The determination of how enzymes achieve their catalytic power requires an understanding of how structural motifs are used to position functional groups of enzymes and substrates within active sites. The recent explosion of RNA crystal structures provides an extraordinary opportunity to delve deeply into the relationship between ribozyme structure and function. The Tetrahymena group I ribozyme provides an attractive system for such studies because of the wealth of structural information, with ten crystal structures of group I introns solved in the past five years,[1–5] and extensive functional information[6] that enables incisive analysis of the energetics of catalysis.
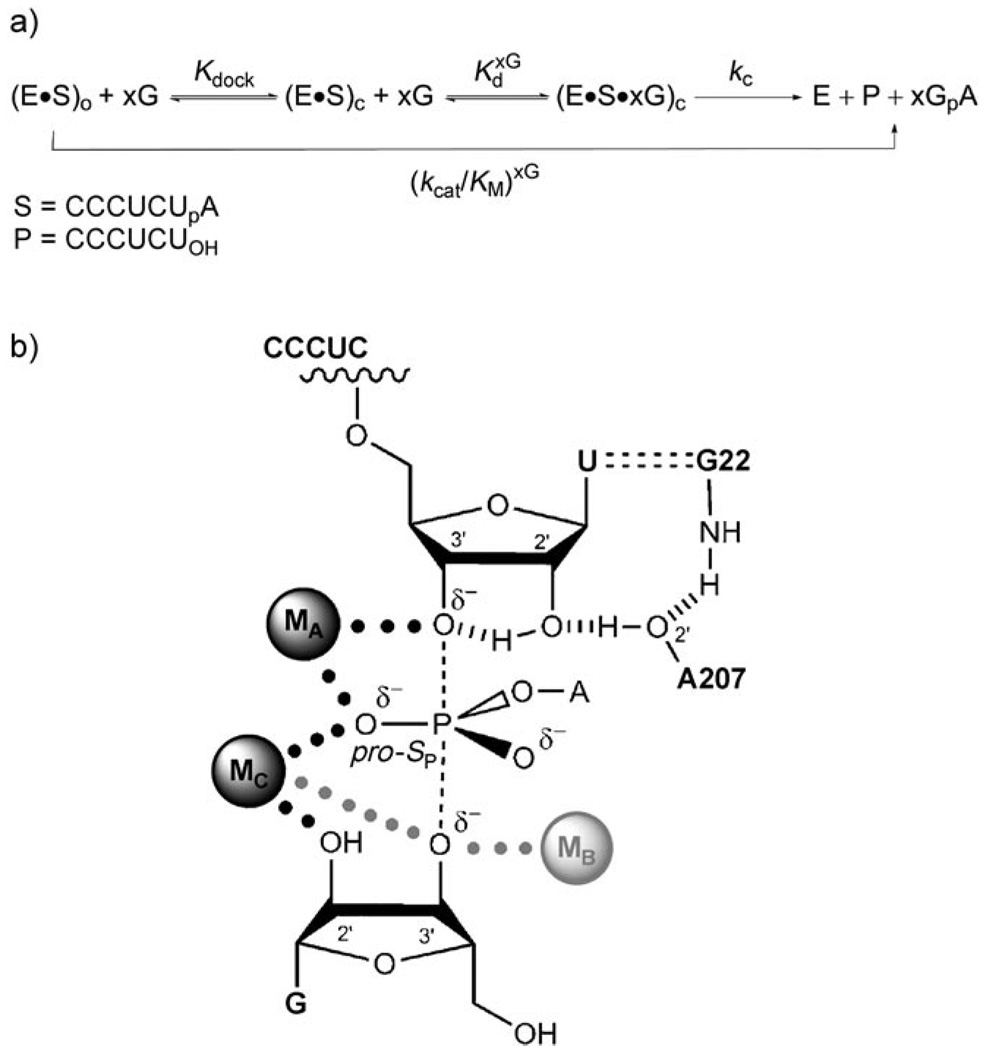
The Tetrahymena group I ribozyme catalyzes a phosphoryl-transfer reaction that mimics the first step of self-splicing of group I introns (Scheme 1).[6, 7] This reaction involves two substrates, an oligonucleotide (S) and an exogenous guanosine molecule. S binds to the ribozyme first by base pairing[8] to form the “open complex” (subscript “o” in Scheme 1), and then docks into the active site of the ribozyme[9, 10] to form tertiary interactions in the “closed complex” (subscript “c” in Scheme 1). Guanosine also binds to the ribozyme in a dedicated, highly conserved site,[1–3, 11] and the substrates can bind in either order. Within the bound complex, the 3′-OH group of guanosine is deprotonated and attacks S at a specific phosphoryl group, thereby transferring the phosphoryl group and the A tail. Functional and structural studies have elucidated an intricate network of active-site interactions involving metal ions and hydrogen bonds, and have partially dissected their contributions to catalysis (Scheme 1b).[6] With this wealth of information about direct catalytic interactions, this system is poised for investigation of how these active-site interactions are established.
