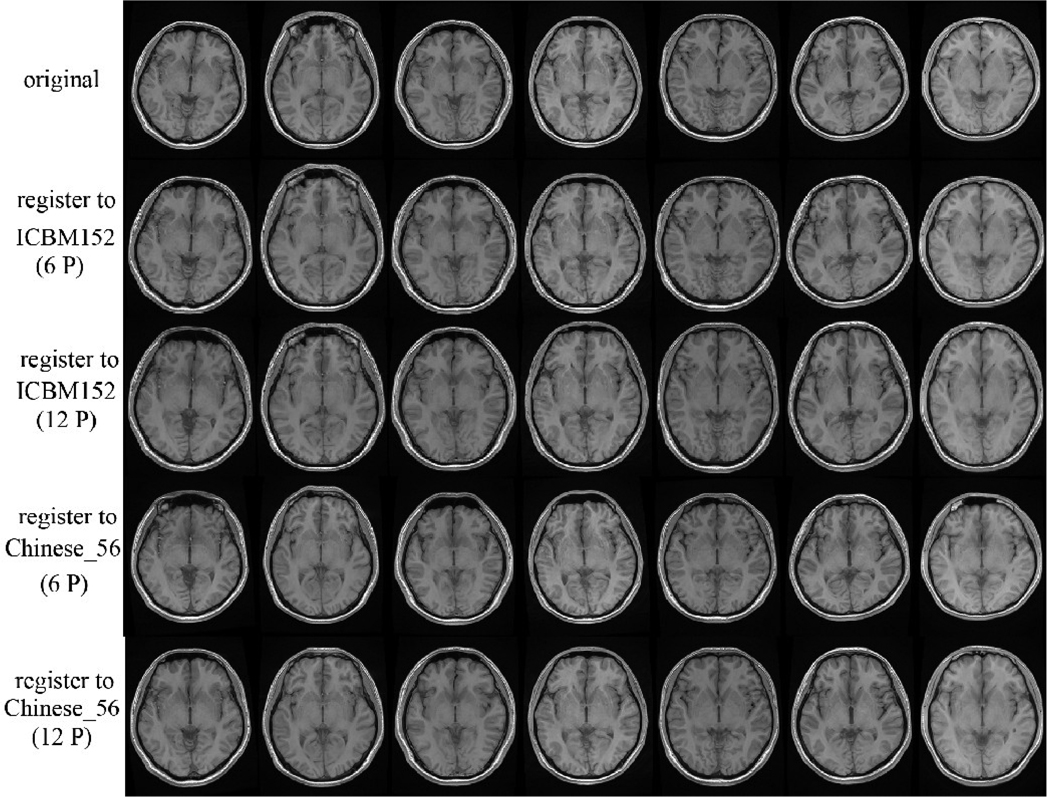
After comparing the image registration to the Chinese brain template (Chinese_56) and to the ICBM152 template (Figure 5) we assessed quantitatively some global brain features (Table 5). We found that more significant deformations were required to register the 7 additional l Chinese brains to the ICBM152 template than to the Chinese_56 atlas using a 12-parameter transformation. This suggests that the Chinese brain template (Chinese_56) better represents the shape and size of the Chinese population.
Fig.5. Validation of the Chinese_56 atlas.
Seven individual Chinese brain MRI volumes were separately aligned to the Chinese brain template (Chinese_56) and ICBM152 template using both 6-parameter and 12-parameter transformations. These results illustrate that 12-parameter transformation significantly reduces the individual brain variability for both target. The Chinese brain template (Chinese_56) is more precise as a registration target than the ICBM152 template, as it reduces the severity of the distortion necessary to co-register all subjects in a common stereotactic space.
Table5.
