Abstract
The prognosis of adult normal karyotype (NK) precursor B cell acute lymphoblastic leukemia (B-ALL) has not improved over the last decade mainly because separation into distinct molecular subsets has been lacking and no targeted treatments are available. We screened the genome of blasts from ten adult NK B-ALL patients for novel genomic alterations by array comparative genomic hybridization and verified our results with fluorescent in situ hybridization and gene expression profile using the same probes. The results demonstrate cryptic deletions of 9q34 involving SET, PKN3, NUP188, ABL1 and NUP214 in three of the samples. The smallest deletion resulted in the likely juxtaposition of the SET and NUP214 genes. This aberration has not been described before in adult NK B-ALL. Larger number of samples is warranted to determine the prognostic significance of this cryptic deletion.
Introduction
The outcome of adult normal karyotype (NK) precursor B cell acute lymphoblastic leukemia (B-ALL) has not improved over the last decade [1, 2]. In addition, our ability to stratify these patients into prognostically different subgroups and develop targeted therapies is lacking. Identifying new molecular aberrations could help both in risk stratification and in finding new therapeutic targets. Using genome-wide analysis of genetic alterations, five groups [3-7] demonstrated hidden gene dose changes in a total of 64 NK B-ALL cases of whom only four were adults. We therefore screened the genome of blasts from ten consecutive adult NK B-ALL patients admitted to Roswell Park Cancer Institute (RPCI) for novel genomic alterations by array comparative genomic hybridization (array CGH).
Materials and Methods
Patient samples
Cryopreserved bone marrow samples from three females and seven males with a median age at diagnosis of 56.5 (range, 21-70) years were studied. All patients underwent induction treatment on or off protocol with a five-drug containing regimen followed by intensive consolidation and maintenance regimens [8]. Six patients achieved complete remission (CR). Three patients relapsed at a median of 21 (range 6-25) months. After a median follow-up of 85 months for the living patients, three are alive in continuous first CR. None of the patients underwent allogeneic transplantation in first CR. The study was approved by RPCI’s Scientific Review Committee and the Institutional Review Board.
Cytogenetic analysis
Standard cytogenetic analysis was performed on Giemsa banded metaphase chromosomes from ALL bone marrow aspirates as previously described [9]. Twenty metaphases were reviewed in each case; none harbored t(9;22)(q34;q11).
Cell Sorting
Two samples with <60% blasts were sorted according to the diagnostic immunophenotypic pattern. The following antibodies were used: anti-CD19 (clone J4.119) phycoerythrin (PE) cyanine 5-conjugated monoclonal antibody (mAb), anti-CD34 (clone D3HL60.251) PE-conjugated mAb, anti-CD38 (clone T16) fluorescein isothiocyanate-conjugated mAb and anti-CD45 (clone J33) allophycocianin-conjugated mAb (Beckman Coulter, Miami, FL). In the bivariate display of forward vs. side scatter, a generous mononuclear sorting region was drawn to exclude debris and aggregates and the leukemic cells were further selected as CD45 negative to dim, and expressing CD19 and CD34 or CD38. Dead cells were excluded with violet fixable live dead reagent (Invitrogen, Carlsbad, CA). Cells were sorted using FACSAria (BD Bioscience, San Jose, CA) and the percentage of blasts following sorting exceeded 95% in the two samples with a viability >90%.
BAC Array CGH
Array CGH was performed on the RPCI bacterial artificial chromosome (BAC) array containing ~19,000 BAC clones as previously described [10-12].
CNV/SNP array analysis
Sample 05-1528 carried the smallest deletion of the 3 samples exhibiting 9q34 rearrangements. In order to refine the breakpoints in this sample a high resolution copy number variation (CNV)/single nucleotide polymorphism (SNP) analysis was performed using the Infinium Human1M-Duov3 (Illumina, Inc., San Diego, CA) as per manufacturer instructions, and analyzed in Genome Studio (Illumina, Inc.).
Interphase Fluorescence in situ hybridization (FISH) validation
DNA from BAC clones was labeled with SpectrumOrange or SpectrumGreen (Abbott Molecular,Inc., Des Plaines, IL) by nick translation using a kit from Abbott Molecular Inc., as per the manufacturer’s instructions. Labeled DNA was then hybridized to archived cytogenetic preparations from the appropriate patients as previously described [13]. Slides were visualized and analyzed on a Nikon Microscope using the CytoVision Program (Applied Imaging, Inc., Monroe Twp, NJ).
