Abstract
Pulmonary function measures are heritable traits that predict morbidity and mortality and define chronic obstructive pulmonary disease (COPD). We tested genome-wide association with forced expiratory volume in 1 s (FEV1) and the ratio of FEV1 to forced vital capacity (FVC) in the SpiroMeta consortium (n = 20,288 individuals of European ancestry). We conducted a meta-analysis of top signals with data from direct genotyping (n ≤ 32,184 additional individuals) and in silico summary association data from the CHARGE Consortium (n = 21,209) and the Health 2000 survey (n ≤ 883). We confirmed the reported locus at 4q31 and identified associations with FEV1 or FEV1/FVC and common variants at five additional loci: 2q35 in TNS1 (P = 1.11 × 10−12), 4q24 in GSTCD (2.18 × 10−23), 5q33 in HTR4 (P = 4.29 × 10−9), 6p21 in AGER (P = 3.07 × 10−15) and 15q23 in THSD4 (P = 7.24 × 10−15). mRNA analyses showed expression of TNS1, GSTCD, AGER, HTR4 and THSD4 in human lung tissue. These associations offer mechanistic insight into pulmonary function regulation and indicate potential targets for interventions to alleviate respiratory disease.
Measures of pulmonary function, such as FEV1 and FEV1/FVC ratio, are important predictors of population morbidity and mortality1-4 as well as forming the basis for the diagnosis of COPD. It is well established that pulmonary function is partially genetically determined. Twin studies in European and US populations give heritability estimates for FEV1 as high as 0.77 (refs. 5,6). Longitudinal studies in families suggest that genetic effects are consistent over time7. Genetic determinants of pulmonary function seem to operate, at least in part, independent of disease status (such as asthma) and smoking status8, suggesting that population-based association studies are a viable way to identify key genetic determinants of lung function.
Adequately powered genome-wide association studies (GWAS) using hundreds of thousands of common SNPs can identify loci associated with common diseases and the quantitative traits that underlie them. Collaborative studies achieving sample sizes in excess of 10,000 have been able to identify associations with common genetic variants with typically modest effect sizes (usually <0.1 s.d.)9. In the past year, GWAS have reported association between an intergenic locus at chromosome 4q31 and FEV1/FVC ratio and COPD, but no large-scale collaborative GWAS have yet been undertaken for lung function10,11.
If common SNPs underlying lung function have modest effects, very large sample sizes will be required to identify them. We therefore established the SpiroMeta consortium to facilitate large-scale meta-analysis of GWAS of lung function. Here we report a meta-analysis of GWAS in the SpiroMeta consortium, comprising 20,288 individuals of European ancestry, that tested association between cross-sectional lung function measures and ~2.5 million genotyped or imputed SNPs (stage 1). We followed up SNPs drawn from the most significantly associated loci in up to 32,184 individuals by direct genotyping (stage 2a) and using in silico summary association data relating to a further 22,092 individuals (stage 2b). These studies confirm the previous reported association at 4q31 and show that five previously unreported loci are robustly associated with lung function.
RESULTS
Genome-wide association with lung function (stage 1)
We included 14 studies of individuals of European ancestry, with sample sizes totaling 20,288 (Table 1). All individuals had measures of FEV1 and FVC and smoking status recorded. FEV1 and (separately) FEV1/FVC measures were adjusted for age, age2, sex, height and ancestry principal components within each study. Genome-wide genotyping was undertaken with a variety of platforms, and standard quality control measures were used (Online Methods and Supplementary Table 1). Genotypes were imputed for ~2.5 million autosomal SNPs from HapMap CEU data and tested for association separately for the inverse-normal transformed residuals of FEV1 and FEV1/FVC under an additive genetic model. We carried out meta-analysis of study-specific test statistics using an inverse variance weighting. We applied genomic control at the study and meta-analysis levels to avoid overinflation of test statistics owing to population structure or relatedness. Test statistic inflation before applying genomic control at the meta-analysis level was modest (λGC = 1.046 for FEV1 and 1.035 for FEV1/FVC). The plots of meta-analysis test statistics against expected values under the null hypothesis showed an excess of extreme values even after exclusion of the previously reported11 4q31 locus near HHIP, indicative of additional loci associated with lung function (Supplementary Fig. 1a,b).
Table 1.
study characteristics
| Study |
N total |
N male |
N female |
Age range (y) at FEV1/FVC measurement |
Mean age, y (s.d.) |
Mean FEV1, l (s.d.) |
Mean FVC, l (s.d.) |
Mean FEV1/FVC (s.d.) |
N neversmokers |
N ever- smokers |