Scheme 1.
The reaction catalyzed by the Tetrahymena ribozyme (E). a) Simplified reaction scheme, in which xG represents guanosine or a guanosine analogue. The subscripts “o” and “c” refer to the open and closed complex, respectively. The chemical step and product release are indicated as a single step. b) Representation of the transition state of the reaction.[6] The oligonucleotide substrate S is shown on top; the guanosine nucleophile is at the bottom. The metal ion interacting with the deprotonated 3′-OH group of the guanosine nucleophile (MB, gray) is shown as a separate metal ion from that interacting with the 2′-OH group of the same residue, as inferred from functional data;[26, 27] structural data suggest that the 3′-OH and 2′-OH groups of the guanosine nucleophile interact with the same metal ion, MC (interaction indicated by the gray dotted line to the oxygen atom at the 3′-position of guanosine).[1,5]
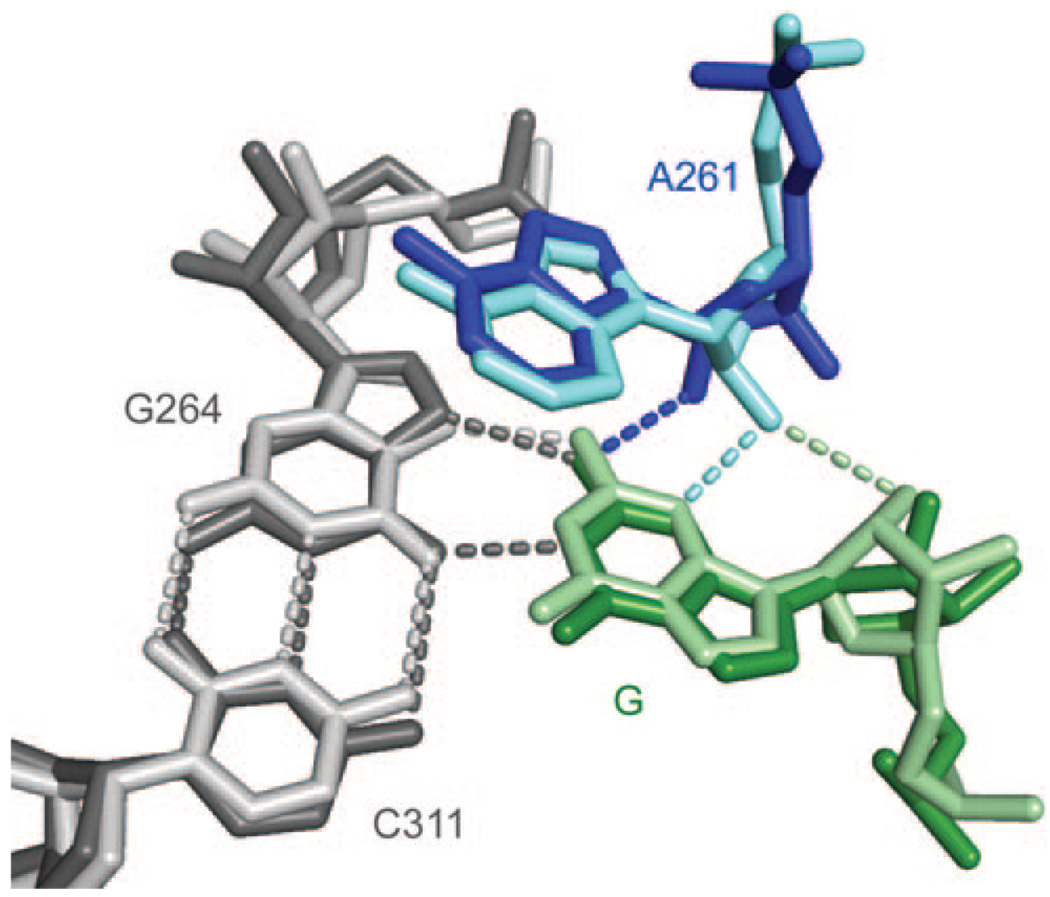
We focus on an interaction involving the 2′-OH group of the most conserved residue in group I introns, A261 (Tetrahymena ribozyme numbering; Figure 1).[11] Previous functional data have shown that the replacement of the 2′-OH group at position A261 with other groups is deleterious.[12–14] Consistent with its importance in catalysis, the 2′-OH group of A261 lies near N3 of guanosine in the structural model derived from crystals of the Twort group I ribozyme (Figure 1, lighter residues).[2] On the basis of this model, a ribose zipper interaction involving the donation of a hydrogen bond from the 2′-OH group of the guanosine nucleophile to the 2′-OH group of A261 (Figure 1, dashed light-green line), which in turn donates a hydrogen bond to N3 of guanosine (Figure 1, dashed light-blue line), has been proposed.[2, 15] A261 is part of one of the three base triples in the guanosine-binding site,[2] and hydrogen bonds between the 2′-OH group of A261 and the guanosine nucleophile may structurally connect two of these layers, presumably fortifying the interactions needed for transition-state stabilization. Because these proposed interactions involve conserved residues in the catalytic core, they have been suggested to be important for all the group I ribozymes.[2, 15] This proposed network of interactions involving the 2′-OH group of A261 is also consistent with functional data obtained subsequently that strongly imply a hydrogen-bond-donor role for the 2′-OH group of guanosine (Figure 1, dashed light-green line)[16] in addition to its well-established role as a ligand for a catalytic metal ion (Scheme 1 b).[17, 18]