Brain shape and size differences of registering separately the 7 additional Chinese subjects into the ICBM152 and the Chinese_56 atlas spaces
| Measurement (Mean±S.D.) |
Original brains |
Registered to ICBM152(12P) |
Registered to Chinese_56(12P) |
P-value |
|
|---|---|---|---|---|---|
| P1 | P2 | ||||
| AC-PC (mm) | 25.52±1.53 | 28.57±1.72 | 26.79±1.34 | 1.74E-03* | 0.03 |
| Length (mm) | 157.99±4.25 | 172.14±6.20 | 167.36±1.88 | 2.83E-03* | 7.98E-04* |
| Width (mm) | 141.44±3.69 | 140.57±2.76 | 144.46±2.97 | 0.42 | 0.02 |
| Height (mm) | 108.80±5.14 | 127.00±4.16 | 109.10±2.90 | 1.45E-04* | 0.87 |
| W/L | 0.90±0.04 | 0.82±0.03 | 0.86±0.02 | 7.40E-03* | 0.05 |
| H/L | 0.69±0.05 | 0.74±0.04 | 0.66±0.02 | 0.07 | 0.06 |
| H/W | 0.77±0.03 | 0.90±0.03 | 0.76±0.03 | 2.89E-05* | 0.25 |
P < 0.01
Seven new Chinese brain MRI volumes were aligned separately to both the Chinese brain template (Chinese_56) and the ICBM152 atlas using a 12-parameter transformation. A quantitative assessment of the brain global features was performed. P1 was the statistical significacet for the measured values of original brains and the brains registering to the ICBM152 atlas. Similarly, P2 was the statistical significance for the measured values of original brains and the brains registering to the Chinese_56. These results show that more deformation was required in brain shape and size when Oriental brains are registered to the ICBM152 template using a 12-parameter transformation.
Discussion
In this paper, we presented the construction, validation and utilization of a new Chinese brain template using high resolution 3.0T T1 structural MR images. The within- and between-slice spatial resolution was chosen to be 0.47×0.47mm and 0.7mm, respectively, to achieve a good spatial resolution for more detailed structural information. The data included a total of 63 Chinese male volunteers ranged in age from 20 to 30yr (mean age=24.49±1.76yr). The Chinese_56 atlas was created using 56 volumes and it was validated on the remaining 7 subjects.
Dissimilarities of genetics and environmental exposures between different populations lead to differences in brain structure and function. Areas of functional differences between Chinese and Caucasian groups have been identified by a rapidly growing body of imaging studies (Kuo et al., 2001; Kuo et al., 2003; Tan et al., 2001a; Tan et al., 2001b; Tan et al., 2003; Tan et al., 2000). Not all regions reported as having functional differences were seen to have anatomical difference. However, each area where anatomical differences were observed has been selectively implicated in Chinese language processing by one or more studies. Further, the areas detected as being anatomically different between the groups have shown robust and highly reproducible functional differences (Kochunov et al., 2003). To explore the anatomical differences between Chinese and Caucasian brains, we selected two comparable samples (35 subjects for each group) from the Chinese and Caucasian populations that are matched for gender and age. Although global brain shape and size can not provide detailed structural information throughout the human brain, these measures are important for comparing different brains. Analysis of these morphometric measurements indicated that the mean values of length, width, height and AC-PC line distance were significantly different (p<0.01) between the Chinese brain and Caucasian brain. Thus, if Caucasian-based brain atlases are employed as reference templates in Oriental neuroimaging studies, some bias, processing errors or localized differences may be observed that are driven by the intrinsic differences between these two cohorts, and not caused by the underlying process investigated in the studies.