|---|---|---|---|---|---|---|---|---|---|---|
| Stage 1: GWAS | ||||||||||
| ALSPAC | 1,391 | 676 | 715 | 8–9 | 8.7 (0) | 1.70 (0.26) | 1.93 (0.31) | 0.88 (0.07) | 1,391 | 0 |
| B58C T1DGC | 2,343 | 1,131 | 1,212 | 44–45 | 44.5 (0) | 3.31 (0.78) | 4.19 (0.96) | 0.79 (0.08) | 692 | 1,651 |
| B58C WTCCC | 1,372 | 691 | 681 | 44–45 | 44.5 (0) | 2.93 (0.75) | 4.18 (0.96) | 0.79 (0.08) | 394 | 978 |
| EPIC obese cases | 1,104 | 476 | 628 | 39–76 | 59.1 (8.8) | 2.35 (0.69) | 2.84 (0.87) | 0.82 (0.17) | 489 | 615 |
| EPIC population based | 2,336 | 1,100 | 1,236 | 39–77 | 59.2 (9.0) | 2.50 (0.72) | 3.04 (0.90) | 0.85 (0.16) | 1,061 | 1,275 |
| FTC | 134 | 13 | 121 | 23–76 | 57.4 (19.3) | 2.69 (0.94) | 2.93 (0.61) | 0.79 (0.09) | 104 | 30 |
| KORA S3 | 555 | 261 | 294 | 29–73 | 47.6 (9.0) | 3.43 (0.78) | 4.18 (0.99) | 0.83 (0.07) | 266 | 289 |
| Korcula | 825 | 300 | 525 | 18–90 | 55.5 (13.5) | 2.84 (0.81) | 3.37 (0.93) | 0.84 (0.09) | 397 | 428 |
| NFBC1966 | 4,556 | 2,182 | 2,374 | 31–31 | 31.0 (0) | 3.96 (0.79) | 4.73 (0.99) | 0.84 (0.06) | 1,648 | 2,908 |
| NSPHS | 549 | 255 | 294 | 18–91 | 50.0 (19.1) | 3.02 (0.95) | 3.68 (1.12) | 0.82 (0.09) | 464 | 85 |
| ORCADES | 692 | 322 | 370 | 19–93 | 54.9 (15.3) | 2.88 (0.84) | 3.58 (0.98) | 0.80 (0.09) | 404 | 288 |
| SHIP | 1,777 | 870 | 907 | 25–85 | 52.3 (13.7) | 3.28 (0.89) | 3.87 (1.03) | 0.87 (0.06) | 773 | 1,004 |
| TwinsUK | 1,885 | 0 | 1,885 | 18–79 | 48.4 (12.2) | 2.73 (0.56) | 3.40 (0.61) | 0.80 (0.08) | 943 | 942 |
| Vis | 769 | 323 | 446 | 18–88 | 56.3 (15.3) | 3.39 (1.22) | 4.38 (1.43) | 0.77 (0.09) | 328 | 441 |
| Stage 1 sample size | 20,288 | |||||||||
| Stage 2a: studies with direct genotyping | ||||||||||
| ADONIX | 1,338 | 635 | 703 | 25–75 | 49.2 (13.6) | 3.35 (0.86) | 4.24 (1.02) | 0.79 (0.07) | 743 | 595 |
| BHS | 4,350 | 1,793 | 2,557 | 18–96 | 50.1 (17.0) | 3.02 (0.97) | 3.89 (1.16) | 0.77 (0.08) | 2,459 | 1,891 |
| BRHS | 3,897 | 3,897 | 0 | 60–79 | 68.7 (5.5) | 2.57 (0.69) | 3.37 (0.84) | 0.77 (0.11) | 1,132 | 2,765 |
| BWHHS | 3,644 | 0 | 3,644 | 59–80 | 68.8 (5.5) | 1.98 (0.52) | 2.82 (0.76) | 0.71 (0.09) | 2,060 | 1,584 |
| Gedling | 1,263 | 632 | 631 | 27–80 | 56.2 (12.3) | 2.85 (0.85) | 3.68 (1.02) | 0.77 (0.07) | 633 | 630 |
| GS:SFHS | 5,474 | 2,254 | 3,220 | 18–89 | 46.0 (14.3) | 3.15 (0.87) | 4.11 (1.03) | 0.77 (0.10) | 3,005 | 2,469 |
| HCS | 2,850 | 1,511 | 1,339 | 59–73 | 66.1 (2.8) | 2.44 (0.68) | 3.42 (0.92) | 0.72 (0.09) | 1,319 | 1,531 |
| KORA F4 | 1,305 | 610 | 695 | 41–61 | 51.6 (5.7) | 3.32 (0.81) | 4.28 (1.00) | 0.78 (0.06) | 499 | 806 |
| NFBC1986 | 4,946 | 2,379 | 2,567 | 15–17 | 16.0 (0.38) | 3.77 (0.70) | 4.30 (0.84) | 0.88 (0.08) | 3,708 | 1,238 |
| Nottingham Smokers | 509 | 280 | 229 | 40–89 | 59.5 (10.4) | 2.00 (0.95) | 3.01 (1.06) | 0.64 (0.16) | 0 | 509 |
| NSHD | 2,608 | 1,308 | 1,300 | 53–53 | 53 (0) | 2.80 (0.70) | 3.50 (0.89) | 0.81 (0.19) | 1,080 | 1,528 |
| Stage 2a sample size | 32,184 | |||||||||
| Stage 2b: studies with in silico data | ||||||||||
| Health 2000 | 883 | 427 | 456 | 30–75 | 50.2 (11.0) | 3.32 (0.91) | 4.19 (1.08) | 0.79 (0.07) | 266 | 617 |
| CHARGEa (ref. 30) | 21,209 | |||||||||
| Stage 2b sample size | 22,092 | |||||||||
| Total sample size | 74,564 | |||||||||
Characteristics are shown for studies analyzed in stage 1 (GWAS meta-analysis), stage 2a (direct genotyping follow-up, 10 SNPs) and stage 2b (in silico follow-up, 30 SNPs). Stage 1 studies: ALSPAC, Avon Longitudinal Study of Parents and Children; B58C-T1DGC, British 1958 Birth Cohort–Type 1 Diabetes Genetics Consortium; B58C-WTCCC, British 1958 Birth Cohort–Wellcome Trust Case Control Consortium; EPIC obese cases, European Prospective Investigation into Cancer and Nutrition, Obese Cases; EPIC population based, European Prospective Investigation into Cancer and Nutrition Cohort; FTC, Finnish Twin Cohort incorporating FinnTwin16 and FITSA; KORA S3, Cooperative Health Research in the Region of Augsburg; the Korcula study; NFBC1966, Northern Finland Birth Cohort of 1966; NSPHS, Northern Sweden Population Health Study; ORCADES, Orkney Complex Disease Study; SHIP, Study of Health in Pomerania; the TwinsUK study; the Vis study. Stage 2a studies: ADONIX, Adult-Onset Asthma and Nitric Oxide; BHS, Busselton Health Study; BRHS, British Regional Heart Study; BWHHS, British Women’s Heart and Health Study; the Gedling study; GS:SFHS, Generation Scotland: Scottish Family Health Study; HCS, Hertfordshire Cohort Study; KORA F4, Cooperative Health Research in the Region of Augsburg; NFBC1986, Northern Finland Birth Cohort of 1986; the Nottingham Smokers study; NSHD, Medical Research Council National Survey of Health and Development (also known as the British 1946 Birth Cohort). Stage 2b studies: Health 2000, Finnish Health 2000 survey; CHARGE, Cohorts for Heart and Aging Research in Genomic Epidemiology.
Characteristics of the constituent studies of the CHARGE Consortium are presented in the companion paper from the CHARGE Consortium30.
