Abstract
In vivo bioluminescence imaging offers the opportunity to study biological processes in living animals, and the study of viral infections and host immune responses can be enhanced substantially through this imaging modality. For most studies of viral pathogenesis and effects of anti-viral therapies, investigators have used recombinant viruses engineered to express a luciferase enzyme. This strategy requires stable insertion of an imaging reporter gene into the viral genome, which is not feasible for many RNA viruses, and provides data on the viral component of pathogenesis but not the host. Genetically-engineered mice with luciferase reporters for specific viral or host genes provide opportunities to overcome these limitations and expand applications of bioluminescence imaging in viral infection and therapy. We review several different types of reporter mice for bioluminescence imaging, including animals that permit in vivo detection of viral replication, trafficking of immune cells, activation of key genes in host immunity to viral infection, and response to tissue damage. By utilizing luciferase enzymes with different emission spectra and/or substrates, it is possible to monitor two different biologic processes in the same animal, such as pathogen replication and sites of tissue injury. Combining imaging reporter viruses with genetically-engineered reporter mice is expected to substantially enhance the power of bioluminescence imaging for quantitative studies of viral and host factors that control disease outcome and effects of established and new therapeutic agents.
Keywords: bioluminescence, imaging, luciferase, transgenic mouse, viral infection, immunity
Introduction
Bioluminescence imaging is emerging as a powerful technology for studies of viral pathogenesis, immune responses to infection, and quantifying effects of therapy in living animals (Mandl et al., 2002; Gross et al., 2007; Hutchens and Luker, 2007). Unlike conventional methods that analyze viral infection and host immunity by euthanizing groups of animals at multiple time points, bioluminescence and other imaging techniques allow longitudinal studies of viral replication and dissemination in the same cohort of mice. Besides accounting for animal-to-animal variations within experimental groups, imaging allows investigators to identify unexpected sites of infection and/or patterns of host response that might be missed by analyzing only selected tissues at predetermined time points. Imaging also reduces numbers of animals needed to generate statistically significant data.
Bioluminescence imaging studies of viral infection typically have been performed with recombinant viruses engineered to express a luciferase enzyme from a viral promoter (Luker et al., 2002; Cook and Griffin, 2003; Hwang et al., 2008; Zaitseva et al., 2009). This strategy allows imaging to be used to track sites of viral infection over time in living animal. Since the luciferase enzyme is regulated by a promoter in the viral genome, bioluminescence can be used to quantify relative levels of changes in viral replication over the course of disease. Such recombinant viruses may be engineered to express a luciferase reporter from a promoter element expressed selectively at defined times in a viral life cycle, allowing imaging to identify specific points in viral replication. Bioluminescence imaging studies of luciferase-expressing viruses also are effective for quantifying effects of anti-viral therapy in vivo (Luker et al., 2002) (Zaitseva et al., 2009).
While recombinant viruses have proven to be highly effective tools for detecting and quantifying viral replication in living animals, there are important limitations to this approach for bioluminescence imaging. For example, it is easier to engineer large DNA viruses with luciferase reporters for imaging because the genomes of these viruses, such as poxviruses and herpes viruses, typically are larger than RNA viruses (Luker et al., 2002; Luker et al., 2005; Zaitseva et al., 2009). DNA viruses allow insertion of foreign DNA, including luciferase imaging cassettes, with minimal or no attenuation of viral replication and pathogenicity in animal models of infection. By comparison, the smaller genomes of most RNA viruses are much less tolerant of insertion of imaging reporter genes. Recombinant Sindbis viruses have been used successfully for in vivo imaging studies, but these viruses were attenuated in mouse models (Cook and Griffin, 2003). Efforts have been made to engineer other RNA viruses, such as West Nile virus, with green fluorescent protein (GFP) as an optical reporter gene. While insertion of GFP markedly attenuated West Nile virus (Pierson et al., 2005), investigators have successfully engineered HIV and other lentiviruses to stably express this fluorescent protein (Mahlknecht et al., 2000; Liddament et al., 2004). It is possible that techniques used to stably integrate GFP into lentiviral genomes can be employed to engineer other RNA viruses to express modified reporter proteins for bioluminescence imaging studies using transgenic mice described in this review.
