Abstract
Embryonic stem (ES) cells are pluripotent cells that can self renew or be induced to differentiate into multiple cell lineages, and thus have the potential to be utilized in regenerative medicine. Key pluripotency specific factors (Oct 4/Sox2/Nanog/Klf4) maintain the pluripotent state by activating expression of pluripotency specific genes and by inhibiting the expression of developmental regulators. Pluripotent ES cells are distinguished from differentiated cells by a specialized chromatin state that is required to epigenetically regulate the ES cell phenotype. Recent studies show that in addition to pluripotency specific factors, chromatin remodeling enzymes play an important role in regulating ES cell chromatin and the capacity to self-renew and to differentiate. Here we review recent studies that delineate the role of ATP dependent chromatin remodeling enzymes in regulating ES cell chromatin structure.
Keywords: Embryonic stem cells, Pluripotency, Self-renewal, Differentiation, Chromatin, Histone modifications, Histone variants, Chromatin remodeling enzymes
Introduction
Embryonic stem (ES) cells are cells derived from the inner cell mass of the mammalian blastocyst that can give rise to the three germ layers and ultimately to different cell types. ES cells have the capacity to self renew and are also pluripotent in that they can differentiate into multiple cell lineages. Because of these characteristics, ES cells are ideal models for studying molecular mechanisms that determine cell fate, and have the potential to be utilized in replacement and regenerative therapy for treating a variety of human diseases.
ES cell pluripotency is characterized by transcriptional profiles that promote self-renewal and silence differentiation specific gene expression. A limited set of transcription factors, designated “core” pluripotency factors, are required to maintain ES cell pluripotency and when introduced in combination can re-program differentiated cells back to a pluripotent state [1–4]. The pluripotency factors, Oct4, Sox2, Nanog, and Klf4 co-occupy target genes and function in an extensive regulatory circuit that silences the expression of key transcription factors required for differentiation and activates the expression of genes important for maintenance of pluripotency [5–7]. Other important factors, including c-Myc, regulate a distinct set of genes and can enhance reprogramming of adult cells to the pluripotent state [6]. The delicate balance between gene activation and repression may be regulated by the extent of promoter co-occupancy by the four different pluripotency factors. Silent promoters are generally bound by a single factor while actively transcribed promoters are simultaneously bound by multiple pluripotency factors [6]. The correlation between promoter occupancy by pluripotency factors and gene expression also coincides with specific chromatin modifications [6]. Thus, in addition to transcriptional regulation by the combinatorial activities of multiple pluripotency factors, there is another level of regulation imposed by epigenetic mechanisms that involve ES cell specialized chromatin states. The activities of chromatin remodeling enzymes have been implicated in maintaining the unique chromatin structure in pluripotent ES cells and for catalyzing the dramatic reorganization of chromatin structure that occurs during differentiation [8].
Embryonic Stem Cell Chromatin
The packaging of DNA into chromatin through associations with histone and non-histone proteins inhibits multiple cellular processes. Chromatin restricts DNA accessibility and generates complex mechanisms for regulation of processes that utilize DNA [9]. The basic unit of chromatin is the nucleosome, formed by the wrapping of 146 base pairs of DNA around a core octamer of two copies each of four histone proteins (H2A, H2B, H3, and H4) [10]. Nucleosomes occur as repeating arrays that cover approximately 75–90% of the genome and are separated by linker DNA that is often associated with a fifth histone, H1 [11]. DNA that is packaged into nucleosomes is less accessible to DNA binding proteins than is linker DNA, thus nucleosome positioning regulates utilization of specific DNA sites by transcription factors and other regulatory proteins [12]. Furthermore, the orientation of DNA around the histone octamer can also influence site accessibility to DNA binding proteins and its utilization [13, 14]. The chromatin state of a particular site is further regulated by the incorporation of variant histones, that differ in amino acid sequence from conventional histones, and by histone post-translational modifications such as acetylation, methylation, phosphorylation, and ubiquitination that can alter DNA-histone interactions and may generate a “code” that can be recognized by specific protein domains to activate or repress gene expression [15–17]. Thus, nucleosome structure provides multiple mechanisms for regulating cellular processes such as transcription: by altering DNA site accessibility and by generating alternative recognition sites through specialized chromatin structure.
In addition to local nucleosome structure, higher order chromatin structure is generated by the wrapping of nucleosome arrays into three dimensional structures [18]. On a genomic level, chromatin can be divided into compact heterochromatic regions and less condensed euchromatic regions [19]. The regulation of higher order chromatin is not well understood but appears to be closely intertwined with that at the nucleosome level, being dependent on nucleosome positioning and H1 association at linker regions, incorporation of particular histone variants, and specific histone post-translational modifications [11, 15, 20].
“Poised” Chromatin in Embryonic Stem Cells
A characteristic feature of ES cell chromatin is that specific regulatory sites, particularly those at lineage specific transcription factor loci appear to be in a silent but poised state of activation. This specialized chromatin state is promoted by the incorporation of histone variants and by covalent histone modifications.