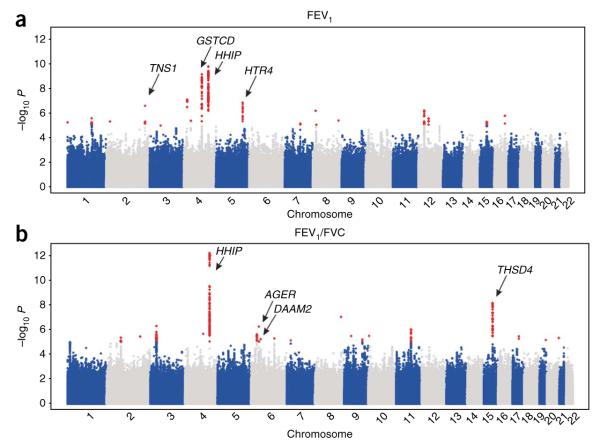
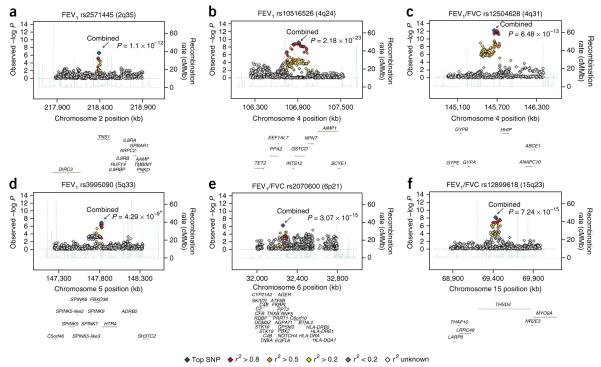
We observed independent regions of association at 17 loci with P < 1 × 10−5 for FEV1 and 23 for FEV1/FVC (Figs. 1a,b and 2), including three regions (4q24 in GSTCD, 4q31 near HHIP and 15q23 in THSD4) that reached P < 5 × 10−8 in the stage 1 GWAS data alone, corresponding to a threshold of P < 0.05 after adjusting for 1 million independent tests12. SNP rs12504628, which was associated with both FEV1/FVC (P = 6.48 × 10−13; Fig. 2c and Table 2) and FEV1 (P = 1.50 × 10−10; Table 3), lies in an intergenic region upstream of HHIP and spanning ~300 kb at 4q31 that has been associated with lung function11, COPD11 and height9. Our top SNP rs12504628 was in strong linkage disequilibrium (LD; r2 = 0.97) with the previously reported SNP associated with lung function, rs13147758 (P = 5.30 × 10−10 for FEV1 and P = 1.11 × 10−12 1 for FEV1/FVC in our data), and with SNPs associated previously with height (rs6854783, r2 = 0.55; rs2055059, r2 = 0.48), suggesting a role in skeletal growth and development. The hedgehog gene family, of which HHIP is a member, encodes signaling molecules involved in regulating lung morphogenesis, suggesting other mechanisms underlying these associations13. This intergenic region also contains multiple ESTs in human fetal lung (UCSC Browser).
Figure 1.
Manhattan plots of association results for FEV1 and FEV1/FVC (analysis stage 1). (a,b) Manhattan plots ordered by chromosome position. SNPs for which −log10 P > 5 are indicated in red. The six loci indicated by arrows showed association with FEV1 (a) or FEV1/FVC (b; P < 5 × 10−8) in the meta-analysis of data from stages 1, 2a and 2b.
Figure 2.
Regional association plots of six lung function–associated loci. (a–f) Statistical significance of each SNP on the −log10 scale as a function of chromosome position (NCBI build 36) in the meta-analysis of stage 1 data alone. The sentinel SNP at each locus is shown in blue; the correlations (r2) of each of the surrounding SNPs to the sentinel SNP are shown in the indicated colors. The six loci included are those that showed association with FEV1 or FEV1/FVC (P < 5 × 10–8) in the meta-analysis of data from stages 1, 2a and 2b. The combined P values for all stages are indicated by arrows. The relevant trait (FEV1 or FEV1/FVC ratio) is indicated for each plot. For rs12504628, the plot shows only the association of FEV1/FVC; this SNP was associated (P < 5 × 10−8) with both FEV1 and FEV1/FVC. Fine-scale recombination rate is plotted in blue39. Combined P value from stages 1 and 2a only; SNP rs3995090 had low imputation quality in the CHARGE Consortium data and so was not included in stage 2b.
Table 2.
Loci associated with lung function
| Chr. | Measure | SNP ID (NCBI36 position), function |
Coded allele |
Stage | Coded allele freq. |
N | Beta (s.e.m.) | P | Joint meta-analysis of all stages |
r 2 |
N total |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (s.e.m.) | P | |||||||||||
| 2 | FEV1 |
rs2571445 (218,391,399), TNS1 (ns) |
G | 1 2a 2b |
0.59 0.60 0.61 |
18,710 29,922 21,697 |
0.057 (0.011) 0.031 (0.008) 0.027 (0.007) |
2.41 × 10−7 9.03 × 10−5 1.94 × 10−4 |
0.035 (0.005) | 1.11 × 10−12 | 0.07% | 70,329 |
| 4 | FEV1 |
rs10516526 (106,908,353), GSTCD (intron) |
G | 1 2a 2b |
0.06 0.06 0.07 |
20,179 31,217 22,092 |
0.135 (0.022) 0.092 (0.013) 0.066 (0.014) |
6.67 × 10−10 8.51 × 10−12 3.37 × 10−6 |
0.089 (0.009) | 2.18 × 10−23 | 0.14% | 73,488 |
| 4 | FEV1/FVC |
rs12504628 (145,655,774), HHIP (upstream) |
T | 1 2a 2b |
0.56 | 19,400 0 0 |
−0.077 (0.011) — — |
6.48 × 10−13 — — |
–0.077 (0.011) | 6.48 × 10−13 | 0.27% | 19,400 |
| 5 | FEV1 |
rs3995090 (147,826,008), HTR4 (intron) |
C | 1 2a 2b |
0.41 0.41 0.36 |
18,792 29,630 883 |
0.058 (0.011) 0.029 (0.008) −0.037 (0.050) |
1.36 × 10−7 3.04 × 10−4 4.62 × 10−1 |
0.038 (0.006) | 4.29 × 10−9 | 0.07% | 49,305 |
| 5 | FEV1 |
rs6889822 (147,826,900), HTR4 (intron) |
G | 1 2a 2b |
0.40 0.38 |
19,652 0 21,442 |
0.057 (0.011) — 0.025 (0.0072) |
1.73 × 10−7 5.76 × 10−4 |
0.034 (0.006) | 8.17 × 10−9 | 0.08% | 41,094 |
| 6 | FEV1/FVC |
rs2070600 (32,259,421), AGER (ns) |
T | 1 2a 2b |
0.05 0.06 0.04 |
19,183 26,229 21,998 |
0.115 (0.023) 0.069 (0.014) 0.130 (0.031) |
5.86 × 10−7 7.90 × 10−7 2.24 × 10−5 |
0.088 (0.011) | 3.07 × 10−15 | 0.09% | 67,410 |
| 6 | FEV1/FVC |
rs2395730 (39,892,343), DAAM2 (intron) |
C | 1 2a 2b |
0.419 0.424 |
19,887 0 21,560 |
0.048 (0.011) — 0.038 (0.013) |
5.86 × 10−6 — 3.22 × 10−3 |
0.044 (0.008) | 7.98 × 10−8 | 0.07% | 41,447 |
| 15 | FEV1/FVC |
rs12899618 (69,432,174), THSD4 (intron) |
G | 1 2a 2b |
0.84 0.85 0.85 |
19,875 26,124 21,050 |
0.082 (0.014) 0.043 (0.011) 0.072 (0.018) |
7.17 × 10−9 7.51 × 10−5 4.21 × 10−5 |
0.060 (0.008) | 7.24 × 10−15 | 0.09% | 67,049 |
Shown are the top SNPs or SNPs for each independent locus associated (P < 5 × 10−7) with FEV1 or FEV1/FVC in a joint analysis of up to 74,564 individuals of European ancestry from the SpiroMeta GWAS (stage 1), follow-up genotyping (stage 2a) and in silico data from the CHARGE Consortium (companion paper in this issue30) and the Health 2000 study (stage 2b). The six genome-wide significant (P < 5 × 10−8) loci are indicated in bold. Results are also shown for rs2395730 (DAAM2), as this locus fell just below genome-wide significance after meta-analysis of stage 1 and 2b data (it was not selected for stage 2a genotyped follow-up). For the GWAS (stage 1 and 2b) data, the sample sizes (N) shown are the effective sample sizes. Effective sample size within each study is the product of sample size and imputation quality metric. Total sample size is the sum of the N effective values across all stage 1 and 2b studies, plus the sample size from the directly genotyped (stage 2a) studies; for each SNP, this is lower than 74,564 individuals owing to missing genotypes and imperfect imputation. Joint meta-analysis includes data from stages 1, 2a and 2b. Beta values reflect effect-size estimates on an inverse-normal transformed scale after adjustments for age, age2, sex, height and ancestry principal components. SNP rs12504628 showed association with both FEV1 and FEV1/FVC (P < 5 × 10−8 ); statistics for the strongest association (FEV1/FVC) are shown. As this association has been previously published11, rs12504628 was not assessed in the follow-up studies, so statistics are not presented for stages 2a and 2b for rs12504628. SNP rs3995090 was genotyped in stage 2a follow-up studies and was available in Health 2000 study data, but owing to low imputation quality for rs3995090 in the CHARGE Consortium data, an alternative SNP in the region, rs6889822, was selected for in silico exchange with the CHARGE Consortium. The estimated proportion of variance explained by each SNP in the joint meta-analysis is shown (r2). ns, nonsynonymous coding SNP.