Gene expression array analysis
Based on material availability, total RNA was extracted and paired with Human Universal Reference Total RNA (Clontech, Inc., Mountain View, CA) for two color gene expression analysis using Agilent Technology 44k Whole Genome Oligo microarrays (Foster City, CA) as per manufacturers’ instructions. The expression data were extracted and processed with BGsub (background correction), Dyenorm (dye normalization) and Ratio (log ratio calculation) algorithms implemented in Agilent Feature Extraction Software.
Results and Discussion
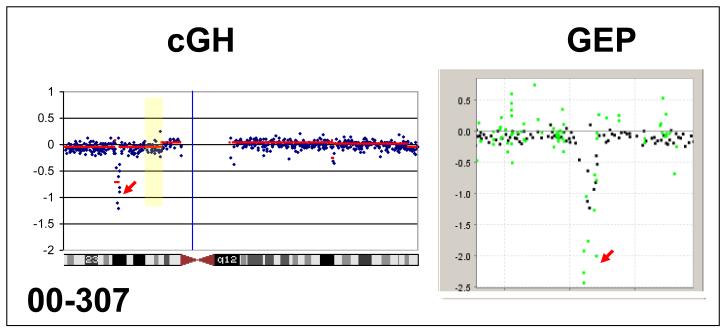
Three samples (Table 1) were found to have cyclin-dependent kinase inhibitor 2A (CDKN2A) deletions and these were confirmed by FISH (Figure 1). Specifically, 99-1069 carried a hemizygous deletion in 35% (35/100), and a homozygous deletion in 9/100 of the interphase nuclei examined while sample 00-307 harbored biallellic or homozygous deletions for CDKN2A in 93% (186/200) of the interphase nuclei examined. This latter sample also demonstrated a 2.0-fold decrease in CDKN2A mRNA expression (Figure 1). Deletions of CDKN2A have been previously reported in adolescents and young adults [14]. In that study, 46% of adolescents and young adults carried CDKN2A deletions, while in our sample set this loss was detected in 33%. Due to the small numbers in our series this difference does not seem to be significant.
Table 1.
| Patient ID | Age/Gender | CR | Relapse | DFS (Months) | Deletion |
|---|---|---|---|---|---|
| 99-350 | 43/F | Yes | No | 115+ | CDKN2A, 9q34 |
| 99-1069 | 52/M | Yes | Yes | 6 | CDKN2A |
| 00-307 | 68/M | No | CDKN2A | ||
| 00-947* | 70/M | Yes | Yes | 25 | 9q34 |
| 01-782 | 56/M | Yes | No | 85+ | |
| 02-231 | 53/M | No | |||
| 02-1252* | 21/F | Yes | Yes | 21 | |
| 06-063 | 68/M | No | |||
| 05-1528 | 42/F | No | 9q34 | ||
| 07-874 | 68/M | Yes | No | 15+ |
Abbreviations: CR, complete remission, DFS, disease-free survival; F, female; M, male
Two samples with <60% blasts were sorted according to the diagnostic immunophenotypic pattern.
Figure 1.
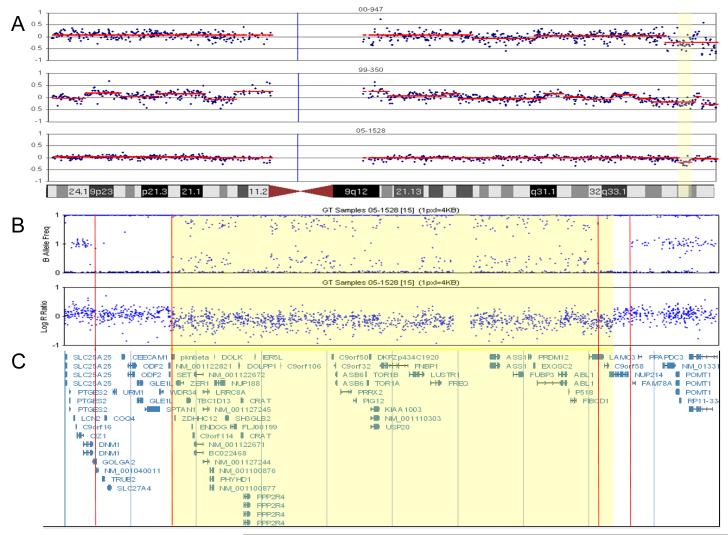
A. BAC array CGH results of three NK B-ALL samples with recurrent deletion on chromosome 9. The yellow region is the minimal critical region of the deletion. Each plot represents the log2 test/control value for each BAC (in blue) as well as the segmented value (in red). This value is equivalent to the LogR ration in panel B.