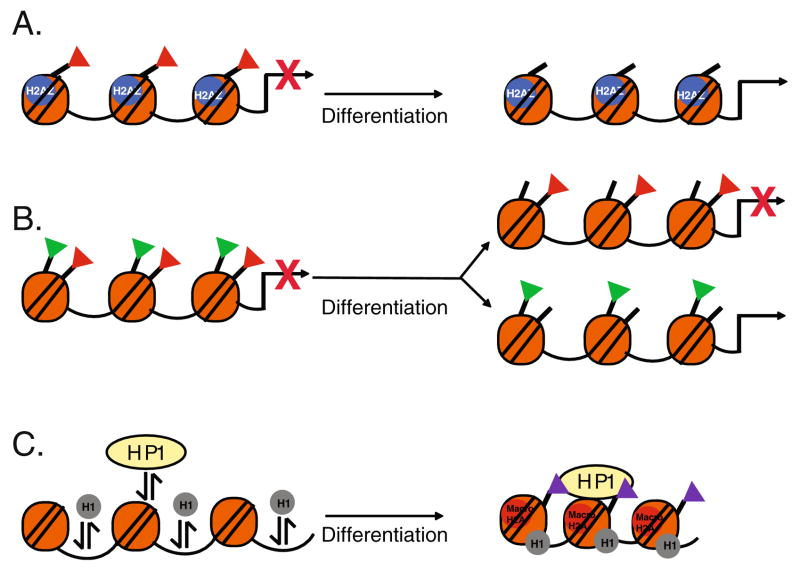
The histone variant, H2AZ is required for early mammalian development and has been implicated in regulating ES cell specific gene expression [21, 22]. H2AZ, incorporated at regions flanking transcriptional start sites, can influence nucleosome positioning, H1 linker binding, chromatin remodeling enzyme activity, and can promote both transcriptional activation and repression [23–25]. In pluripotent ES cells, H2AZ is enriched at silenced developmentally regulated promoters and is correlated with binding of repressive proteins as well as repressive covalent histone modifications [22]. Down-regulation of H2AZ results in de-repression of H2AZ occupied promoters, indicating that H2AZ plays an important role in silencing developmental regulators. Interestingly, upon differentiation, H2AZ becomes enriched at actively transcribed promoters. These data suggest that H2AZ incorporation may be one of several mechanisms (see below) by which developmental regulators are kept silent but poised for activation in ES cells (Fig. 1a).
Fig. 1.
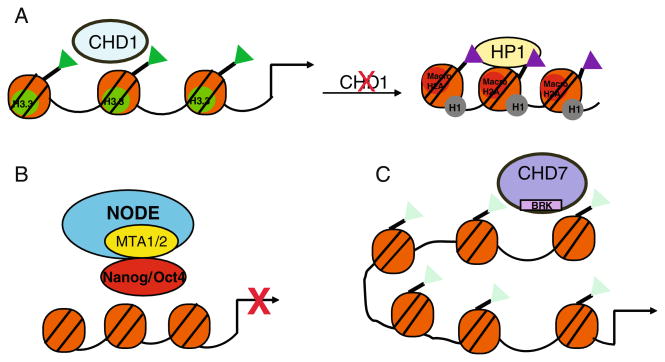
Embryonic Stem Cell Chromatin. The specialized state of chromatin structure in ES cells is promoted by incorporation of histone variants and by specific histone modifications and is dramatically remodeled upon differentiation. a In pluripotent ES cells, incorporation of the histone variant H2AZ is associated with the repressive covalent modification, H3K27me3 (red triangles) and is required to repress developmentally regulated promoters. Upon differentiation, H2AZ is redistributed to active promoters. b The chromatin structure at developmentally regulated sites is characterized by bivalent domains that have both repressive, H3K27me3 (red triangles) and activating, H3K4me3 (green triangles) covalent modifications. Upon differentiation, these bivalent modifications are resolved depending on transcriptional activity. c ES cell chromatin is characterized by hyperdynamic binding and rapid exchange of heterochromatin protein 1 (HP1) and histone H1. Upon differentiation, HP1 and H1 association with chromatin is stabilized and contributes to heterochromatin formation. There is also an increase in the repressive histone modifications, H3K9me2 (purple triangles) and incorporation of macroH2A at heterochromatin foci
The amino termini of the core histones extend from the nucleosome core and are sites for multiple modifications that have profound effects on gene expression. These modifications may act as “marks” that signal activation or repression of transcription. Acetylation of histones H3 and H4 and tri-methylation of histone H3 at lysine 4 (H3K4me3) are marks that are generally associated with activation of transcription, whereas, tri-methylation of histone H3 at lysine 9 (H3K9me3) and lysine 27 (H3K27me3) are marks that are repressive [26]. Covalent modifications regulate transcription by altering chromatin structure and by promoting the binding of regulatory proteins.
The epigenetic landscape characteristic of pluripotent ES cells displays features that may promote plasticity in gene expression. For example, lineage-specific genes are simultaneously enriched for repressive modifications (H3K27me3) and for activating marks (H3 acetylation and H3K4me3) [27, 28]. These “primed” genes are associated with less condensed chromatin structure and thus replicate earlier in ES cells than in differentiated cells [27]. During differentiation, repressive marks (H3K27me3) are erased from activated lineage specific promoters whereas activating marks (H3K4me3) are erased from promoters that remain silent [28]. Thus, “bivalent”marks provide signals to maintain the expression of distinct differentiation specific factors in a silent but poised state for activation and are an important feature that may promote ES cell plasticity (Fig. 1b).
Hyperdynamic Transcriptionally Active Chromatin in ES Cells
Pluripotent ES cells are characterized by higher order chromatin structure that is generally dynamic and permissive to the transcriptional machinery. In the pluripotent state, ES cell chromatin is enriched in euchromatin, and upon differentiation there is accumulation of highly condensed, transcriptionally inactive heterochromatic foci [29, 30]. Multiple mechanisms promote heterochromatin formation during ES cell differentiation. Heterochromatin protein (HP1), initially diffuse in undifferentiated ES cells, becomes concentrated into discret foci upon differentiation. Differentiation is also associated with changes in the residence time that HP1 and histone proteins are bound to chromatin. In undifferentiated ES cells, there is rapid exchange of HP1, as well as linker histone H1, indicating that they are hyper-dynamically bound [30] (Fig. 1c).
On a global level, ES cell chromatin is associated with chromatin marks indicative of active transcription and deficient in repressive marks [31]. Differentiation results in increased levels of repressive H3K9me2/3 and increased global incorporation of the histone variant, macroH2A, associated with silencing of gene expression [30, 32, 33] (Fig. 1c). Increases in H3K9me2 occur over large blocks of chromatin termed “LOCKs” (large organized chromatin K9 modifications) that overlap with silenced nuclear lamina associated genes [33]. Activated genes may be looped out of repressive “LOCK” regions.