Table 3.
Relation of SNPs at genome-wide significant loci to FEV1, FVC and FEV1/FVC, and impact of adjustment for smoking in stage 1 (spiroMeta GWAs) data
| SNP ID (NCBI36 position), function |
Chr. | Coded allele |
Noncoded allele |
N | Measure | Unadjusted stage 1 analyses |
Smoking-adjusted stage 1 analyses |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | s.e.m. | P | Beta | s.e.m. | P | ||||||
| rs2571445 (218391399), TNS1 (ns) |
2 | G | A | 18,710 | FEV1 FVC FEV1/FVC |
0.057 0.041 0.026 |
0.011 0.011 0.011 |
2.41 × 10−7 1.84 × 10−4 0.018 |
0.055 0.038 0.031 |
0.012 0.011 0.012 |
2.12 × 10−6 8.94 × 10−4 0.0073 |
| rs10516526 (106908353), GSTCD (intron) |
4 | G | A | 20,179 | FEV1 FVC FEV1/FVC |
0.135 0.112 0.038 |
0.022 0.022 0.022 |
6.67 × 10−10 2.53 × 10−7 0.084 |
0.138 0.119 0.030 |
0.023 0.023 0.023 |
2.06 × 10−9 1.29 × 10−7 0.19 |
| rs12504628 (145655774), HHIP (upstream) |
4 | T | C | 19,400 | FEV1 FVC FEV1/FVC |
−0.068 −0.021 −0.077 |
0.011 0.011 0.011 |
1.50 × 10−10 0.046 6.48 × 10−13 |
−130.069 −130.020 −130.080 |
0.011 0.011 0.011 |
7.18 × 10−10 0.066 8.25 × 10−13 |
| rs3995090 (147826008), HTR4 (intron) |
5 | C | A | 18,792 | FEV1 FVC FEV1/FVC |
0.058 0.023 0.045 |
0.011 0.011 0.011 |
1.36 × 10−7 0.034 1.92 × 10−5 |
0.059 0.031 0.038 |
0.011 0.011 0.011 |
1.47 × 10−7 0.0048 7.0 × 10−4 |
| rs6889822 (147826900), HTR4 (intron) |
5 | G | A | 19,652 | FEV1 FVC FEV1/FVC |
0.057 0.025 0.046 |
0.011 0.011 0.011 |
1.73 × 10−7 0.021 2.15 × 10−5 |
0.062 0.034 0.038 |
0.011 0.011 0.011 |
5.23 × 10−8 0.0025 7.3 × 10−4 |
| rs2070600 (32259421), AGER (ns) |
6 | T | C | 19,183 | FEV1 FVC FEV1/FVC |
0.027 −0.043 0.115 |
0.023 0.023 0.023 |
0.24 0.063 5.86 × 10−7 |
0.044 −0.035 0.126 |
0.024 0.024 0.024 |
0.072 0.14 2.42 × 10−7 |
| rs12899618 (69432174), THSD4 (intron) |
15 | G | A | 19,875 | FEV1 FVC FEV1/FVC |
0.028 −0.024 0.082 |
0.014 0.014 0.014 |
0.047 0.085 7.17 × 10−9 |
0.033 −0.024 0.092 |
0.015 0.015 0.015 |
0.025 0.11 6.56 × 10−10 |
Each SNP included in table showed genome-wide significant association (P < 5 × 10−8) with either FEV1 or FEV1/FVC in data from stages 1, 2a and 2b. Beta values shown reflect effect-size estimates on an inverse-normal transformed scale in the stage 1 data. Effective sample size was calculated within each study as the product of sample size and imputation quality metric and were summed across studies to calculate total effective sample size (N). Estimates from analyses unadjusted for smoking status and adjusted for ever-smoking versus never-smoking status are shown.
Follow-up analyses (stage 2)
To validate potential associations with lung function, we selected 10 SNPs for further genotyping in additional studies of European ancestry (stage 2a, 32,184 individuals; Supplementary Table 2) and 30 SNPs for in silico follow-up (stage 2b; Supplementary Table 3). We obtained the in silico association results from the Health 2000 study (883 individuals) and from the CHARGE Consortium (21,209 individuals). Meta-analysis of the association results across stages 1, 2a and 2b showed five novel loci reaching genome-wide significance (P < 5 × 10−8): 2q35 in TNS1, 4q24 in GSTCD, 5q33 in HTR4, 6p21 in AGER and 15q23 in THSD4 (Table 2 and Fig. 2). A further locus, 6p21 in DAAM2, which was not selected for further genotyping follow-up in stage 2a, fell just below the threshold for genome-wide significance for association with FEV1/FVC after meta-analysis across stages 1 and 2b (rs2395730, P = 7.98 × 10−8; Supplementary Table 3 and Table 2).
The strongest association of FEV1 was at 4q24 in GSTCD (rs10516526, P = 2.18 × 10−23; Table 2 and Fig. 2b). Relatively little is known about GSTCD, but the presence of the C-terminal α-helical domain common to the glutathione S-transferase (GST) family of enzymes suggests this protein is involved in cellular detoxification by catalyzing conjugation of glutathione to products of oxidative stress14. GST enzymes also show glutathione peroxidase activity regulating the synthesis of prostaglandins and leukotrienes14. To explore the potential function of GSTCD, we conducted a protein homology search and identified homology with chloride intracellular channels 1, 3, 4, 5 and 6, suggesting a role for GSTCD beyond the GST enzyme family. Genes in the region also include INTS12 and NPNT. INTS12 associates with RNA polymerase II and mediates 3′-end processing of small nuclear RNA15.