Figure 1.
Superposition of structural models of the Twort[2] (lighter colors, PDB ID: 1Y0Q) and Azoarcus[4] (darker colors, PDB ID: 3BO3) group I ribozymes. The G–C base pair involved in a base-triple interaction is depicted in gray; A261 is in blue; the guanosine nucleophile (G) is in green. Hydrogen bonds in the base triple are indicated in gray. Interactions involving the 2′-OH group of A261 are indicated in light blue and light green (Twort) or dark blue (Azoarcus). The residues are numbered according to the Tetrahymena group I ribozyme. The figure was generated with PyMOL (W. L. DeLano, The PyMOL Molecular Graphics System (2002) on World Wide Web http://www.pymol.org).
Nevertheless, the environment surrounding the 2′-OH group of A261 varies somewhat in structural models of different group I ribozymes. In particular, recent models of the Azoarcus group I ribozyme[4] suggest a different network of interactions (Figure 1, darker residues), involving a contact between the 2′-OH group of A261 and the exocyclic amino group of guanosine (dashed blue line). This amino group has also been shown to be important for catalysis: guanosine binding is about 200 times as strong as the binding of inosine, which has a hydrogen atom instead of an amino group at this position.[19, 20]
The structural models that lead to these two different proposals show identical positioning of the base triple that involves the guanosine nucleophile (C311–G264–G)[11] and of the adenine ring of A261 above this base triple. However, the models differ in the positioning of the 2′-OH group of A261 as a result of different sugar puckering (Figure 1). Nevertheless, the structure factors from the structural models of Twort and Azoarcus ribozymes at a resolution of 3.6 and 3.3 Å, respectively, are consistent with either sugar pucker (see Figure 1 in the Supporting Information); thus, they do not resolve the functional interactions of the 2′-OH group of A261.