Permissive chromatin structure is also evident at specific genomic regions and these regions become more condensed after ES cell differentiation. As female ES cells differentiate, inactivation of the X chromosome is associated with deposition of repressive H3K27me3 and decreased levels of activating H3K4me3 [34]. Furthermore, ES cell telomere chromatin is highly enriched in the histone variant, H3.3, which is phosphorylated at Ser 31 (H3.3S31P) during metaphase. Upon differentiation, there is a decrease in Ser31 (H3S31P), an increase in repressive marks, decreased chromatin accessibility at the telomere ends, and an increase in H3.3S31P at pericentric heterochromatin [35]. Thus, ES cell pluripotency is most likely maintained not only by an open global arrangement of chromatin structure that promotes high levels of gene expression but also by a unique arrangement of chromatin structure at telomeres that promotes ES cell specific telomere function. Chromatin remodeling enzymes play an important role in regulating chromatin structure in ES cells. In this review, we will focus on a class of chromatin remodeling enzymes that utilize the hydrolysis of ATP to regulate ES cell chromatin structure in the pluripotent state and during differentiation.
The Role of Chromatin Remodeling Enzymes in Embryonic Stem Cells
Chromatin remodeling enzymes fall into two basic categories: those that add or remove histone covalent modifications and those that utilize the energy of ATP hydrolysis to disrupt chromatin structure. Enzymes that post-translationally modify the amino termini of histone proteins, by adding acetyl, methyl, phosphate, sumo, or ubiquitin moieties have been referred to as “writers” of a “code” that signals activation or repression of gene expression [26]. These enzymes include histone acetyl transferases (HATs) which comprise a large family of enzymes that add acetyl groups to specific amino acids on histone proteins and generally function to activate transcription. Histone methyl transferases (HMTs) add methyl groups to histone proteins and can be activating (e.g. H3K4me3) or repressive for transcription (e.g. HK9me3 and H3K27me3), depending on the methylated residue. The activities of HATs and HMTs are dynamically opposed by the activities of histone deacetylases (HDACs) and histone demethylases which remove these groups and are often referred to as “erasers”. For example, the Polycomb proteins play a critical role in repressing expression of developmental regulators in pluripotent ES cells by catalyzing histone methylation at H3K27 while the activities of members of the Jumonji domain family of demethylases remove critical methyl groups from histones during ES cell differentiation [36–41]. A newly discovered mechanism for “erasing” histone covalent modifications in differentiating ES cells involves removing histone tails by proteolytic cleavage [42]. Thus, enzymatic activities that add and remove covalent modifications dynamically regulate ES cell chromatin structure in the pluripotent state and during differentiation. Posttranslational modifications can form a code that is then “read” by the second category of chromatin remodeling enzymes.
The second category of chromatin remodeling enzymes consists of enzymes that utilize the energy from the hydrolysis of ATP to disrupt contacts between histone proteins and DNA and induce changes in nucleosome conformation, positioning and higher order chromatin structure [43, 44]. ATP dependent chromatin remodeling enzymes have been shown to increase DNA accessibility, allowing gene specific regulators or general transcription factors to bind in order to activate or repress gene expression [45]. It is thought that re-distribution of nucleosome positions in response to extracellular signals or during development requires the activities of chromatin remodeling enzymes. Because chromatin remodeling enzymes generally lack sequence specific binding ability, they are thought to be recruited to specific sites by interactions with gene specific transcriptional regulators. In ES cells, ATP dependent chromatin remodeling enzymes have been shown to either regulate the expression of pluripotency factors or to cooperate with pluripotency factors to regulate chromatin structure and gene expression [8].
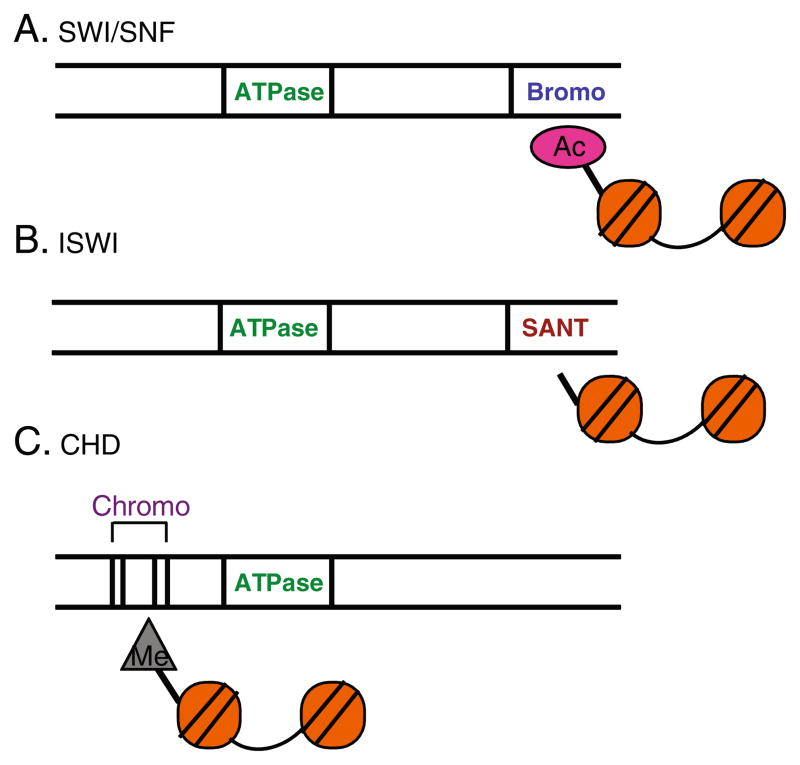
Three well characterized families of ATP dependent chromatin remodeling enzymes (SWI/SNF, ISWI, CHD) have been implicated in regulating aspects of embryonic stem cell pluripotency. The enzymes in these families are multi-subunit complexes containing a catalytic subunit with a conserved ATPase domain as well as additional components to form large multiprotein-complexes. The three families are distinguished by unique features within their catalytic subunits (Fig. 2) that allow them to “read” specific histone post-translational modifications that stabilize their interaction with chromatin. However, the ATP dependent chromatin remodeling enzymes are more than just “readers” because they have been shown to promote specific histone post-translational modifications and to promote incorporation of histone variants (see below). Components of these three families of ATP dependent chromatin remodeling complexes have been demonstrated to play important roles in early development and in regulating ES cell chromatin. In addition, the TIP60-p400, a complex that contains a related ATP dependent catalytic subunit and a histone acetyl transferase, also plays an important role in regulating ES cell chromatin.