The second locus associated with FEV1 was at 2q35, localized to the TNS1 gene (nonsynonymous coding SNP rs2571445, P = 1.11 × 10−12; Table 2 and Fig. 2a). The protein this encodes, tensin-1, is an actin-binding protein that contains Src homology 2 domains, suggesting a role in linking cytoskeletal changes with signal transduction16. Tensin-1 may be functionally involved in cell migration17.
Multiple genes potentially underlie the third locus associated with FEV1 at 5q33. The most strongly associated SNPs in this region, rs3995090 and rs6889822 (P = 4.29 × 10−9 and P = 8.17 × 10−9; Table 2 and Fig. 2d), are located in an intron in HTR4 and are part of a cluster of associated SNPs also spanning a SPINK5-like gene, SPINK7, SPINK9 and FBXO38. HTR4, which encodes 5-hydroxytryptamine receptor-4, is expressed in neurons in the respiratory pre-Bötzinger complex. Activation of this G protein–coupled receptor protects spontaneous respiratory activity18. Notably, selective antagonism of HTR4 in human bronchial strips in vitro attenuates the facilitation of electric field–stimulated cholinergic contraction by 5-hydroxytryptamine, suggesting a role for HTR4 in mediating airway caliber19. HTR4 expression has recently been confirmed in airway epithelial type II cells, where receptor stimulation seems to regulate cytokine responses20. The SPINK family of serine protease inhibitors may have a role in antimicrobial protection of mucous epithelia21. F-box protein-38 (encoded by FBXO38) is a member of a family of proteins that are believed to mediate protein-protein interactions and protein degradation22.
The strongest association with FEV1/FVC was at 6p21, a gene-rich region of the major histocompatibility complex (MHC). The extended LD in this region of the MHC prevented accurate localization of the association signal. However, we observed the peak of association for a nonsynonymous coding SNP in AGER (rs2070600, P = 3.07 × 10−15; Table 2 and Fig. 2e), which is a plausible candidate for causal association. AGER, also known as RAGE, is a multiligand receptor of the immunoglobulin superfamily23. AGER is highly expressed in the lung, in particular alveolar epithelial cells24, with a potential role in epithelium–extracellular matrix interactions. Reduced AGER expression has been identified in individuals with idiopathic pulmonary fibrosis25, and Ager-/- mice develop age-related pulmonary fibrosis26. Another candidate in this region is the nearby gene NOTCH4, a member of the family of transmembrane receptors involved in cell fate decisions27. Notch4 is expressed in endothelial cells of the adult mouse lung, where it is believed to regulate angiogenesis28.
The second identified association with FEV1/FVC was at 15q23, encompassing the THSD4 gene (rs12899618, P = 7.24 × 10−15; Table 2 and Fig. 2f). THSD4 shows homology with members of the thrombospondin family of extracellular calcium-binding proteins that modulate cellular attachment, proliferation and migration and have been implicated in wound healing, inflammation and angiogenesis29.
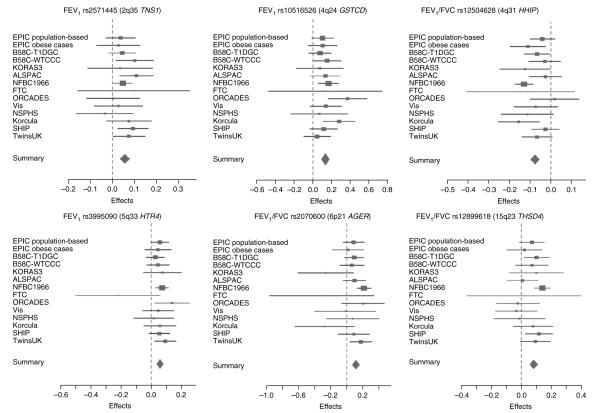
For each of the loci we reported, the estimated effect sizes were broadly consistent across the GWAS (Fig. 3).
Figure 3.
Forest plots of the stage 1 meta-analysis for the six lung function–associated loci. Each of the SNPs included in the figure showed genome-wide significant association (P < 5 × 10−8) with either FEV1 or FEV1/FVC in the data from stages 1, 2a and 2b. The plots show the meta-analysis of the stage 1 data for each sentinel SNP. The contributing effect (transformed beta) from each study is shown by a square, with confidence intervals indicated by horizontal lines. The contributing weight of each study to the meta-analysis is indicated by the size of the square. The combined meta-analysis estimate in the stage 1 data is shown at the bottom of each graph.
Association of variants with FVC
We tested the top SNP at each of the loci showing genome-wide significant association (P < 5 × 10−8) with FEV1 or FEV1/FVC for association with the other of the two traits, and with FVC in the stage 1 studies (Table 3). In addition to being associated with FEV1, rs10516526 in GSTCD was associated with FVC (P = 2.53 × 10−7) but showed no discernible effect on FEV1/FVC.
Effect of smoking on SNP associations
Adjustment for ever-smoking status in the stage 1 data (Table 3) did not show materially different effect-size estimates for the associations with the sentinel SNPs in TNS1, GSTCD, HTR4, AGER, THSD4 or HHIP. Similarly, adjustments for a quantitative measure of lifetime smoking exposure (pack-years) did not show substantially different effect-size estimates for the identified SNP associations (data not shown). We also tested the associations of the top SNPs in TNS1, GSTCD, HTR4, AGER and THSD4 separately in ever-smokers and never-smokers (Supplementary Table 4); all P values were >0.05 for tests of interaction between smoking status and these SNPs on lung function.
Gene expression
We determined the mRNA expression profiles of GSTCD, HHIP, THSD4, TNS1, HTR4, AGER and NOTCH4 in human lung tissue and a series of primary cells. We detected all transcripts in lung tissue (Supplementary Fig. 2a) and bronchial epithelial cells (Supplementary Fig. 2b); six transcripts (excluding NOTCH4) were present in human airway smooth muscle cells. We also detected GSTCD, TNS1, HTR4, AGER and NOTCH4 transcripts in peripheral blood mononuclear cells (Supplementary Fig. 2b). For AGER, we noted the presence of two PCR products suggesting an unreported splice variant; we confirmed the presence of the splice variant by sequencing.
DISCUSSION
Our study reports a meta-analysis of GWAS results from 20,288 participants and follow-up analyses in 54,276 participants, identifying five novel genome-wide significant loci for pulmonary function. The regions identified were 4q24 (GSTCD), 2q35 (TNS1) and 5q33 (HTR4) for FEV1, and 6p21 (AGER) and 15q23 (THSD4) for FEV1/FVC. In addition, we identified a region suggestive of association with FEV1/FVC at 6p21 in DAAM2. The companion manuscript from the CHARGE Consortium, which reports a GWAS of lung function in 20,890 participants, also identifies genome-wide significant associations at GSTCD, HTR4 and AGER30. Both SpiroMeta and CHARGE confirmed the previously reported association between FEV1 and FEV1/FVC and the 4q31 locus upstream of HHIP11.