We used double-mutant cycles[21, 22] to determine which of these interactions contributes to catalysis by the Tetrahymena group I ribozyme. Relative to proteins, RNA molecules are readily amenable to conservative substitutions;[23] double-mutant cycles can often be constructed with atomic-level substitutions. Specifically, we asked whether replacement of the 2′-OH group at A261 with a group no longer capable of making the proposed interaction has the same functional effect, or a diminished effect, in reactions with guanosine and with a guanosine analogue lacking the proposed partner of the 2′-OH group at A261. A diminished effect would indicate a functional interdependence and would suggest, given the structural information, a direct interaction.
To test the proposed hydrogen-bond donation from the 2′-OH group of A261 to the N3 atom of guanosine, as suggested by the Twort structural model,[2, 15] we used a ribozyme containing a 2′-OMe group in the residue at position A261 (A261OMe). This construct is not capable of donating a hydrogen bond through the substituent at the 2′-position. It was used in combination with 3-deazaguanosine (3dNG), a guanosine analogue that lacks the nitrogen atom proposed to accept a hydrogen bond from the 2′-OH group of A261. To test the possible hydrogen-bond donation from the exocyclic amine group of guanosine to the 2′-OH group of A261, as suggested by the Azoarcus structural model,[4] we used a ribozyme containing a 2′-H atom, which is neither capable of accepting nor capable of donating a hydrogen bond, in the residue at position A261 (A261H), and inosine (I), a guanosine analogue that lacks the exocyclic amino group proposed to donate a hydrogen bond to A261. In the following, we use xG to refer to guanosine or guanosine analogues and G to refer solely to the cognate guanosine nucleophile.
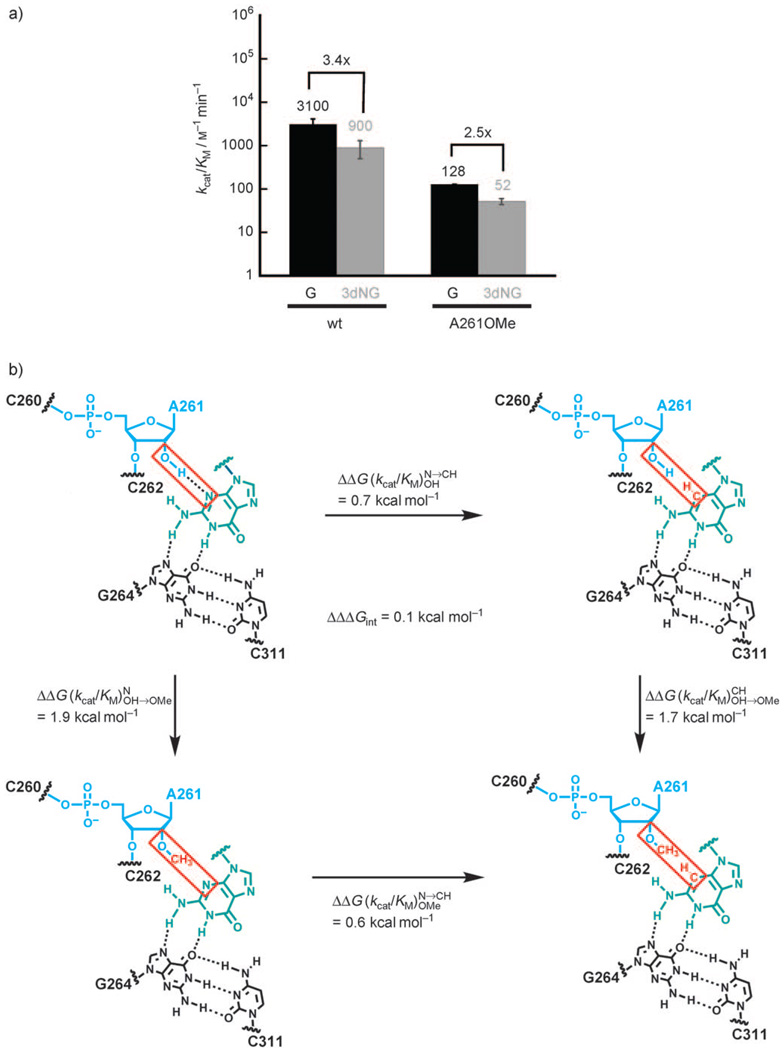
Figure 2 summarizes our test of the proposed interaction between the 2′-OH group of A261 and N3 of G.We measured (kcat/KM)xG (the second-order rate constants for the reaction (E·S)o+xG→products, Scheme 1) for two ribozymes, the wild-type (wt) ribozyme and the A261OMe ribozyme; xG represents either G or 3dNG. In the wt ribozyme, the reaction rates with G and 3dNG differ by less than a factor