Fig. 2.
Three ATP Dependent Chromatin Remodeling Families are distinguished by signature motifs that recognize specific histone modifications. The different ATP dependent chromatin remodeling enzymes have a conserved ATPase domain and additional motifs that confer functional specificity. a The SWI/SNF ATPases contain a bromodomain that has high affinity for acetylated histones. b The ISWI ATPases contain a SANT domain that has high affinity for unmodified histones. c The CHD ATPases have chromodomains that have high affinity for methylated histones
SWI/SNF
Mammalian SWI/SNF enzymes are multisubunit complexes of 1–2 MDa and consist of 9–12 subunits, one of which has an ATPase that is either Brg1 or Brm [46]. The distinguishing feature of this family of chromatin remodeling enzymes is that the catalytic subunits, Brg1 and Brm, have a bromodo-main that preferentially interacts with acetylated histones [46]. In vitro, a core complex consisting of one of the two ATPses, Baf47, and Baf155/170 are required for optimum nucleosome remodeling activity [47]. In vivo, heterogeneous SWI/SNF complexes are generated by the presence of either ATPase and a varying composition of associated subunits (Bafs). SWI/SNF complexes are often classified into two major subfamilies: Baf and Pbaf (polybromo) [48]. Pbaf complexes are distinguished from Baf complexes by the incorporation of two additional subunits, Baf180 and Baf200 and the lack of Baf250 [49]. Many studies suggest that Baf subunits are important for protein-protein interactions and for responding to signaling pathways, thus they impart functional specificity to the SWI/SNF complex [50–54]. SWI/SNF enzymes are well known for playing crucial roles in cellular differentiation. In fact, SWI/SNF enzymes are required for almost every differentiation event looked at to date [46]. Recent studies also implicate SWI/SNF components as important regulators of ES cell pluripotency.
Most SWI/SNF components are required for early mouse development [55–60]. Brm is the only SWI/SNF subunit that has been disrupted and does not result in embryonic lethality [61]. Disruption of the alternative SWI/SNF ATPase, Brg 1 results in lethality at the blastocyst stage, thus ES cells cannot be derived from Brg1 null zygotes [55]. Moreover, maternally derived Brg1 is required for zygotic genome activation, a transcriptional reprogramming event that establishes totipotency and is required for continued development [62]. Interestingly, zygotes derived from Brg1 depleted oocytes exhibit reduced levels of transcription and a decrease in H3K4me2 levels, suggesting that Brg1 is required for downstream epigenetic changes associated with transcriptional activation that are needed for further embryonic development.
Components of the SWI/SNF complex are required for ES cells to self renew and to maintain a pluripotent state as well as for the process of differentiation. Several studies suggest that Brg1 is expressed earlier in development than Brm and is the predominant SWI/SNF ATPase in pluripo-tent ES cells [63–65]. Pluripotent ES cells are also characterized by high expression of Baf155 and Baf60a and low expression of the paralogous Baf170 and Baf60c [65]. Furthermore, ES cell specific PBAF complexes contain a novel component, BRD7. BRD7 containing PBAF complexes have different biochemical properties and differentially regulate gene expression as ES cell BAF complexes [64]. Thus, heterogeneous SWI/SNF complexes may play specialized roles in regulating ES cell chromatin.
A large scale RNAi screen directed against chromatin structural and regulatory proteins identified Brg1 as being required to maintain ES colony morphology [66]. Knockdown of Brg1 in undifferentiated ES cells leads to loss of self renewal [65]. Brg1 interacts with the pluripotency factors, Nanog, Oct4, and Sox2, and co-localizes with these factors at target genes, suggesting that the requirement for Brg1 in maintaining a proliferative ES cell state is programmatic rather than a general requirement for cell viability [65, 67, 68]. Brg1 depleted ES cells are impaired in ectodermal and mesodermal differentiation, suggesting that Brg1 is also required for ES cell pluripotency [65]. Thus, Brg1 is required both for ES cell proliferation and the capacity to differentiate into different cell lineages.
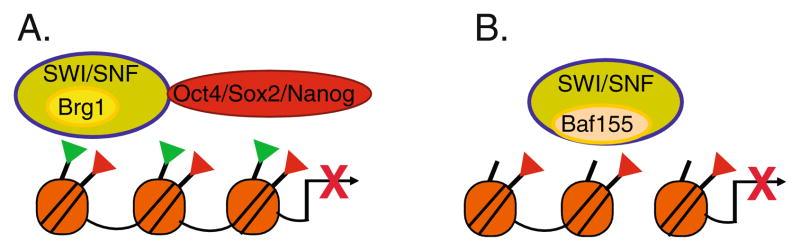
Brg1 is highly enriched at actively transcribed promoters but knockdown of Brg1 mostly has a de-repressive effect on gene expression, particularly on many genes that regulate ES cell pluripotency as well as on developmental regulators [68, 69]. Oct4 and Sox2 maintain a pluripotent state by activating pluripotency specific genes and by repressing differentiation specific genes. Brg1 was found, at least initially, to repress Oct4 and Sox2 target genes that promote pluripotency, suggesting that the SWI/SNF role in promoting pluripotency is to refine the activities of Oct 4 and Sox2 by preventing over-expression of pluripotency specific genes [68]. In contrast to the opposing role Brg1 plays with Oct4 and Sox2 in repressing targets that promote pluripotency, Brg1 cooperates with Oct4 and Sox2 to repress differentiation specific gene expression [68, 69]. Interestingly, Brg1 binds to a significant number of genes that have bivalent chromatin marks, suggesting that Brg1 may regulate the silent but “poised” state of chromatin on developmentally regulated genes in ES cells [69]. In combination, these data suggest that in pluripotent ES cells, the Brg1 component of the SWI/SNF complex interacts with pluripotency factors and thereby plays an important role in the repression of gene expression (Fig. 3a). This repressive function is in contrast to the generally positive role Brg1 plays in regulating gene expression during zygotic genome activation [62]. Thus, BRG1 based SWI/SNF complexes play a dynamic role in regulating gene expression in early development. Studies suggest that SWI/SNF function may be modulated by the incorporation of specific BAFs.