Our findings have several important implications. First, the loci identified were observed in the whole population studied and were not specific to smokers. The presence of genetic determinants of lung function that do not depend on prior smoking exposure has been suggested by previous studies of heritability8. This does not rule out a possible subset of genetic determinants with effects on lung function that are partially or wholly dependent on smoking exposure.
We have also attempted to address the issue of genetic factors that influence smoking behavior. We did not observe any association with the CHRNA3-CHRNA5-CHRNB4 locus previously reported to be associated with cigarette smoke exposure, lung cancer, peripheral arterial disease31 and COPD10 (rs1051730, P = 0.23 for FEV1 and 0.56 for FEV1/FVC). The associations we show in GSTCD, TNS1, HTR4, THSD4 and AGER do not seem to be attenuated by adjustment for qualitative or quantitative adjustments for smoking exposure. None of these loci have been implicated in published GWAS of smoking quantity, although a recent report suggested a role for TSHD4 variants in smoking cessation32.
SNPs showing association with height could also show association with lung function measures because of incomplete adjustment for height, or because of SNP effects on skeletal growth with consequences for both height and lung function. The 4q31 locus near HHIP has shown convincing association with height33. An association was recently reported between height and rs185819 at 6p21 (ref. 34). Although this association signal was broad, reflecting the extended LD across this region of the MHC, rs185819 was in weak LD (r2 = 0.069) with rs2070600 (the sentinel SNP we reported for FEV1/FVC in AGER). These findings leave open the possibility of shared genetic determinants of growth of pulmonary function and height, but they do not suggest that our findings are primarily accounted for by inadequate adjustment for height.
The level of FEV1 at a given time point in an individual depends on two potentially independent processes: the maximum lung function obtained during development, and the rate of decline of lung function with age. Lung function reaches a maximum by age 25–35 years35. The populations studied in SpiroMeta cover a wide range of ages except the very elderly; as expected, FEV1 and FVC values were much lower in children. At least for the loci we identify, there was little evidence for age-specific effects, suggesting that the genetic risk factors identified operate across the age ranges; these findings again are in keeping with those of previous epidemiological studies7. Our analyses were based on cross-sectional measures of lung function; additional studies in cohorts with longitudinal data will be needed to identify determinants of the gradients of development and decline in lung function with age.
The magnitude of the estimated effect on untransformed FEV1 of rs10516526 in GSTCD was 52 ml per copy of the G allele (frequency, 0.06). This is equivalent to about 3 years of FEV1 decline in the nonsmoking population35. Allelic effect sizes on FEV1 of the more common variants (minor allele frequencies ~0.4) were 19–23 ml for rs3995090 in HTR4 and rs2571445 in TNS1. Individually, the five loci we describe account for a small proportion (0.07%–0.14%) of the variance in FEV1 and in FEV1/FVC (Table 2) in the general population.
After exclusion of the locus near HHIP and the five reported regions, meta-analysis test statistics still showed an excess of extreme values compared with expected values under the null, particularly for FEV1. Although we cannot rule out the possibility of residual population stratification, this indicates the potential to detect further loci associated with lung function (Supplementary Fig. 1a,b). We have provided a list of the top 2000 associations for FEV1 and for FEV1/FVC (Supplementary Table 5) as a resource to other investigators.
We imputed nongenotyped SNPs using two software implementations36,37 that share similar underlying population genetic models38. This methodology facilitates meta-analysis across different marker sets and improves coverage across the genome, and its utility has been empirically shown in several large GWAS. However, the power to detect associations with rare alleles is limited. The loci we report include two relatively infrequent SNPs, GSTCD (rs10516526, minor allele frequency 0.06) and AGER (rs2070600, minor allele frequency 0.05); these SNPs were directly genotyped in the majority of stage 1 subjects (16,514 and 15,386 individuals, respectively).
The associations we report relate to the general population but were of comparable magnitude after the exclusion of documented cases of asthma or COPD (data not shown). Although pulmonary function is an important predictor of morbidity and mortality per se, it will be important to investigate, in appropriately powered studies, whether the risk alleles in the genes identified in this study act as independent susceptibility markers for COPD or influence the development of airway obstruction in other diseases, such as asthma.
Our expression profiling studies identified expression of all of the candidate genes in relevant tissues. Further work is required to elucidate the mechanisms underlying the novel association signals we describe. In broad terms, however, it is notable that the most probable candidate genes in the regions identified seem to be involved either in developmental pathways important for lung growth or in tissue remodeling pathways that might be expected to alter airway architecture.
In conclusion, the results presented here from the SpiroMeta consortium, together with those reported by the CHARGE Consortium30, provide strong evidence for newly identified genetic loci that act as important determinants of pulmonary function.
ONLINE METHODS
Study design
The study consisted of two stages. In stage 1, a meta-analysis was conducted on directly genotyped and imputed SNPs from 14 studies of individuals of European ancestry, with a total sample size of 20,288. Details of these studies are given in Table 1. This meta-analysis provided loci for further genotyping in up to 32,184 individuals of European origin (stage 2a) and in silico comparisons in 22,092 individuals of European origin (stage 2b).
Stage 1 samples
The SpiroMeta consortium consists of 14 GWAS studies: ALSPAC, B58C-T1DGC, B58C-WTCCC, EPIC (obese and population-based substudies), the EUROSPAN studies (Korcula, NSPHS, ORCADES and Vis), FTC (incorporating the FinnTwin16 and Finnish Twin Study on Aging), KORA S3, NFBC1966, SHIP and TwinsUK (see Table 1 for definitions of acronyms). The primary analyses on FEV1 and FEV1/FVC included 20,288 individuals of European descent. The measurements of FEV1 and FVC are described in the Supplementary Note.
Genome-wide genotyping and quality control
The platforms used were Affymetrix 500K GeneChip array (four studies), Illumina HumanHap 550 Beadchip (one study), Illumina 317K (four studies), Affymetrix Genome-Wide SNP6.0 (one study), Illumina Hap370cnv (one study), Illumina Hap300 v1 (one study) and Illumina Hap300 v2 (two studies). Each individual study applied quality-control criteria as described in Supplementary Table 1.
Imputation
Imputation of nongenotyped SNPs was undertaken with MACH36 or IMPUTE37 with preimputation filters and parameters as shown in Supplementary Table 1. SNPs were excluded if the imputation information, assessed using r2.hat (MACH) or .info (IMPUTE), was <0.3. In total, 2,705,257 autosomal SNPs were analyzed.
Transformation of data and genotype-phenotype association analysis
Linear regression of age, age2, sex, height and ancestry principal components was undertaken on FEV1 (milliliters) and FEV1/FVC (percentage). The residuals were transformed to ranks and subsequently to normally distributed z scores, and were then used as the phenotype for association testing under an additive genetic model using software specified in Supplementary Table 1. Appropriate tests for association in related individuals were applied where necessary, as described in the Supplementary Note.