We used the LONI BrainParser software (Tu et al., 2008) to measure the regional volumes of 56 brain structures for all 70 subjects. For each subject we obtained 56 contiguously labeled structures in the subject native delineation space. After analyzing the volumes of all the 56 structures, we found that some brain structures were significantly different (p<0.01) between the Chinese and Caucasian brains. For instance, some structures of the Chinese brains (e.g., the middle orbitofrontal gyrus, the gyrus rectus, the left superior parietal gyrus, the superior temporal gyrus, the middle temporal gyrus, the inferior temporal gyrus, the left parahippocampal gyrus, the lingual gyrus, the left cingulated gyrus, the putamen) are larger than their counterparts in the Caucasian brains. Whereas some structures (e.g., the right superior frontal gyrus, the right precentral gyrus, the lateral orbitofrontal gyrus, the right postcentral gyrus, the right superior parietal gyrus, the right angular gyrus, the precuneus, the right superior occipital gyrus, the middle occipital gyrus, the right parahippocampal gyrus, the insular cortex, the caudate) are smaller compared to the corresponding regions in Caucasian brains. We also found the hemispheric asymmetries of the brain structures in both of the two population groups. This confirmed previous brain-asymmetry studies (Kuo et al., 2001; Kuo et al., 2003; Tan et al., 2001a; Tan et al., 2001b; Tan et al., 2003; Tan et al., 2000). These results demonstrate the need for lateral and population-specific data processing and analysis (including atlas-based spatial normalization) in modern computational neuroimaging studies of brain structure and function.
Our findings show that global and regional anatomical brain measurements are significantly different between the Chinese and Caucasian populations. Thus, widely popular Caucasian atlas templates (Evans AC, 1993; Mazziotta et al., 2001; Talairach J, 1988; Toga and Thompson, 2001) may not provide an optimal reference framework for processing brain images from the Chinese population. This implies that appropriate population-specific atlases (e.g., ethnic, gender, age, or disease) of average brain anatomy need to be employed in neuroimaging studies of well-stratified cohorts. To address this need for Oriental populations, we developed an average brain atlas specific to the Chinese population. As shown in Table 4, the Chinese brain template is about 168.77mm in length, 144.39mm in width, 110.64mm in height and AC-PC distance is 26.25mm. Compared to the widely-used ICBM152 brain template, the Chinese brain template is relatively shorter but wider, and its height is notably smaller. Among the 3 dimensional ratios, only the width/length ratio of the Chinese brain template is greater than that of the ICBM152 counterpart. This implies that the Chinese brain template is smaller but flatter. In the previous reports, the length, width and height of the Korean standard man brain template were estimated to be 16.50cm, 14.30cm and 12.10cm, respectively (Lee et al., 2005), and Japanese hemispheres were relatively shorter but wider than European hemispheres (Zilles et al., 2001). The shape and size measurements of the new Chinese brain atlas support these previous studies of regional brain differences between Asian and Occidental populations. In addition, the new Chinese brain template is composed of high quality data originated from subjects scanned using 3.0T MRI scanner, while the ICBM152 was constructed using the data originated from 1.5T MRI scanner. As a result, the resolution of the Chinese brain template is 0.47×0.47×0.47 mm3, which is much higher than ICBM152 with the resolution of 1×1×1 mm3. Thus, the Chinese brain template may capture more detailed and precise regional-based anatomical information about Oriental brains. The accuracy of the registration to the Chinese brain atlas was evaluated and the achieved results were compared with analogous co-registration results based on the ICBM152 target. For spatial normalization of individual Chinese brains, higher deformations were required to align these subjects into the ICBM152 template, compared to overall lower deformations necessary to register Chinese brains to the new Chinese_56 atlas. This suggests that the Chinese brain atlas is more precise for 12-parameter registration of Chinese cohorts into a common stereotaxic space.