Fig. 3.
SWI/SNF complexes regulate ES cell pluripotency. a Brg1 interacts with pluripotency factors to down-modulate pluripotency specific gene expression and to inhibit expression of developmental regulators. BRG1 is significantly enriched at loci that have bivalent domains with H3K27me3 (red triangles) and H3K4me3 (green triangles). b Baf155 is required to compact chromatin and promote increased H3K27me3 (red triangles) at the Nanog promoter during differentiation. These modifications lead to repression of Nanog expression
The incorporation of the two alternative Baf specific subunits, Baf250a or Baf250b into the SWI/SNF complex is modulated during ES cell differentiation. Baf250a is incorporated to a greater extent prior to differentiation and Baf250b is incorporated to a greater extent after differentiation [64]. Baf250a has previously been shown to repress proliferation specific gene expression, in opposition to Baf250b, which activates expression of these genes [70]. Thus, the expression of the repressive Baf250a or the activating Baf250b subunit correlates with SWI/SNF function in mediating transcriptional repression in pluripo-tent ES cells and gene activation upon differentiation. However, depletion of either Baf250a or Baf250b severely compromises ES cell pluripotency [60, 71].
Baf250a and Baf250b are both required to maintain ES cell pluripotency, but differentially regulate the potential of ES cells to develop into specific lineages. ES cells deficient in Baf250a cannot differentiate into mesoderm derived cardio-myocytes but form primitive endoderm like cells and ectoderm derived neurons [60]. In contrast, disruption of Baf250b activates the mesodermal marker, Gata2 [71]. Thus, distinct SWI/SNF complexes containing either Baf250a or Baf250b differentially mediate cell specification during ES cell differentiation. This indicates that aspects of SWI/SNF specificity are intrinsically defined by subunit composition in order to allow SWI/SNF function to change at different stages of development. Interestingly, the BAF155 subunit plays a role in regulating ES cell differentiation by a mechanism that may be distinct from that of Baf250a and Baf250b [72].
A large scale screen for repressors of Nanog expression in differentiating ES cells identified Baf155, a core component of the SWI/SNF complex [72]. Nanog levels remain aberrantly high in differentiating ES cells depleted of Baf155 and differentiation is compromised. Depletion of Baf155 also results in the failure to condense chromatin structure at the Nanog promoter and interferes with deposition of the repressive H3K27me3 mark [72]. Furthermore, on a global level, down-regulation of Baf155 results in reduced H3K9me3 foci associated with hetero-chromatin formation in differentiated ES cells [72]. Thus, in addition to maintaining important features of ES cell pluripotency through interactions with the ES cell transcriptional circuitry, SWI/SNF complexes are required to halt the activities of this circuitry and to promote the dramatic changes in chromatin structure characteristic of a differentiated cell (Fig. 3b).
ISWI
The mammalian ISWI complexes contain one of two alternative ATPase subunits, Snf2h and Snf2l, each of which has a SANT domain that preferentially interacts with unmodified histones [73]. Each of the ATPases can associate with different factors to generate distinct ISWI complexes. In mammalian cells, Snf2h is a component of RSF [74], WICH [75], NoRC [76], CHRAC [77] and ACF [78]. The different ISWI complexes containing Snf2h display distinct modes for interacting with nucleosomes and have unique biological functions that include replication of heterochromatin and regulation of transcription [79–82]. Disruption of Snf2h in mice is embryonic lethal (prior to implantation), however the precise role that Snf2h plays in early development is difficult to define because of its incorporation into multiple complexes with diverse functions [83]. Snf2h was recently identified as a component of a large chromatin remodeling network that is highly expressed in pluripotent oocytes and ES cells compared to somatic cells [84].
The alternative ISWI ATPAse, Snf2l, is a component of NURF and CERF, both of which have been implicated in early development [85, 86]. NURF was first characterized in Drosophila where it can activate and repress transcription and plays an important role in fly development [87, 88]. Mammalian NURF is composed of Snf2l, RbAp46/48, and BPTF (Bromodomain PHD-finger Transcription Factor). The PHD finger of BPTF preferentially interacts with H3K4me3 marks, thus coupling the ATP dependent chromatin remodeling activities of NURF with histone methylation [89].
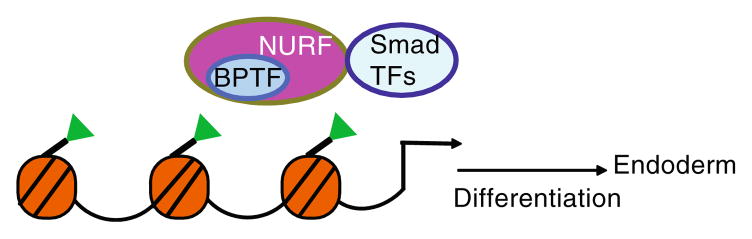
BPTF has recently been shown to play an important role in early embryonic development with disruption leading to lethality just after the implantation stage and failure to form distal visceral endoderm [90]. In ES cells, depletion of BPTF inhibits expression of genes required for development of mesoderm, endoderm, and ectoderm but has only mild effects on proliferation. The requirement for BPTF in differentiation of endoderm is linked to interactions with transcription factors of the Smad signaling pathway and activation of Smad responsive genes [90] (Fig. 4).