Meta-analysis of stage 1 data
All stage 1 study effect estimates were corrected using genomic control40 and were oriented to the forward strand of the NCBI build 36 reference sequence of the human genome, consistently using the alphabetically higher allele as the coded allele. Study-specific lambda estimates are shown in Supplementary Table 1. The pooled effect-size estimate and s.e.m. were computed using inverse variance weighting, and genomic control was applied to the pooled effect-size estimates. To describe the effect of imperfect imputation on power, we report ‘N effective’, the sum of the study-specific products of the sample size and the imputation quality metric. Meta-analysis statistics and figures were produced using R version 2.7.0.
Selection of SNPs for stage 2
Ten leading SNPs were selected for stage 2a genotyping follow-up (Supplementary Table 2). Thirty leading SNPs were selected for stage 2b in silico exchange, according to P value (under the threshold of 5 × 10−5), N effective (≥70% of the total sample size) and evidence from supporting SNPs (Supplementary Table 3).
Stage 2a samples (follow-up genotyped data)
We genotyped 10 SNPs in up to 32,184 individuals from the ADONIX, BHS, BRHS, BWHHS, Gedling, GS:SFHS, HCS, KORA F4, NFBC1986, Nottingham Smokers and NSHD studies. The characteristics of the studies are summarized in Table 1, and stage 2a study information is provided in the Supplementary Note.
Stage 2b samples (in silico data)
The CHARGE Consortium includes four population-based studies with data on FEV1 and FEV1/FVC: the Atherosclerosis Risk in Communities (ARIC) study, the Cardiovascular Health Study (CHS), the Framingham Heart Study (FHS) and the Rotterdam Study (RS). Details are provided in the companion paper in this issue from the CHARGE Consortium30. Given differences between the analysis approaches for GWAS adopted by the SpiroMeta and CHARGE consortia, the CHARGE analyses were undertaken using the analysis approach adopted by the SpiroMeta consortium (21,209 individuals; larger than the sample in the companion paper, which excluded subjects with missing or incomplete pack-years data). We also included 883 population-based subjects from the Health 2000 study in the stage 2b analysis.
Combined analysis of stage 1 and 2 samples
Meta-analysis of data from stages 1, 2a and 2b was conducted using inverse variance weighting. We described associations as genome-wide significant if P < 5 × 10−8.
Secondary analyses
To examine the effect of smoking on the causal pathway between the SNPs and the traits of interest, an adjustment for smoking was applied. The subgroups of ‘ever-smokers’ and ‘never-smokers’ were analyzed separately, and the stratum-specific estimated effects were combined within each individual study using inverse variance weights before meta-analyzing over studies. Additional adjustments were undertaken by adjusting for pack-years among the ever-smokers with these data available, and repeating the analyses.
PCR expression profiling
The mRNA expression profiles of GSTCD, HHIP, THSD4, TNS1, HTR4, AGER and NOTCH4 were determined in human lung tissue and primary cell samples using RT-PCR, including RNA from lung (Ambion/ABI), brain, airway smooth muscle cells41 and human bronchial epithelial cells (Clonetics42). Peripheral blood mononuclear cells were isolated from whole blood using 6% (w/v) dextran and 42%–51% (v/v) Percoll gradients (Sigma). Ethical approval for the use of primary cells was obtained from the local ethics committees. Total RNA was extracted from samples using an RNeasy kit (Qiagen) as directed by the manufacturer. cDNA was generated from 1 μg of RNA template using random hexamers and a SuperScript kit (Invitrogen) as directed by the manufacturer. PCR assays were designed to cross intron-exon boundaries and where splice variation was known, in order to detect all variants. Primer sequences are given in Supplementary Table 6. All PCR was done using Platinum Taq High Fidelity (Invitrogen) with 100 ng of cDNA template in a 25-μl reaction. Cycling conditions were as follows: 94 °C for 3 min, 35 cycles of 94 °C for 45 s, 55 °C for 30 s, and 72 °C for 90 s.
Supplementary Material
ACKNOWLEDGMENTS
We thank the many colleagues who contributed to collection and phenotypic characterization of the clinical sampling, genotyping and analysis of the GWAS data. We especially thank those who kindly agreed to participate in the studies.
Major funding for this work is from the following sources (in alphabetical order): Academy of Finland (including project grants 104781, 120315 and 1114194) and Center of Excellence in Complex Disease Genetics; Arthritis Research Campaign; Asthma UK; AstraZeneca; Biocenter Oulu, University of Oulu; Biocentrum Helsinki; Biotechnology and Biological Sciences Research Council project grant; British Heart Foundation (including project grants PG/06/154/22043 and PG/97012 and Senior Research Fellowship FS05/125); British Lung Foundation; Cancer Research United Kingdom; Chief Scientists Office, part of the Scottish Government Health Directorate (including grant CZD/16/6); Department of Health Air Pollution PRP (ref. no. 0020029); ENGAGE project (HEALTH-F4-2007-201413); European Commission (EURO-BLCS, FP-5/QLG1-CT-2000-01643, FP-7/2007-2013, FP-6 LSHB-CT-2006-018996 (GABRIEL), FP-6 LSHG-CT-2006-01947 (EUROSPAN), HEALTH-F2-2008-201865-GEFOS and FP-5 GenomEUtwin project QLG2-CT-2002-01254); Finnish Ministry of Education; German Federal Ministry of Education and Research (BMBF, including grants 01ZZ96030, 01ZZ0701 and 01GI0883 and German Asthma and COPD Network (COSYCONET) grant 01GI0883); German Ministry for Education, Research and Cultural Affairs; German National Genome Research Network (NGFN-2 and NGFN-plus); Healthway, Western Australia; HEFCE Science Research Investment Fund; Helmholtz Zentrum München; German Research Center for Environmental Health, Neuherberg, Germany; International Osteoporosis Foundation; Juvenile Diabetes Research Foundation International; Leicester Biomedical Research Unit in Cardiovascular Science (NIHR); Medical Research Council UK (including grants G0500539, G0501942, G0000943 and G990146); Medical Research Fund of the Tampere University Hospital; Ministry for Social Affairs of the Federal State of Mecklenburg-West Pomerania; MRC Human Genetics Unit; Munich Center of Health Sciences, as part of LMUinnovativ; National Human Genome Research Institute; National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London; National Institute for Health Research Cambridge Biomedical Research Centre; National Institute of Allergy and Infectious Diseases; National Institute of Child Health and Human Development; National Institute of Diabetes and Digestive and Kidney Diseases; National Heart, Lung, and Blood Institute (grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01)); Oulu University Hospital; PHOEBE (FP6, LSHG-CT-2006-518418); Public Population Project in Genomics (Genome Canada and Genome Quebec), Republic of Croatia Ministry of Science, Education and Sports (research grant 108-1080315-0302); Royal Society; Siemens Health Care Sector; Swedish Heart and Lung Foundation (grant 20050561); Swedish Medical Research Council (project no. K2007-66X-20270-01-3); Swedish Research Council for Working Life and Social Research (FAS, grants 2001-0263 and 2003-0139); the Great Wine Estates of the Margaret River region of Western Australia; UBS Wealth Foundation (grant BA29s8Q7-DZZ); UK Department of Health Policy Research Programme; University of Nottingham; University of Bristol; US National Institutes of Health (U01 DK062418); US National Institutes of Health–National Institute of Mental Health (5R01MH63706:02); Wellcome Trust (including grants 068545/Z/02, 076113/B/04/Z, 079895, 077016/Z/05/Z, 075883 and 086160/Z/08/A); and Zentren für Innovationskompetenz (BMBF grant 03ZIK012).