Relying solely on bioluminescent reporter viruses for imaging studies also limits effective use of reagents developed previously for studies of viral infection. For essentially all viruses, researchers have made a large number of mutants designed to analyze effects of various genes on processes including immune evasion, replication, and dissemination. These viruses commonly were made without reporter genes for bioluminescence imaging. Such mutant viruses are a valuable resource for in vivo imaging studies of infection and host immune responses. Re-engineering multiple viral mutants with an imaging reporter gene or to introduce multiple new mutations in an imaging reporter virus would require a large effort that may be prohibitive for imaging studies. However, an alternative strategy in which an imaging reporter is present in a genetically-engineered animal would allow existing mutant viruses to be used for bioluminescence imaging studies.
In this review, we briefly will describe luciferase enzymes used for bioluminescence imaging. We then will highlight several different genetically-engineered mouse models with bioluminescence imaging reporters for in vivo studies. These genetically-engineered mice incorporate a variety of promoters driving luciferase expression, including promoters that respond directly to infection with native or recombinant viruses, quantify activation of various immune response genes, and enable cell trafficking studies. In many cases, these mice have not yet been used for studies of viral infection. However, there is tremendous potential to substantially advance knowledge of viral infection anti-viral therapeutics by incorporating bioluminescence imaging reporter mice into studies of viral pathogenesis.
Bioluminescence imaging overview
Bioluminescence imaging detects photons emitted from luciferase enzymes in vivo to detect, localize, and quantify specific biologic processes, such as viral replication and trafficking of immune cells to sites of infection. This imaging technique is highly sensitive, allowing detection of as few as 1 × 102 pfu of luciferase-expressing viruses in vivo (Luker et al., 2002). Instruments for bioluminescence imaging are available at most academic centers and pharmaceutical companies, making this technique readily accessible to many researchers. As a result, bioluminescence imaging studies are commonly presented in publications by investigators in a wide variety of scientific disciplines.
While bioluminescence imaging has several advantages for small animal imaging studies, persons unfamiliar with this technique should note some limitations of this modality. Diffusion and scattering of light in tissues limits spatial resolution of bioluminescence imaging to 2–3 mm. Since light is attenuated approximately 10-fold per cm of tissue, light from superficial sites is detected to a greater extent than light emitted from deeper organs and tissues (Contag and Bachmann, 2002). Attenuation of light in tissues also is relatively greater for luciferase enzymes that emit blue light, such as Gaussia, Renilla, and bacterial luciferases (see below). While there are methods to generate three-dimensional images, bioluminescence images typically are two-dimensional. For more detailed descriptions of the strengths and limitations of bioluminescence imaging, readers are referred to recent reviews on this topic (Contag and Bachmann, 2002; Hutchens and Luker, 2007).
Bioluminescence imaging studies have been performed with several different luciferase enzymes. Firefly luciferase (Photinus pyralis) is used most commonly for in vivo imaging studies because of the sustained kinetics of light emission, favorable pharmacokinetics of the luciferin substrate, and relatively red-shifted emission spectrum (Berger et al., 2008). Renilla (Renilla reniformis) and Gaussia (Gaussia princeps) luciferases use coelenterazine as a substrate, distinguishing these enzymes from firefly luciferase. Relative to luciferin, coelenterazine has disadvantages for in vivo imaging, including more limited biodistribution and oxidation in serum (Pichler et al., 2004). Oxidation in serum produces light and further decreases bioavailability of substrate, contributing to reduced detection of Renilla and Gaussia luciferases in vivo as compared with the combination of firefly luciferase and luciferin. Renilla and Gaussia luciferases have flash kinetics with rapid onset and diminution of bioluminescence, which necessitates that imaging begin rapidly after injecting substrate. Blue-green light emitted by these enzymes also has reduced penetration through tissues relative to firefly luciferase, although investigators have developed a more red-shifted mutant Renilla luciferase (Loening et al., 2007). Despite some limitations of Renilla and Gaussia luciferases, a key benefit of these enzymes is that their optical properties and substrate allow discrimination from firefly luciferase. In vivo imaging studies also have been performed with bioluminescent bacteria engineered to express a lux operon, such as the operon from Photorhabdus luminescence (Kadurugamuwa and Francis, 2008). While this operon has not yet been integrated successfully into mammalian cells, the blue light emitted from bacterial luciferase can be distinguished from light from firefly luciferase, allowing multi-spectral imaging. As highlighted in this review, imaging with dual bioluminescent reporters allows two different biologic processes to be monitored in the same animal. For further information on luciferase enzymes, readers are reviewed to recent reviews on this subject (Massoud et al., 2007; Luker and Luker, 2008).