Fig. 4.
BPTF is required for endoderm differentiation. The BPTF subunit of the NURF complex interacts with H3K4me3 (green triangles) and with SMAD transcription factors to activate genes important for distal visceral endoderm differentiation
CHD
The ATPase subunits (CHDs) of the chromodomain heli-case DNA-binding (CHD) family of chromatin remodeling enzymes are distinguished by having two chromodomains that have affinity for methylated histones [91]. There are nine CHD proteins that can be divided into three subfamilies based on the presence of other conserved domains and interacting factors: I. [CHD1 and 2], II. [CHD3 and 4], III. [CHD 5–9]. Members of each sub-family have been shown to play important roles in ES cells.
CHD1 is a transcriptional activator that has an ATPase domain, two chromodomains which recognize H3K4me2/3, and a DNA binding domain that interacts with AT rich sequences [92, 93]. Down-regulation of CHD1 in pluripo-tent ES cells compromises self-renewal and results in decreased expression of Oct-4 and approximately 25 other genes [94]. Although CHD1 depleted cells retain aspects of the pluripotent ES cell state, a large number of genes involved in neurogenesis become up-regulated and the cells become prone to spontaneous neural differentiation. Upon differentiation to embryoid bodies, there is an increase in neural differentiation but loss of endoderm and mesoderm formation. Despite a primarily negative role in transcription, genome wide binding studies showed that CHD1 binding correlates with that of RNA Polymerase II and H3K4me3 and that upon differentiation, CHD1 is relocalized to activated promoters of differentiation specific genes [94].
The generally permissive chromatin structure characteristic of pluripotent ES cells is dramatically altered in CHD1 depleted ES cells [94]. There is striking accumulation of highly condensed, heterochromatic foci with enrichment in H3K9me3 and HP1γ. Importantly, H3K9 histone methyl-transferase or H3K9 demethylase levels do not change with CHD1 depletion, indicating that CHD1 is directly needed to counteract H3K9 methylation in pluripotent ES cells in order to prevent heterochromatin accumulation. How does a chromatin remodeling enzyme that associates with euchro-matin prevent heterochromatin formation? In Drosophila, CHD1 mediates incorporation of the histone variant, H3.3 [95]. H3.3 is associated with active transcription and is less readily methylated at H3K9 [96]. If CHD1 promotes incorporation of H3.3 in ES cells, this activity would help prevent formation of heterochromatin foci and maintain the open chromatin structure that characterizes the ES cell pluripotent state (Fig. 5a).
Fig. 5.
The CHD chromatin remodelers regulate chromatin structure in ES cells. a CHD1 is associated with the activating histone mark, H3K4me3 (green triangles) required to maintain permissive chromatin structure in pluripotent ES cells, possibly by promoting the incorporation of H3.3. Down-regulation of CHD1 results in formation of heterochromatin foci. b The ES cell specific complex called NODE interacts with Nanog and Oct4 to repress expression of developmentally regulated genes. C. CHD7 is enriched at distal regulatory elements at actively expressed loci and co-localizes with H3K4me2 (light green triangles). CHD7 binding at distal regions may promote enhancer-promoter interactions
The CHD3 (Mi2α) and CHD4 (Mi2β) sub-family of CHD proteins have, in addition to the ATPase and chromodomains, PHD fingers that interact with methylated histones [97]. The CHD ATPases are components of the Nucleosome Remodeling Deacetylase (NuRD) complexes. NURD complexes uniquely contain both ATP dependent chromatin remodeling and histone deacetylase activities [98]. NuRD complexes are composed of at least seven components that include CHD3 or CHD4, deacetylase subunits Hdac1/2 and associated subunits including methyl-CpG-binding proteins, Mbd 1/2/3 and metastasis associated, Mta1/2/3 [98]. These complexes play important roles in transcriptional repression and have been shown to antagonize SWI/SNF mediated activation of gene expression during an inflammatory response and during B cell development [99, 100]. CHD4 is also involved in activation of gene expression and is required for hematopoi-etic stem cell self renewal and differentiation [101–103]. Components of NuRD have been implicated in the regulation of ES cell pluripotency and differentiation via histone deacetylation.
Mbd3 is required for post-implantation development of the mouse embryo [104] for maintainance of ES cell pluripotency, but not self renewal [105]. Loss of MBD3 results in hyperacetylation and de-repression of select genes such as Pramel6 and Pramel7 but does not have any effect on the expression of pluripotency factors such as OCT4 or Nanog. MBD3 also represses trophoectoderm specific gene expression and MBD3 deficient ES cells are prone to trophoectoderm differentiation [106]. Thus MBD3 mediated repression of gene expression is required to maintain ES cell pluripotency and for full cell lineage development.
Interestingly, a NURD complex called NODE that lacks MBD3 but contains Mta1/2 and Hdac1/2 has been detected in ES cells [67]. NODE interacts with Nanog and Oct4 in ES cells and is recruited to Nanog/Oct4 target genes, independently of Mbd3. Knockdown of Mta1 has different effects on gene expression when compared to depletion of Mbd3. Depletion of Mta1 causes up-regulation of lineage specific factors, such as Gata6 and Foxa2 [67]. This requirement for Mta1 in repressing markers associated with endoderm differentiation is similar to that for Nanog. Thus, a unique repressive complex containing a subset of NuRD components plays a differential role in maintaining ES cell pluripotency by directly interacting with Nanog and Oct4 (Fig. 5b).