The Chinese brain template (Chinese_56) is based on 56 subjects, while the ICBM 152 is based on 152 subjects chosen from a database with more than 7000 ICBM subjects. It is not known how many subjects are needed to build an optimal average brain template. But all the subjects used in the construction of Chinese_56 template were randomly selected young men aged from 20yr to 30yr. As age and gender are important factors in the delineation of brain structures and functions, and population-specific atlases are necessary for modern computational neuroimging, the sample size of 56 is appropriate to observe global and local patterns of group anatomical difference and to construct a brain template for Chinese populations. Cohort-specific brain templates are important for multi-subject structural or functional brain studies. Although this study focused only on ethnicity-specific brain atlases, it demonstrates that further research on other phenotypic characteristics such as gender, age, and disease should be taken into account for optimal and powerful analyses of regional brain morphometry. Such group-specific templates may replace the static atlases that are currently provided as default with many research tools. A population-specific brain atlas may increase the accuracy of the results by improving the statistical power and decreasing type I and type II error rates. For example, Oriental neuroimaging studies may replace the ICBM Caucasian brain atlas by the Chinese_56 template, as there are structural differences between these two populations (Kim et al., 2005) . Therefore, it is necessary to make and distribute novel cohort-specific brain templates of well-stratified populations in the future. We will continue to collect and augment our Chinese database using different sub-populations (i.e., ethnic, gender, age, or disease), extend the Chinese brain atlas and establish smaller customized phenotypic brain templates for different sub-groups. There are also some potential limitations of the Chinese_56 atlasing framework. For instance, the Chinese_56 template is constructed using 3.0T MRI scans, whereas the 1.5T ICBM152 template and some future Oriental subjects studies using the Chinese_56 atlas may use 1.5T MRI acquisition protocols. These differences in the strength of the magnetic fields may introduce important variations between the imaging characteristics of the data and the Chinese_56 template (e.g., field effects, tissue intensity distributions, image contrasts, etc.). In the construction of the Chinese_56 atlas we averaged the set of 56 initially-aligned brains. This may cause anatomical detail to be lost, due to the reslicing interpolation. To mitigate this limitation we used sinc interpolation, which introduces the smallest (aliasing) artifacts. The final registration step employed a polynomial warp of 5th degree. The decision of the complexity of the final registration step may also affect the final atlas, as 5th degree may not be the optimal complexity for this population. An alternative is to geometrically average the warping fields, instead of the resliced volumes, and avoid the anatomical intensity averaging of the interpolation step.
Acknowledgments
This work was funded by the National Natural Science Foundation of China through the NSFC Roadmap for Basic Medical Research, Grant number: 30470905 (PI: Shuwei Liu). This work was funded in part by the National Institutes of Health through the NIH Roadmap for Medical Research, Grants U54 RR021813, NIH/NCRR 5 P41 RR013642 and NIH/NIMH 5 R01 MH71940. We are also indebted to the members of the Laboratory of Neuro Imaging, our collaborators for providing useful feedback about Pipeline usability.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahsan RL, Allom R, Gousias IS, Habib H, Turkheimer FE, Free S, Lemieux L, Myers R, Duncan JS, Brooks DJ, Koepp MJ, Hammers A. Volumes, spatial extents and a probabilistic atlas of the human basal ganglia and thalamus. Neuroimage. 2007;38:261–270. doi: 10.1016/j.neuroimage.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Altaye M, Holland SK, Wilke M, Gaser C. Infant brain probability templates for MRI segmentation and normalization. Neuroimage. 2008;43:721–730. doi: 10.1016/j.neuroimage.