B. CNV/SNP array (Illumina 1M Duo) results defining the breakpoints in sample 05-1528. Top plot is the B Allele Frequency (BAF). The BAF is the percent of the sample that has the B Allele for a given SNP. A BAF of 0 indicates the SNP is AA and BAF of 1 means the SNP is BB and a BAF of 0.5 is AB. Note, the BAF in 05-1528 deviates from the normal three band pattern at 9q34.11. The double heterozygous band is indicative of a deletion with ~25% normal cell DNA contamination, and is supported by the decrease in the LogR ratio in panel B. The Log R ratio value is the log2 of test/normal for each SNP. Note the loss of material at 9q34.11. The two sets of red lines indicate regions of uniparental disomy (UPD) observed as loss of heterozigosity flanking the rearrangement. The lack of contaminating normal suggests that these UPD regions are in the germline. The yellow highlighted region defines the region of copy number loss. C. Genomic description of the deleted area.
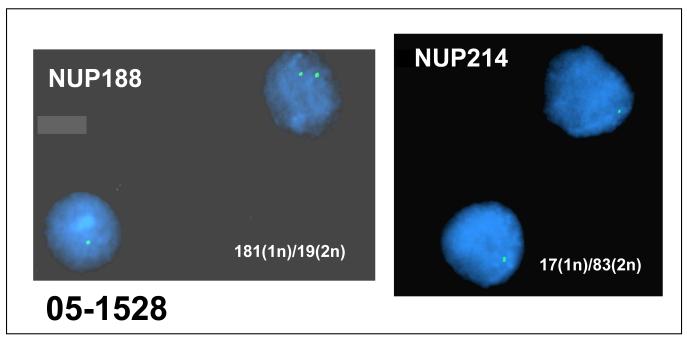
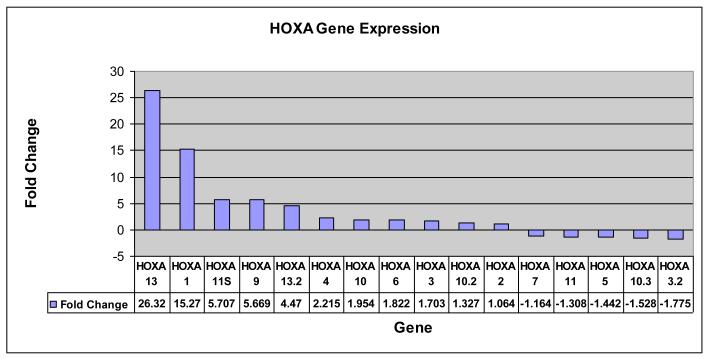
Three samples were found to have overlapping deletions of 9q34 based on BAC array CGH results (Figure 2A). These deletions included two nucleoporin NUP188 and NUP 214. Sample 99-350 exhibited a hemizygous deletion of NUP214 in 27% (27/100) nuclei and 1.7 and 1.9-fold decrease in NUP188 and NUP214 gene by expression profiling. Further, sample 05-1528 carried hemizygous deletions in 181/200 nuclei for NUP188 and a hemizygous deletion of NUP214 in 17% (17/100) nuclei (Figure 3). A 1.3- and 1.2-fold decrease in NUP188 and NUP214 mRNA was detected in this sample. A comparison of the gene expression data also revealed increased gene expression of the HOXA gene family in 05-1528 which has been previously observed in pediatric T cell ALL [15] and one case of adult acute myeloid leukemia (AML) [16] with deletions of 9q34 that result in a fusion gene from the juxtaposition of the telomeric region of the SET gene with NUP214 (Figure 4).
Figure 2.
CDKN2A/p16 homozygous deletion by array CGH and concordance of gene expression illustrated by an overlay of array CGH profiles and gene expression profiles (GEP) at CDKN2A. Red arrows point to CDKN2A.
Figure 3.
FISH analysis demonstrating deletion of NUP188 (left panel) and NUP214 (right panel) in NK B-ALL. FISH was performed using the same BAC probe used in the array CGH.
Figure 4.
Activation of HOXA gene cluster. The figure demonstrates an increased expression of genes in the HOXA cluster in sample 05-1528. Note that nine of the 16 HOXA genes with probes in Agilent Platform show at least 1.5-fold increased expression; in contrast to that only two are at least 1.5-fold down-regulated.