Footnotes
METHODS Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
URLs. UCSC browser, http://genome.ucsc.edu/.
References
- 1.Myint PK, et al. Respiratory function and self-reported functional health: EPIC-Norfolk population study. Eur. Respir. J. 2005;26:494–502. doi: 10.1183/09031936.05.00023605. [DOI] [PubMed] [Google Scholar]
- 2.Schünemann HJ, Dorn J, Grant BJ, Winkelstein W., Jr. Trevisan, Pulmonary function is a long-term predictor of mortality in the general population: 29-year follow-up of the Buffalo Health Study. Chest. 2000;118:656–664. doi: 10.1378/chest.118.3.656. [DOI] [PubMed] [Google Scholar]
- 3.Strachan DP. Ventilatory function, height, and mortality among lifelong non-smokers. J. Epidemiol. Community Health. 1992;46:66–70. doi: 10.1136/jech.46.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young RP, Hopkins R, Eaton TE. Forced expiratory volume in one second: not just a lung function test but a marker of premature death from all causes. Eur. Respir. J. 2007;30:616–622. doi: 10.1183/09031936.00021707. [DOI] [PubMed] [Google Scholar]
- 5.Hubert HB, Fabsitz RR, Feinleib M, Gwinn C. Genetic and environmental influences on pulmonary function in adult twins. Am. Rev. Respir. Dis. 1982;125:409–415. doi: 10.1164/arrd.1982.125.4.409. [DOI] [PubMed] [Google Scholar]
- 6.McClearn GE, Svartengren M, Pedersen NL, Heller DA, Plomin R. Genetic and environmental influences on pulmonary function in aging Swedish twins. J. Gerontol. 1994;49:264–268. doi: 10.1093/geronj/49.6.m264. [DOI] [PubMed] [Google Scholar]
- 7.Lewitter FI, Tager IB, McGue M, Tishler PV, Speizer FE. Genetic and environmental determinants of level of pulmonary function. Am. J. Epidemiol. 1984;120:518–530. doi: 10.1093/oxfordjournals.aje.a113912. [DOI] [PubMed] [Google Scholar]
- 8.Palmer LJ, et al. Familial aggregation and heritability of adult lung function: results from the Busselton Health Study. Eur. Respir. J. 2001;17:696–702. doi: 10.1183/09031936.01.17406960. [DOI] [PubMed] [Google Scholar]
- 9.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilk JB, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 13.Miller L-AD, et al. Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Dev. Dyn. 2004;231:57–71. doi: 10.1002/dvdy.20105. [DOI] [PubMed] [Google Scholar]
- 14.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 15.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Weigt C, Gaertner A, Wegner A, Korte H, Meyer HE. Occurrence of an actin-inserting domain in tensin. J. Mol. Biol. 1992;227:593–595. doi: 10.1016/0022-2836(92)90915-7. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Duncan IC, Bozorgchami H, Lo SH. Tensin1 and a previously undocumented family member, tensin2, positively regulate cell migration. Proc. Natl. Acad. Sci. USA. 2002;99:733–738. doi: 10.1073/pnas.022518699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manzke T, et al. 5–HT4(a) receptors avert opioid-induced breathing depression without loss of analgesia. Science. 2003;301:226–229. doi: 10.1126/science.1084674. [DOI] [PubMed] [Google Scholar]
- 19.Dupont LJ, et al. The effects of 5-HT on cholinergic contraction in human airways in vitro. Eur. Respir. J. 1999;14:642–649. doi: 10.1034/j.1399-3003.1999.14c26.x. [DOI] [PubMed] [Google Scholar]
- 20.Bayer H, et al. Serotoninergic receptors on human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2007;36:85–93. doi: 10.1165/rcmb.2006-0151OC. [DOI] [PubMed] [Google Scholar]
- 21.Mägert HJ, et al. LEKTI, a novel 15-domain type of human serine proteinase inhibitor. J. Biol. Chem. 1999;274:21499–21502. doi: 10.1074/jbc.274.31.21499. [DOI] [PubMed] [Google Scholar]
- 22.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sparvero LJ, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J. Transl. Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fehrenbach H, et al. Receptor for advanced glycation endproducts (RAGE) exhibits highly differential cellular and subcellular localisation in rat and human lung. Cell. Mol. Biol. 1998;44:1147–1157. [PubMed] [Google Scholar]
- 25.Konishi K, et al. Gene expression profiles of acute exacerbations of Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2009;180:167–175. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Englert JM, et al. A role for the receptor for advanced glycation end products in idiopathic pulmonary fibrosis. Am. J. Pathol. 2008;172:583–591. doi: 10.2353/ajpath.2008.070569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Favre CJ, et al. Expression of genes involved in vascular development and angiogenesis in endothelial cells of adult lung. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H1917–H1938. doi: 10.1152/ajpheart.00983.2002. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Herndon ME, Lawler J. The cell biology of thrombospondin-1. Matrix Biol. 2000;19:597–614. doi: 10.1016/s0945-053x(00)00107-4. [DOI] [PubMed] [Google Scholar]
- 30.Hancock DB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 2009 Dec 13; doi: 10.1038/ng.500. advance online publication, doi:10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhl GR, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch. Gen. Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 35.Kohansal R, et al. The natural history of chronic airflow obstruction revisited: an analysis of the framingham offspring cohort. Am. J. Respir. Crit. Care Med. 2009;180:3–10. doi: 10.1164/rccm.200901-0047OC. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 37.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 38.Guan Y, Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 40.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 41.Sayers I, Swan C, Hall IP. The effect of beta2-adrenoceptor agonists on phospholipase C (beta1) signalling in human airway smooth muscle cells. Eur. J. Pharmacol. 2006;531:9–12. doi: 10.1016/j.ejphar.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 42.Wadsworth SJ, Nijmeh HS, Hall IP. Glucocorticoids increase repair potential in a novel in vitro human airway epithelial wounding model. J. Clin. Immunol. 2006;26:376–387. doi: 10.1007/s10875-006-9029-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.