Luciferase enzymes have several advantages as reporter genes for imaging studies of viral infection as compared with fluorescent proteins or β-galactosidase. While in vivo imaging has been performed successfully with several different fluorescent proteins (Yang et al., 2007; Lin et al., 2009), signal-to-background is notably greater for luciferases because of tissue autofluorescence. Luciferases have higher sensitivity and a larger dynamic range than direct imaging of β-galactosidase (Tung et al., 2004), although β-galactosidase also has been imaged with a caged luciferin substrate that is cleaved sequentially by β-galactosidase and firefly luciferase to produce bioluminescence (Wehrman et al., 2006). The latter strategy is difficult to implement in transgenic mice because it requires co-expression of both β-galactosidase and firefly luciferase in the same cell. Relative to luciferase enzymes, fluorescent proteins and β-galactosidase also have the disadvantage of relatively long protein half-lives, which limits the ability to monitor kinetics of viral infection by in vivo imaging. Unlike radiotracer imaging techniques (positron emission tomography, PET; or single photon emission computed tomography, SPECT), bioluminescence imaging does not require imaging probes with shortlived radioisotopes, making bioluminescence imaging accessible to more investigators. However, bioluminescence imaging may be combined with other imaging techniques, such as PET and MRI, by using fusion reporters or bicistronic reporter constructs (Maggi and Ciana, 2005; Ray et al., 2007). Developing transgenic mice with multi-modality imaging reporter genes would greatly increase the value of such mice for studies of viral infection and response to therapy.
Genetically-engineered mice for bioluminescence imaging
Reporter mice used for bioluminescence imaging experiments are designed with a promoter of interest driving expression of a luciferase enzyme. Essentially all of these mice use firefly luciferase as the imaging reporter gene, capitalizing on the favorable properties of the enzyme itself and the luciferin substrate. Since these reporter mice regulate luciferase at the level of transcription, there is an inherent delay between activation of the promoter and expression of sufficient amounts of luciferase for detection by imaging. Similarly, there also is a delay between cessation of promoter activity and loss of bioluminescence signal, dependent upon relative stability of the luciferase reporter. While the relatively slow kinetics of promoter-firefly luciferase mice potentially may limit investigations of rapid responses to viral infection, these reporter constructs are similar to those engineered in recombinant viruses used for bioluminescence imaging studies. Sustained expression of firefly luciferase beyond the period of promoter activation also increases amounts of enzyme available for imaging, which improves sensitivity for in vivo imaging. While bioluminescence imaging allows sensitive detection of firefly luciferase in whole animals, light from luciferase enzymes cannot be detected readily by microscopy or flow cytometry. This limitation of luciferase enzymes can be overcome by fusing a fluorescent protein, such as GFP, to luciferase (Wang et al., 2002; Wilson et al., 2008). Although not utilized in the mice described in this review, transgenic reporter constructs including both a luciferase enzyme and fluorescent protein would integrate microscopic imaging, cellular analyses, and whole animal imaging techniques.
Another potential limitation of transgenic mice for imaging studies is unintentional variations in expression of the reporter construct in various organs and tissues. For several of the transgenic reporter mice described in this review, investigators have shown activation of the firefly luciferase reporter gene in multiple anatomic sites. However, detailed studies generally have not been performed to quantify cell-type specific variations in bioluminescence in response to the same activation event or to determine that expression of the reporter is stable over several generations of mice. To achieve more uniform, stable expression of transgenes in mice, investigators have used insulator sequences (matrix-attachment regions, MAR; HS4 from the β-globin gene) to reduce effects of adjacent genes or targeted transgenes to ubiquitously-expressed sites, such as ROSA26 (McKnight et al., 1992; Zambrowicz et al., 1997; Bell et al., 2001). These types of strategies may benefit investigators designing new transgenic mice for bioluminescence imaging.