of four (Figure 2a; (kcat/KM)G=3100 m−1min−1; (kcat/KM)3dNG=900 m−1min−1). This difference corresponds to (Figure 2b). This small effect is evidence against a critical functional role of the N3 atom. G reacts 24 times more slowly with the A261OMe ribozyme than with the wt ribozyme (). This result is consistent with hydrogen-bond donation from the 2′-OH group of A261, in agreement with prior functional experiments,[12, 13] and with the proposal that an important contact is mediated by this 2′-OH group.[2, 15, 16]
Figure 2.
Testing the proposed contact between the 2′-OH group of A261 and N3 of guanosine. a) Values of kcat/KM for the reaction of the oligonucleotide substrate d(CCCUC)UdA5 with G (black) or 3dNG (gray) for the wt and A261OMe ribozymes (see the Supporting Information for experimental details). b) A double-mutant cycle suggests no functional interaction between the 2′-OH group of A261 and the N3 atom of guanosine. The contact tested is highlighted by a red box and corresponds to the light-blue dashed line in Figure 1. Horizontal arrows: difference in reactivity between G and 3dNG for the wt ribozyme (top) and the A261OMe ribozyme (bottom). Vertical arrows: difference in reactivity between the wt and A261OMe ribozymes when G (left) or 3dNG (right) was used as the nucleophile. Values of ΔΔG were calculated from the relationship ΔΔG=RTln(ratio (kcat/KM)), with ratio (kcat/KM) defined as the ratio between the kcat/KM value of the species at the beginning of the arrow and that of the species at the end of the arrow; the values were rounded to a single decimal figure to take into account the experimental errors (see errors bars in (a)).
We found that 3dGN reacts 2.5 times more slowly than G with the A261OMe ribozyme (Figure 2a), a difference which corresponds to (Figure 2 b). This effect is the same as that measured with the wt ribozyme (3.4- and 2.5-fold rate decreases, Figure 2a; ΔΔG = 0.7 and 0.6 kcal mol−1, Figure 2b). Thus, despite the significant deleterious effect of the 2′-OMe substitution at position A261, no coupling to the effect of the removal of the N3 atom from the guanosine nucleophile was observed (ΔΔΔGint = 0.1 kcal mol−1, Figure 2b). This result strongly suggests that the 2′-OH group of A261 and the N3 atom of G do not interact.