Sample 05-1528 defined the smallest region of overlap for the recurrent 9q34 deletion. The region in 05-1528 extends from 9q34 centromere-130517102-133053582 base pairs (bp) telomere. The deleted region was further validated and refined using high resolution 1 million element CNV/SNP arrays (Figure 2B and C). This analysis revealed that the 9q34 breakpoints in this sample occur in regions of homozygosity positioned centromerically at 9q34.4.11 from 130,080,000-130,522,000 bp and telomerically at 9q34.13 from 132,936,000-133,125,000 bp. The deletion breakpoints reside within these regions of homozygosity and extend from centromere-130,499,000 -133,020,000 bp-telomere. The breakpoints for the adult NK B-ALL deletion can best be positioned based on the CNV/SNP array data in the untranslated region of SET or within exon 8 and between exon 17 and 18 of NUP214. This is in agreement with the breakpoints described for T cell ALL [15, 16].
Cryptic deletions, such as MLL and ETV6/CBFA2 [17] in B-ALL, resulting in the formation of oncogenic fusion genes have been described for a subset of hematologic malignancies. While there are several genes within the 9q34 region, SET, PKN3, ABL1 and NUP214 have previously been implicated for a role in malignancy with SET, ABL, and NUP214 being implicated in leukemogenesis. An oncogenic SET/NUP214 fusion gene has been reported in a case of NK adult AML as a result of a cryptic deletion of 9q34 [18]. This same cryptic deletion has been identified to also occur in pediatric T cell ALL with the centromeric breakpoint lying in SET with deletion of PKN3 just distal telomerically to SET [15]. Similarly, ABL1 and NUP214 have also been shown to create an oncogenic fusion gene in T-ALL [19]. NUP188 like NUP214 is a nucleoporin, and this family of proteins allows the transport of biomacromolecules such as mRNA across the nuclear membrane [20]. Recent studies have shown that the SET-NUP214 fusion gene results in upregulation of the HOXA gene cluster, and this is reflected in the current case [15]. Functional studies resulting in down regulation of the SET-NUP214 fusion gene have shown a decrease in HOXA gene cluster expression, and differentiation of cells carrying the fusion gene [15]. Thus it appears that SET-NUP214 blocks hematopoietic cells from proceeding through the process of differentiation. The remaining two deletions cover larger genomic distances: 99-350 (125088354 – 133756224 bp); 00-947 (127908459 – 139926251 bp). Their exact role is still to be discovered.
In summary, we describe, for the first time, recurrent deletion of 9q34 in adult NK B ALL. We did not attempt to correlate our results with outcome due to the small number of cases. Further, we concentrated on 9q34 as the only recurrent aberration in three of ten cases and presented our data on CDKN2A to illustrate our ability to replicate what is already known in the literature about this aberration. Finally, a larger number of cases is needed to assess the prognostic significance of this deletion.
Acknowledgements
We would like to thank Jeffery M. Conroy and Devin E. McQuaid for their technical support.
Supported partially by grants from the National Cancer Institute Grant CA16056 (NJN, SNJS, GD, DG, SL, LAF, PKW, ESW, MW), the Szefel Foundation (ESW), the Heidi Leukemia Research Fund (MW) and the Nancy C. Cully Endowment for Leukemia Research (MW), Buffalo, New York.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–43. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- [2].Larson S, Stock W. Progress in the treatment of adults with acute lymphoblastic leukemia. Curr Opin Hematol. 2008;15:400–7. doi: 10.1097/MOH.0b013e3283034697. [DOI] [PubMed] [Google Scholar]
- [3].Kristensen TD, Wesenberg F, Jonsson OG, Carlsen NT, Forestier E, Kirchhoff M, Lundsteen C, Schmiegelow K. High-resolution comparative genomic hybridisation yields a high detection rate of chromosomal aberrations in childhood acute lymphoblastic leukaemia. Eur J Haematol. 2003;70:363–72. doi: 10.1034/j.1600-0609.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- [4].Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–66. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- [5].Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- [6].Kuchinskaya E, Heyman M, Nordgren A, Schoumans J, Staaf J, Borg A, Soderhall S, Grander D, Nordenskjold M, Blennow E. Array-CGH reveals hidden gene dose changes in children with acute lymphoblastic leukaemia and a normal or failed karyotype by G-banding. Br J Haematol. 2008;140:572–7. doi: 10.1111/j.1365-2141.2007.06917.x. [DOI] [PubMed] [Google Scholar]