CHD7 is a member of a less well characterized sub-family of CHD proteins that each possesses BRK domains. The function of the BRK domain is unclear but it has been implicated in binding CTCF, a protein involved in insulator activity [107]. Disruption of CHD7 is embryonic lethal, suggesting an important role in early development [108]. In humans, mutations in CHD7 are associated with a genetic disorder called CHARGE. CHARGE causes a wide range of birth defects that affects multiple cell types [109]. Moreover, one of the mutations that causes CHARGE syndrome disrupts one of the two BRK domains of CHD7, suggesting that this region has an important function [110]. Genome wide mapping of CHD7 binding in ES cells and other cell types showed that CHD7 binding coincides with di-methylated H3K4 and is associated with active gene expression [111]. Highly expressed genes are associated with CHD7 binding and binding is strongest at sites distal to promoters that have enhancer-like properties. Furthermore, CHD7 is re-localized to newly activated genes upon ES cell differentiation into neural progenitors. The finding that CHD7 is highly enriched at distal regulatory regions suggests that CHD7 may activate gene expression by mediating long range communication between distal regions and promoters through chromatin looping (Fig. 5c). In CHARGE patients, disruption of CHD7 function during development mostly likely inhibits gene expression and this could impair differentiation into multiple cell types, thus explaining the wide spectrum of symptoms.
TIP60-p400
The mammalian p400/Domino contains a bipartite SWI/SNF like ATPase, characteristic of the SWR1 class of remodelers, and is part of a complex containing the TIP60 acetyltransferase. These enzymes play an important role in histone variant exchange in both yeast and Drosophila [112, 113]. In yeast, SWR1 deposits the yeast orthologue of H2AZ at euchromatic regions [112]. In Drosophila, the TIP6-p400 complex acetylates the fly orthologue of phosphoH2AX at double strand breaks and exchanges them for unmodified histones [113]. Disruption of p400 in mouse is embryonic lethal and is characterized by severe defects in early embryonic hematopoiesis [114]. TIP60 is also required for early embryonic development [115].
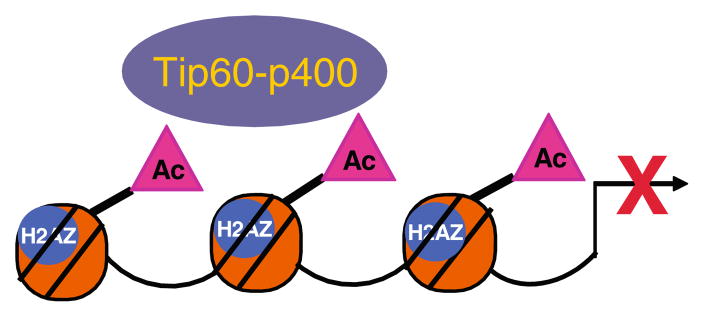
ES cells depleted of Tip60-p400 subunits exhibit altered morphology and are impaired in their ability to self renew [66]. Down-regulation of either TIP-60 or p400 has a mostly de-repressive effect on gene expression, particularly on genes that encode developmental regulators. However, p400 binding is strongest at highly expressed promoters and is correlated with the H3K4me3 mark and with bivalent marks. There is significant overlap between genes affected by Tip60-p400 depletion and those affected by depletion of the pluripotency transcription factor Nanog, however, Nanog does not directly recruit Tip60-p400 to target promoters. Interestingly, Tip60-p400 catalyzes histone H4 acetylation on repressed promoters as well as activated promoters. Histone acetylation is predominantly a mark of active transcription, but in the context of other repressive marks, Tip60-p400 mediated histone acetylation is associated with gene repression (Fig. 6).
Fig. 6.
The Tip60-p400 complex represses gene expression in pluripotent ES cells. In ES cells, down-regulation of Tip60-p400 has a de-repressive effect on the expression of many genes. This introduces the possibility that Tip60-p400 mediated histone acetylation can be associated with gene repression rather than activation depending on the chromatin context
Conclusions
ES cells are characterized by two properties, the ability to self-renew and the ability to differentiate into multiple cell lineages (pluripotency). These properties are associated with different states of chromatin structure, which are established, maintained, and eventually altered by the activities of chromatin remodeling enzymes. Chromatin structure in pluripotent ES cells is generally permissive, characterized by incorporation of specific histone variants, and the presence of bivalent covalent modifications at specific sites. Upon differentiation, there is a dramatic re-organization of chromatin structure that results in formation of heterochromatic foci and redistribution of histone variants and altered histone covalent modifications. Two classes of enzymes have been implicated in establishing, maintaining, and remodeling chromatin structure in ES cells. Enzymes that add and remove post-translational modifications to histones and enzymes that utilize the energy of ATP hydrolysis to disrupt histone-DNA contacts play important roles in regulating ES cell chromatin structure.
Recent studies have shown that several ATP dependent chromatin remodeling enzymes are required for ES cell self-renewal, for pluripotency, and for differentiation into specific lineages. A general theme that emerges from these studies is that the activities of ATP dependent chromatin remodelers is dynamically regulated to maintain a plurip-otent ES cell state and that their function can change upon differentiation. For example, in pluripotent ES cells, SWI/SNF enzymes interact with pluripotency factors to inhibit expression of developmental regulators. Upon ES cell differentiation, SWI/SNF activity turns off pluripotency specific genes, such as Nanog, and activates expression of developmental regulators. How is SWI/SNF function altered from a role as a repressor of differentiation to that of an activator? Clearly, SWI/SNF subunit composition plays an important role in modulating SWI/SNF function, but the mechanisms that trigger alterations in SWI/SNF subunit composition have not been defined nor have the mechanisms by which they alter SWI/SNF function. Also, how does the specialized chromatin structure that characterizes distinct ES cell developmental states modulate chromatin remodeling at specific loci and on a genome wide basis? Lastly, how the activities of the different chromatin remodeling enzymes are integrated to regulate chromatin structure in ES cells is not well understood. A better understanding of the mechanisms that regulate ATP dependent chromatin remodeling activity in ES cells, will clarify the mechanisms by which chromatin is prepared to achieve a developmentally appropriate gene expression program. We will then be better able to utilize ES cells in regenerative medicine.