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Hum Brain Mapp. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage. 2004;23:724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Carmack PS, Spence J, Gunst RF, Schucany WR, Woodward WA, Haley RW. Improved agreement between Talairach and MNI coordinate spaces in deep brain regions. Neuroimage. 2004;22:367–371. doi: 10.1016/j.neuroimage.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Chau W, McIntosh AR. The Talairach coordinate of a point in the MNI space: how to interpret it. Neuroimage. 2005;25:408–416. doi: 10.1016/j.neuroimage.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Chee MW, Chen KH, Zheng H, Chan KP, Isaac V, Sim SK, Chuah LY, Schuchinsky M, Fischl B, Ng TP. Cognitive function and brain structure correlations in healthy elderly East Asians. Neuroimage. 2009;46:257–269. doi: 10.1016/j.neuroimage.2009.01.036. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Demetriades AK. Functional neuroimaging in Alzheimer's type dementia. J Neurol Sci. 2002;203–204:247–251. doi: 10.1016/s0022-510x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- Dinov ID, Van Horn JD, Lozev KM, Magsipoc R, Petrosyan P, Liu Z, Mackenzie-Graham A, Eggert P, Parker DS, Toga AW. Efficient, Distributed and Interactive Neuroimaging Data Analysis Using the LONI Pipeline. Front Neuroinformatics. 2009;3:22. doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ACCD, Mills SR, Brown ED, Kelly RL, Peters TM. 3D statistical neuroanatomical models from 305 MRI volumes. Proc IEEE Nucl Science Symp Medl Imaging Conf; 1993. pp. 1813–1817. [Google Scholar]
- Fox PT, Perlmutter JS, Raichle ME. A stereotactic method of anatomical localization for positron emission tomography. J Comput Assist Tomogr. 1985;9:141–153. doi: 10.1097/00004728-198501000-00025. [DOI] [PubMed] [Google Scholar]
- Friston KJHA, Worsley KJ, Poline JP, Frith CD, Frackowiak RS. Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp. 1995:189–210. [Google Scholar]
- Giraud AL, Truy E, Frackowiak R. Imaging plasticity in cochlear implant patients. Audiol Neurootol. 2001;6:381–393. doi: 10.1159/000046847. [DOI] [PubMed] [Google Scholar]
- Goldstein RZ, Volkow ND. Drug addiction and its underlying neurobiological basis: neuroimaging evidence for the involvement of the frontal cortex. Am J Psychiatry. 2002;159:1642–1652. doi: 10.1176/appi.ajp.159.10.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FL. Race and ethnicity as biological constructs. Ethn Dis. 1992;2:120–125. [PubMed] [Google Scholar]
- Kasai K, Iwanami A, Yamasue H, Kuroki N, Nakagome K, Fukuda M. Neuroanatomy and neurophysiology in schizophrenia. Neurosci Res. 2002;43:93–110. doi: 10.1016/s0168-0102(02)00023-8. [DOI] [PubMed] [Google Scholar]
- Kim HP, Lee JM, Lee DS, Koo BB, Kim JJ, Kim IY, Kwon JS, Yoo TW, Chang KH, Kim SI. Development of a group_specific average brain atlas: a comparison study between Korean and Occidental groups. J Biomed Eng Res. 2005;26:7–13. [Google Scholar]
- Kochunov P, Fox P, Lancaster J, Tan LH, Amunts K, Zilles K, Mazziotta J, Gao JH. Localized morphological brain differences between English-speaking Caucasians and Chinese-speaking Asians: new evidence of anatomical plasticity. Neuroreport. 2003;14:961–964. doi: 10.1097/01.wnr.0000075417.59944.00. [DOI] [PubMed] [Google Scholar]
- Kuo WJ, Yeh TC, Duann JR, Wu YT, Ho LT, Hung D, Tzeng OJ, Hsieh JC. A left-lateralized network for reading Chinese words: a 3 T fMRI study. Neuroreport. 2001;12:3997–4001. doi: 10.1097/00001756-200112210-00029. [DOI] [PubMed] [Google Scholar]
- Kuo WJ, Yeh TC, Lee CY, Wu YT, Chou CC, Ho LT, Hung DL, Tzeng OJ, Hsieh JC. Frequency effects of Chinese character processing in the brain: an event-related fMRI study. Neuroimage. 2003;18:720–730. doi: 10.1016/s1053-8119(03)00015-6. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Fox PT, Downs H, Nickerson DS, Hander TA, El Mallah M, Kochunov PV, Zamarripa F. Global spatial normalization of human brain using convex hulls. J Nucl Med. 1999;40:942–955. [PubMed] [Google Scholar]
- Lee DS, Lee JS, Oh SH, Kim SK, Kim JW, Chung JK, Lee MC, Kim CS. Cross-modal plasticity and cochlear implants. Nature. 2001;409:149–150. doi: 10.1038/35051653. [DOI] [PubMed] [Google Scholar]