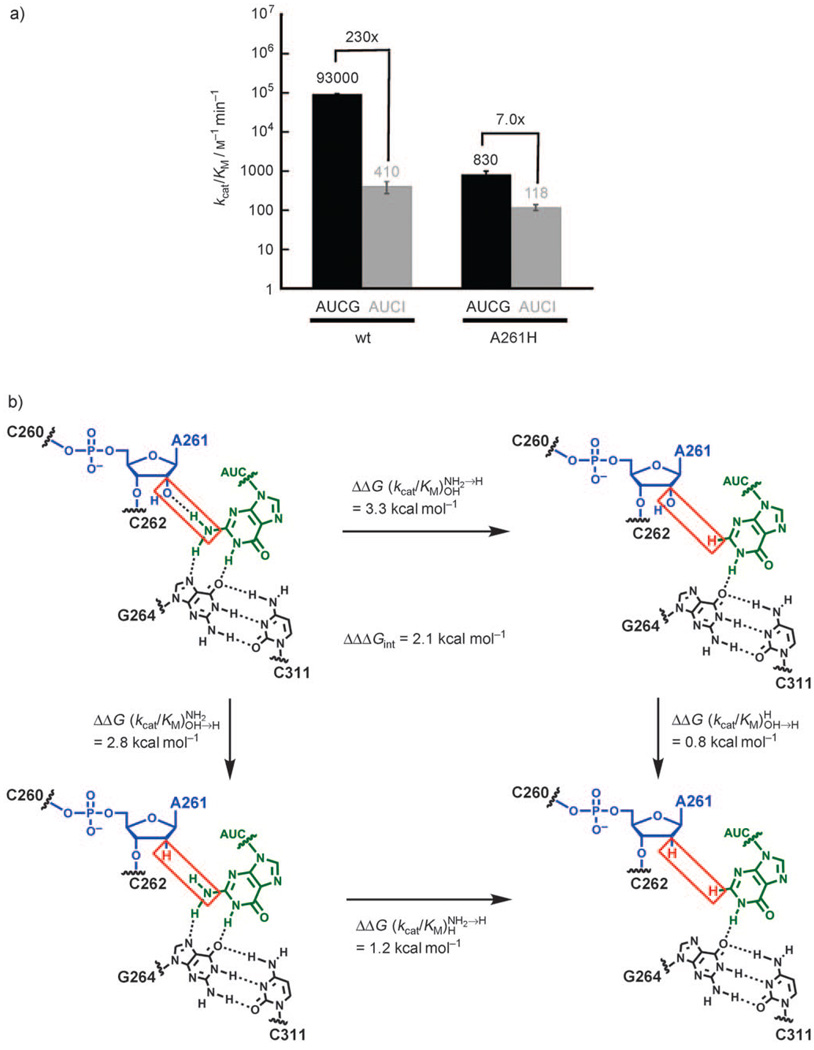
We next tested the possible interaction between the exocyclic amine group of G and the 2′-OH group of A261 (Figure 3). In this case, we measured (kcat/KM)xG (Scheme 1) for G and I, within the context of the wt and A261H ribozymes. To minimize a G-independent reaction, which proceeds faster than the G-dependent reaction at low G concentrations in the A261H ribozyme, we used AUCG and AUCI instead of G and I. The extra residues form additional base pairs with the ribozyme and strengthen binding without changing the reaction details.[24, 25] In agreement with previous results obtained with the wt ribozyme,[19, 20] the reaction of AUCI was about 230 times slower than that of AUCG (, Figure 3a). This result alone is not sufficient to identify the cause of such a decrease in reactivity. Indeed, the exocyclic amine group can potentially donate two hydrogen bonds; a hydrogen bond between the exocyclic amine group and the N7 atom of G264 has been proposed[11] and is supported by all structural models of the group I ribozymes (Figure 1 and Figure 3b).[1–5] The double-mutant cycle shown in Figure 3b is crucial for determining whether there is an additional interaction.
Figure 3.
Testing the possible contact between the 2′-OH group of A261 and the exocyclic amine group of guanosine. a) Values of kcat/KM for the reaction of the oligonucleotide substrate d(CCCUC)UdA5 with AUCG (black) or AUCI (gray) for the wt and A261H ribozymes (see the Supporting Information for experimental details). b) A double-mutant cycle suggests a functional interaction between the 2′-OH group of A261 and the exocyclic amine group of guanosine. The contact tested is highlighted by a red box and corresponds to the dark-blue dashed line in Figure 1. Horizontal arrows: difference in reactivity between AUCG and AUCI for the wt ribozyme (top) and the A261H ribozyme (bottom). Vertical arrows: difference in reactivity between the wt and A261H ribozymes when AUCG (left) or AUCI (right) was used as the nucleophile. ΔΔG is defined as in Figure 2. ΔΔG values were rounded to a single decimal figure to take into account the experimental errors (see errors bars in (a)).