Acknowledgments
Financial Support ILD was supported by the National Institute of Environmental Health Sciences; Grant number: 5K22ES12981, Ohio Cancer Research Associates, American Cancer Society, Ohio Division
References
- 1.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Jiang J, Chan YS, Loh YH, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 8.Keenen B, de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- 9.Hayes JJ, Wolffe AP. The interaction of transcription factors with nucleosomal DNA. Bioessays. 1992;14:597–603. doi: 10.1002/bies.950140905. [DOI] [PubMed] [Google Scholar]
- 10.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 11.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson RT. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 13.Simpson RT. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 14.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleo-somal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- 16.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 17.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature Reviews Molecular Cell Biology. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekker J. A closer look at long-range chromosomal interactions. Trends Biochem Sci. 2003;28:277–280. doi: 10.1016/S0968-0004(03)00089-6. [DOI] [PubMed] [Google Scholar]
- 19.Dillon N. Heterochromatin structure and function. Biol Cell. 2004;96:631–637. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Fan Y, Nikitina T, Zhao J, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Faast R, Thonglairoam V, Schulz TC, et al. Histone variant H2A. Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 22.Creyghton MP, Markoulaki S, Levine SS, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thakar A, Gupta P, Ishibashi T, et al. H2A. Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochem. 2009 doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 24.Li B, Pattenden SG, Lee D, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 29.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nature Reviews Molecular Cell biology. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 30.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developments in Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efroni S, Duttagupta R, Cheng J, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25:2567–2574. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 33.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marks H, Chow JC, Denissov S, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong LH, Ren H, Williams E, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 37.Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan EM, Muratore-Schroeder TL, Cook RG, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutierrez JL, Chandy M, Carrozza MJ, Workman JL. Activation domains drive nucleosome eviction by SWI/SNF. European Molecular Biology Organization journal. 2007;26:730–740. doi: 10.1038/sj.emboj.7601524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biol. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 46.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nature Reviews Genetics. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 47.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 48.Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–661. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Zhai W, Richardson JA, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Link KA, Burd CJ, Williams E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–2215. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 53.Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem. 2008;283:11924–11934. doi: 10.1074/jbc.M705401200. [DOI] [PubMed] [Google Scholar]
- 54.Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. Crucial roles for interactions between Mll3/4 and Ini1 in Nuclear Receptor Transactivation. Molecular Endocrinology. 2009 doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 56.Guidi CJ, Sands AT, Zambrowicz BP, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. European Molecular Biology Organization Reports. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lickert H, Takeuchi JK, Von Both I, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 60.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muchardt C, Bourachot B, Reyes JC, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. European Molecular Biology Organization Journal. 1998;17:223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dauvillier S, Ott MO, Renard JP, Legouy E. BRM (SNF2alpha) expression is concomitant to the onset of vasculogenesis in early mouse postimplantation development. Mech Dev. 2001;101:221–225. doi: 10.1016/s0925-4773(00)00560-8. [DOI] [PubMed] [Google Scholar]
- 64.Kaeser MD, Aslanian A, Dong MQ, Yates JR, Emerson BM. Brd7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. Journal of Biological Chemistry. 2008 doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 68.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 70.Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. European Molecular Biology Organization Journal. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan Z, Wang Z, Sharova L, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schaniel C, Ang YS, Ratnakumar K, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009 doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nature Reviews Molecular Cell Biology. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 74.LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 75.Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. European Molecular Biology Organization Journal. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strohner R, Nemeth A, Jansa P, et al. NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. European Molecular Biology Organization Journal. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poot RA, Dellaire G, Hulsmann BB, et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. European Molecular Biology Organization Journal. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bochar DA, Savard J, Wang W, et al. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci USA. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He X, Fan HY, Garlick JD, Kingston RE. Diverse regulation of SNF2h chromatin remodeling by non-catalytic subunits. Biochemist. 2008;47:7025–7033. doi: 10.1021/bi702304p. [DOI] [PubMed] [Google Scholar]
- 80.Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- 81.Cavellan E, Asp P, Percipalle P, Farrants AK. The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J Biol Chem. 2006;281:16264–16271. doi: 10.1074/jbc.M600233200. [DOI] [PubMed] [Google Scholar]
- 82.Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA. 2003;100:14097–14102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Assou S, Cerecedo D, Tondeur S, et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banting GS, Barak O, Ames TM, et al. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum Mol Genet. 2005;14:513–524. doi: 10.1093/hmg/ddi048. [DOI] [PubMed] [Google Scholar]
- 86.Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. European Molecular Biology Organization Journal. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Badenhorst P, Xiao H, Cherbas L, et al. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 90.Landry J, Sharov AA, Piao Y, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genetics. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flanagan JF, Mi LZ, Chruszcz M, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 93.Sims RJ, 3rd, Millhouse S, Chen CF, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaspar-Maia A, Alajem A, Polesso F, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Konev AY, Tribus M, Park SY, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 98.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 99.Gao H, Lukin K, Ramirez J, Fields S, Lopez D, Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci USA. 2009;106:11258–11263. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramirez-Carrozzi VR, Nazarian AA, Li CC, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 102.Yoshida T, Hazan I, Zhang J, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Williams CJ, Naito T, Arco PG, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 104.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 106.Zhu D, Fang J, Li Y, Zhang J. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS ONE. 2009;4:e7684. doi: 10.1371/journal.pone.0007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Allen MD, Religa TL, Freund SM, Bycroft M. Solution structure of the BRK domains from CHD7. J Mol Biol. 2007;371:1135–1140. doi: 10.1016/j.jmb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 108.Bosman EA, Penn AC, Ambrose JC, Kettleborough R, Stemple DL, Steel KP. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 109.Vissers LE, van Ravenswaaij CM, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 110.Lalani SR, Safiullah AM, Fernbach SD, et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schnetz MP, Bartels CF, Shastri K, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A. Z into euchromatin. PLoS Biology. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kusch T, Florens L, Macdonald WH, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 114.Ueda T, Watanabe-Fukunaga R, Ogawa H, et al. Critical role of the p400/mDomino chromatin-remodeling ATPase in embryonic hematopoiesis. Genes Cells. 2007;12:581–592. doi: 10.1111/j.1365-2443.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- 115.Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]