Bioluminescence imaging mice with viral promoter controlling luciferase: Herpes simplex virus (HSV) reporter mice
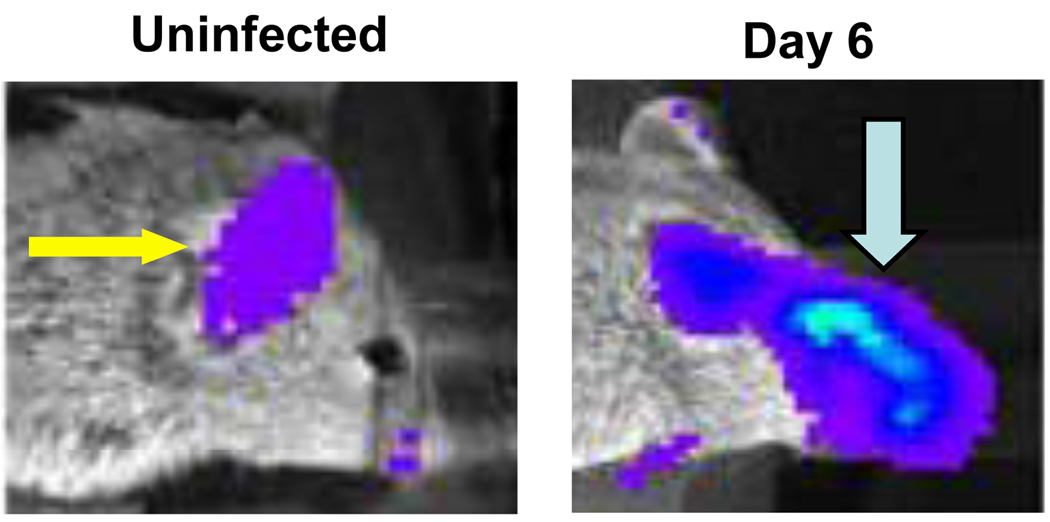
To enable imaging studies of Herpes simplex virus infection without the need for bioluminescent reporter viruses, we generated a transgenic reporter mouse with firefly luciferase regulated by the promoter for HSV-1 thymidine kinase (TK), an early gene in viral replication (Luker et al., 2006). In cell culture studies, the TK promoter had lower basal activity and greater induction of firefly luciferase in response to viral infection than several other HSV-1 promoters, including ICP6 and ICP8. In the absence of infection, mice from four different transgenic founder animals showed low to moderate levels of bioluminescence predominantly detected in paws, ears, and mouth regions. Following ocular infection with HSV-1, firefly luciferase activity in the eye and peri-ocular tissues increased by > 10-fold above basal levels at the peak of infection on days 5–6 (Fig 1). Amounts of bioluminescence quantified by imaging were proportionate to the input titer of virus, establishing that the reporter mouse could be used to detect relative differences in viral burden. Bioluminescence increased in response to ocular infection with 3 different strains of HSV-1, and amounts of luciferase activity correlated directly with relative increases in virulence of various strains. The TK-firefly luciferase reporter also was activated following cutaneous infection with HSV-1. Infection with vaccinia, an unrelated DNA virus, did not change bioluminescence in reporter mice, showing specificity for HSV-1. Collectively, these results demonstrated that the TK-firefly luciferase reporter mouse could be used to image distribution and replication of HSV-1 infection, providing an in vivo model for studying infection with a wide range of existing mutant viruses via multiple routes.
Figure 1. HSV-1 transgenic reporter mouse.
Transgenic mouse with firefly luciferase controlled by the viral promoter for HSV-1 thymidine kinase was infected with 1 × 106 pfu of HSV-1 strain McKrae by corneal scarification. Images prior to infection and on day 6 post-infection show infection-dependent firefly luciferase bioluminescence in the infected eye and peri-ocular tissues (blue arrow). Luciferase activity in the ear (yellow arrow) is present constitutively.
In addition to the HSV-1 TK-firefly luciferase reporter mouse, we also generated transgenic mice with the ICP6 promoter controlling expression of firefly luciferase. ICP6, which encodes the large subunit of the HSV-1 ribonucleotide reductase, had been used successfully to generate a reporter cell line to detect HSV-1 infection in cell culture (Olivo, 1994). Several different founder lines of ICP6-FL mice had constitutive bioluminescence in paws, ears, and mouth regions that was substantially greater than the basal luciferase activity detected in the TK-firefly luciferase reporter mice. Unlike the TK-firefly luciferase mice, infection with different strains of HSV-1 TK did not alter bioluminescence in the ICP6-firefly luciferase animals.