- [7].Paulsson K, Cazier JB, Macdougall F, Stevens J, Stasevich I, Vrcelj N, Chaplin T, Lillington DM, Lister TA, Young BD. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: Unexpected similarities with pediatric disease. Proc Natl Acad Sci U S A. 2008;105:6708–13. doi: 10.1073/pnas.0800408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Larson RA, Dodge RK, Burns CP, Lee EJ, Stone RM, Schulman P, Duggan D, Davey FR, Sobol RE, Frankel SR, et al. A five-drug remission induction regimen with intensive consolidation for adults with acute lymphoblastic leukemia: cancer and leukemia group B study 8811. Blood. 1995;85:2025–37. [PubMed] [Google Scholar]
- [9].Benekli M, Xia Z, Donohue KA, Ford LA, Pixley LA, Baer MR, Baumann H, Wetzler M. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood. 2002;99:252–7. doi: 10.1182/blood.v99.1.252. [DOI] [PubMed] [Google Scholar]
- [10].Nowak NJ, Snijders AM, Conroy JM, Albertson DG, Haines Jonathan L, et al. Current protocols in human genetics / editorial board. 2005. The BAC resource: tools for array CGH and FISH. Chapter 4:Unit 4 13. [DOI] [PubMed] [Google Scholar]
- [11].Nowak NJ, Miecznikowski J, Moore SR, Gaile D, Bobadilla D, Smith DD, Kernstine K, Forman SJ, Mhawech-Fauceglia P, Reid M, Stoler D, Loree T, Rigual N, Sullivan M, Weiss LM, Hicks D, Slovak ML. Challenges in array comparative genomic hybridization for the analysis of cancer samples. Genet Med. 2007;9:585–95. doi: 10.1097/gim.0b013e3181461c4a. [DOI] [PubMed] [Google Scholar]
- [12].Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–4. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- [13].Lee J, Sait SN, Wetzler M. Characterization of dendritic-like cells derived from t(9;22) acute lymphoblastic leukemia blasts. Int Immunol. 2004;16:1377–89. doi: 10.1093/intimm/dxh139. [DOI] [PubMed] [Google Scholar]
- [14].Usvasalo A, Savola S, Raty R, Vettenranta K, Harila-Saari A, Koistinen P, Savolainen ER, Elonen E, Saarinen-Pihkala UM, Knuutila S. CDKN2A deletions in acute lymphoblastic leukemia of adolescents and young adults: an array CGH study. Leuk Res. 2008;32:1228–35. doi: 10.1016/j.leukres.2008.01.014. [DOI] [PubMed] [Google Scholar]
- [15].Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, Stubbs A, Cools J, Nagata K, Fornerod M, Buijs-Gladdines J, Horstmann M, van Wering ER, Soulier J, Pieters R, Meijerink JP. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111:4668–80. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Quentmeier H, Schneider B, Rohrs S, Romani J, Zaborski M, Macleod RA, Drexler HG. SET-NUP214 fusion in acute myeloid leukemia- and T-cell acute lymphoblastic leukemia-derived cell lines. Journal of hematology & oncology. 2009;2:3. doi: 10.1186/1756-8722-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Harrison CJ. Acute lymphoblastic leukaemia. Best Pract Res Clin Haematol. 2001;14:593–607. doi: 10.1053/beha.2001.0156. [DOI] [PubMed] [Google Scholar]
- [18].Rosati R, La Starza R, Barba G, Gorello P, Pierini V, Matteucci C, Roti G, Crescenzi B, Aloisi T, Aversa F, Martelli MF, Mecucci C. Cryptic chromosome 9q34 deletion generates TAF-Ialpha/CAN and TAF-Ibeta/CAN fusion transcripts in acute myeloid leukemia. Haematologica. 2007;92:232–5. doi: 10.3324/haematol.10538. [DOI] [PubMed] [Google Scholar]
- [19].Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, Stul M, Dutta B, Boeckx N, Bosly A, Heimann P, Uyttebroeck A, Mentens N, Somers R, MacLeod RA, Drexler HG, Look AT, Gilliland DG, Michaux L, Vandenberghe P, Wlodarska I, Marynen P, Hagemeijer A. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- [20].Miller BR, Powers M, Park M, Fischer W, Forbes DJ. Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay. Molecular biology of the cell. 2000;11:3381–96. doi: 10.1091/mbc.11.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]