- Lee JS, Lee DS, Kim J, Kim YK, Kang E, Kang H, Kang KW, Lee JM, Kim JJ, Park HJ, Kwon JS, Kim SI, Yoo TW, Chang KH, Lee MC. Development of Korean standard brain templates. J Korean Med Sci. 2005;20:483–488. doi: 10.3346/jkms.2005.20.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi Y, Ohnishi T, Kawachi T, Mori T, Hirakata M, Yamada M, Matsuda H, Komaki G. Specific brain activation in Japanese and Caucasian people to fearful faces. Neuroreport. 2005;16:133–136. doi: 10.1097/00001756-200502080-00012. [DOI] [PubMed] [Google Scholar]
- Munte TF, Altenmuller E, Jancke L. The musician's brain as a model of neuroplasticity. Nat Rev Neurosci. 2002;3:473–478. doi: 10.1038/nrn843. [DOI] [PubMed] [Google Scholar]
- Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Rex DE, Ma JQ, Toga AW. The LONI Pipeline Processing Environment. Neuroimage. 2003;19:1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sterr A, Szameitat A. A template effect study on voxel-based morphometry in statistic parametric mapping. Conf Proc IEEE Eng Med Biol Soc; 2005. pp. 3051–3054. [DOI] [PubMed] [Google Scholar]
- Smith CD, Chebrolu H, Wekstein DR, Schmitt FA, Markesbery WR. Age and gender effects on human brain anatomy: a voxel-based morphometric study in healthy elderly. Neurobiol Aging. 2007;28:1075–1087. doi: 10.1016/j.neurobiolaging.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J,TP. Co-planar stereotaxic atlas of the human brain: 3-dimensional proportional system-an approach to cerebral imaging. New York: Thieme Medical Publishers; 1988. [Google Scholar]
- Tan LH, Feng CM, Fox PT, Gao JH. An fMRI study with written Chinese. Neuroreport. 2001a;12:83–88. doi: 10.1097/00001756-200101220-00024. [DOI] [PubMed] [Google Scholar]
- Tan LH, Liu HL, Perfetti CA, Spinks JA, Fox PT, Gao JH. The neural system underlying Chinese logograph reading. Neuroimage. 2001b;13:836–846. doi: 10.1006/nimg.2001.0749. [DOI] [PubMed] [Google Scholar]
- Tan LH, Spinks JA, Feng CM, Siok WT, Perfetti CA, Xiong J, Fox PT, Gao JH. Neural systems of second language reading are shaped by native language. Hum Brain Mapp. 2003;18:158–166. doi: 10.1002/hbm.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LH, Spinks JA, Gao JH, Liu HL, Perfetti CA, Xiong J, Stofer KA, Pu Y, Liu Y, Fox PT. Brain activation in the processing of Chinese characters and words: a functional MRI study. Hum Brain Mapp. 2000;10:16–27. doi: 10.1002/(SICI)1097-0193(200005)10:1<16::AID-HBM30>3.0.CO;2-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore WH, Gaillard WD. Neuroimaging and the progression of epilepsy. Prog Brain Res. 2002;135:305–313. doi: 10.1016/S0079-6123(02)35028-3. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Hum Brain Mapp. 2000;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Maps of the brain. Anat Rec. 2001;265:37–53. doi: 10.1002/ar.1057. [DOI] [PubMed] [Google Scholar]
- Tu Z, Narr KL, Dollar P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans Med Imaging. 2008;27:495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Toga AW. Is it time to re-prioritize neuroimaging databases and digital repositories? Neuroimage. 2009;47:1720–1734. doi: 10.1016/j.neuroimage.2009.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C. Template-O-Matic: a toolbox for creating customized pediatric templates. Neuroimage. 2008;41:903–913. doi: 10.1016/j.neuroimage.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998a;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998b;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- Yoon U, Fonov VS, Perusse D, Evans AC. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45:769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Zhang K, Ginzburg I, McNaughton BL, Sejnowski TJ. Interpreting neuronal population activity by reconstruction: unified framework with application to hippocampal place cells. J Neurophysiol. 1998;79:1017–1044. doi: 10.1152/jn.1998.79.2.1017. [DOI] [PubMed] [Google Scholar]
- Zilles K, Kawashima R, Dabringhaus A, Fukuda H, Schormann T. Hemispheric shape of European and Japanese brains: 3-D MRI analysis of intersubject variability, ethnical, and gender differences. Neuroimage. 2001;13:262–271. doi: 10.1006/nimg.2000.0688. [DOI] [PubMed] [Google Scholar]