If the deleterious effect measured when AUCI was used in place of AUCG for the reaction of the wt ribozyme were entirely due to interactions that do not involve the 2′-OH group of A261, we would expect this effect to remain the same in the A261H ribozyme, as observed for 3dNG in the wt and A261OMe ribozymes. However, in the A261H ribozyme, AUCI reacts only seven times more slowly than AUCG, instead of 230 times more slowly as observed for the wt ribozyme (Figure 3a). This effect corresponds to (Figure 3b), a value much smaller than that for the wt ribozyme (). The large value of ΔΔΔGint=2.1 kcal mol−1 indicates thermodynamic coupling between the 2′-OH group of A261 and the exocyclic amine group of G. The simplest model to explain this observation is that these two residues interact directly, as shown in dark blue in Figure 1.[4] As noted above, the exocyclic amine group of G is proposed to be involved in an additional interaction with residue G264 (Figure 1, Figure 2, and Figure 3),[11] and the residual sevenfold difference in reactivity between AUCG and AUCI in the A261H ribozyme is consistent with the loss of such an additional interaction.[19] However, the existence of hydrogen-bond donation from the exocyclic amine group of the guanosine nucleophile to the 2′-OH group of A261 indicates that a possible hydrogen bond from the 2′-OH group of G to the 2′-OH group of A261[2, 15, 16] is unlikely, given the conformational and packing constraints at this site.
In conclusion, we have used functional data based on site-specific chemical modifications to distinguish between different models derived from structural data. In doing so, we have provided evidence for an important hydrogen-bond interaction between the 2′-OH group of A261 and the exocyclic amine group of the guanosine nucleophile in the reaction catalyzed by the group I ribozyme. Although we have not investigated whether A261 adopts different conformations in different ribozymes, the high level of conservation in this site suggests that the interaction observed in this study will be universal. Nevertheless, the close arrangement of multiple hydrogen-bond donors and acceptors within this compact, highly structured RNA suggests that alternative structures that involve subtle rearrangements may be accessible for this and other RNA molecules. Indeed, RNA is prone to misfolding and often adopts alternative structures.[25] This ability, although it has the potential to limit structural homogeneity, may have helped ribozymes evolve rapidly and acquire the chemical complexity needed for the development of life, in spite of their rather simple array of functional groups, and may help RNA molecules involved in complex, multistep processes in current-day biology.
Supplementary Material
Footnotes
This research was supported by a grant from the NIH (GM 49243) to D.H. and by a grant from the HHMI to J.A.P. We thank Dr. Nan-Sheng Li for oligonucleotide syntheses, Dr. James L. Hougland for preliminary results, Dr. Barbara Golden for the structure factors associated with the Twort ribozyme crystals, Dr. Niket Shah and Dr. Jonathan K. Lassila for helpful discussions, and members of the Herschlag and Piccirilli research groups for comments on the manuscript.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200903006.
Contributor Information
Dr. Marcello Forconi, Department of Biochemistry, Beckman Center, B400, Stanford University, Stanford, CA 94305-5307 (USA), Fax: (+1) 650-723-6783, Homepage: http://cmgm.stanford.edu/biochem/herschlag/.
Raghuvir N. Sengupta, Department of Biochemistry and Molecular Biology, Department of Chemistry, and Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, CIS W406, Chicago IL 60637 (USA), Fax: (+1) 773-702-0271.
Dr. Mao-Chin Liu, Department of Pharmacology and Developmental Therapeutics Program Cancer Center, Yale University School of Medicine (USA)
Prof. Dr. Alan C. Sartorelli, Department of Pharmacology and Developmental Therapeutics Program Cancer Center, Yale University School of Medicine (USA)
Prof. Dr. Joseph A. Piccirilli, Email: jpicciri@uchicago.edu, Department of Biochemistry and Molecular Biology, Department of Chemistry, and Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, CIS W406, Chicago IL 60637 (USA), Fax: (+1) 773-702-0271.
Prof. Dr. Daniel Herschlag, Email: herschla@stanford.edu, Department of Biochemistry, Beckman Center, B400, Stanford University, Stanford, CA 94305-5307 (USA), Fax: (+1) 650-723-6783, Homepage: http://cmgm.stanford.edu/biochem/herschlag/.
References
- 1.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Nature. 2004;430:45. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 2.Golden BL, Kim H, Chase E. Nat. Struct. Mol. Biol. 2005;12:82. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 3.Guo F, Gooding AR, Cech TR. Mol. Cell. 2005;16:351. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Lipchock SV, Strobel SA. Proc. Natl. Acad. Sci. USA. 2008;105:5699. doi: 10.1073/pnas.0712016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahley MR, Strobel SA. Science. 2005;309:1587. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 6.Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. In: The RNA World. 3rd ed. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. p. 133. [Google Scholar]
- 7.Zaug AJ, Cech TR. Science. 1986;231:470. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 8.Herschlag D, Cech TR. Biochemistry. 1990;29:10159. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 9.Bevilacqua PC, Turner DH. Biochemistry. 1991;30:10632. doi: 10.1021/bi00108a005. [DOI] [PubMed] [Google Scholar]
- 10.Herschlag D. Biochemistry. 1992;31:1386. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 11.Michel F, Hanna M, Green R, Bartel DP, Szostak JW. Nature. 1989;342:391. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- 12.Strauss-Soukup JK, Strobel SA. J. Mol. Biol. 2000;302:339. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- 13.Ortoleva-Donnelly L, Szewczak AA, Gutell RR, Strobel SA. RNA. 1998;4:498. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwans JP. The University of Chicago. 2003 [Google Scholar]
- 15.Golden BL. In: Ribozymes and RNA Catalysis. Lilley DMJ, Eckstein F, editors. Cambridge: The Royal Society of Chemistry; 2008. [Google Scholar]
- 16.Hougland JL, Sengupta RN, Dai Q, Deb SK, Piccirilli JA. Biochemistry. 2008;47:7684. doi: 10.1021/bi8000648. [DOI] [PubMed] [Google Scholar]
- 17.Shan S, Herschlag D. Biochemistry. 1999;38:10958. doi: 10.1021/bi990388e. [DOI] [PubMed] [Google Scholar]
- 18.Sjögren A-J, Petterson E, Sjöberg B-M, Strömberg R. Nucleic Acids Res. 1997;25:648. doi: 10.1093/nar/25.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bass BL, Cech TR. Nature. 1984;308:820. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- 20.McConnell TS, Cech TR. Biochemistry. 1995;34:4056. doi: 10.1021/bi00012a024. [DOI] [PubMed] [Google Scholar]
- 21.Fersht A. Structure and Mechanism in Protein Science. New York: W.H. Freeman; 1999. [Google Scholar]
- 22.Horovitz A, Fersht AR. J. Mol. Biol. 1990;214:613. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 23.Das SR, Fong R, Piccirilli JA. Curr. Opin. Chem. Biol. 2005;9:585. doi: 10.1016/j.cbpa.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Moran S, Kierzek R, Turner DH. Biochemistry. 1993;32:5247. doi: 10.1021/bi00070a037. [DOI] [PubMed] [Google Scholar]
- 25.Russell R, Herschlag D. RNA. 1999;5:158. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan S, Kravchuk AV, Piccirilli JA, Herschlag D. Biochemistry. 2001;40:5161. doi: 10.1021/bi002887h. [DOI] [PubMed] [Google Scholar]
- 27.Shan S, Yoshida A, Piccirilli JA, Herschlag D. Proc. Natl. Acad. Sci. USA. 1999;96:12299. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.