Results with the HSV-1 transgenic reporter mice highlight both the strengths and potential limitations of this strategy for bioluminescence imaging of viral infection. The ability to detect the extent of infection with wild-type viruses is a clear advantage of this method. Developing reporter mice with a luciferase enzyme under control of promoter elements from different viruses may enable studies of other viruses for which bioluminescence imaging has not been possible, such as many RNA viruses. We also note that the TK-firefly luciferase reporter mouse was less sensitive than our recombinant HSV-1-firefly luciferase virus for detecting HSV-1 in vivo. We detected viral infection within 1 day of ocular inoculation of the HSV-1 firefly luciferase reporter virus, while infection could not be identified until day 2 in the TK-firefly luciferase reporter mouse. Results from the ICP6-firefly luciferase mice also highlight potential limitations of using viral promoter elements outside the context of the endogenous viral genome and emphasize careful selection of promoters based on cell culture studies. We also note that the promoter-imaging reporter gene strategy is likely to be more effective for viruses that normally replicate in the nucleus, such as HSV-1. Although not validated experimentally, we anticipate that transgenic mice with promoter-luciferase imaging reporters would be less effective for detecting infection with viruses that replicate exclusively in the cytoplasm, such as poxviruses. Despite the limitations, these data establish feasibility of using a transgenic reporter mouse for bioluminescence imaging of infection with wild-type viruses in vivo.
Gal4/Upstream activation sequence reporter mice
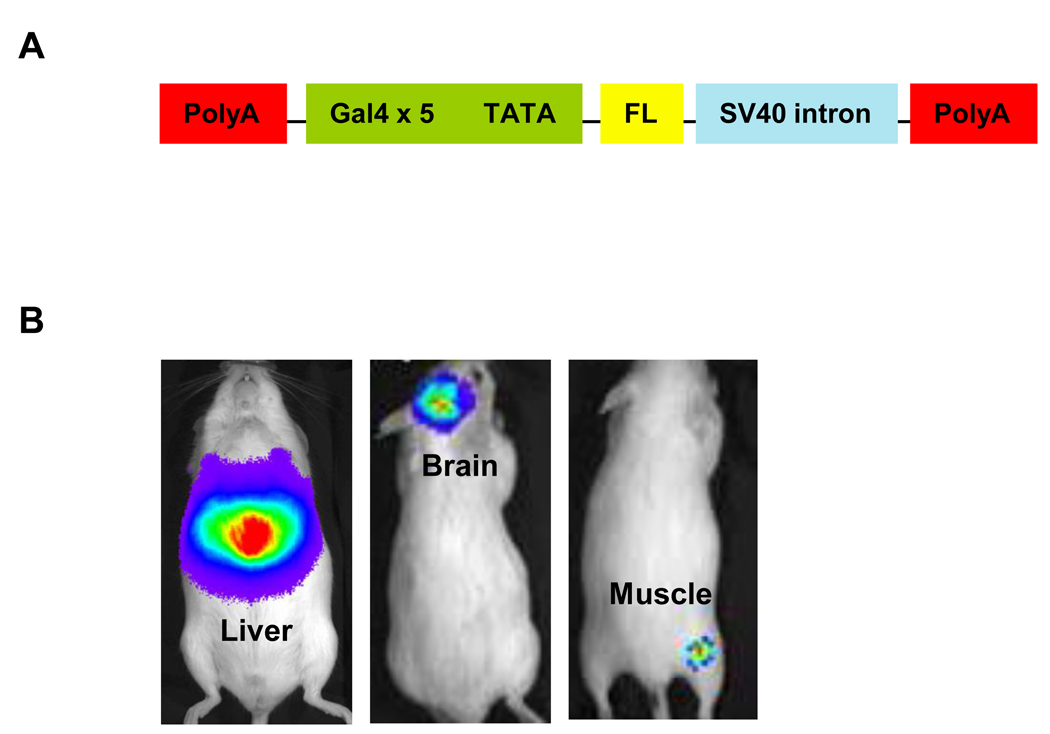
One of the powerful tools available for studies of gene expression is the Gal4/upstream activation sequence (UAS) technology. Although used most commonly for genetic studies in Drosophila melanogaster, Gal4/UAS functions in other heterologous systems including mice. Gal4/UAS uses a promoter of interest to drive expression of Gal4, a yeast transcription factor comprised of a DNA binding domain and transactivation domain. Binding of Gal4 to its target DNA binding sequence (the UAS) activates expression of a second gene, which may be luciferase or other reporter protein (Fig 2). Since the two-step process provides substantial amplification of the second gene, Gal4/UAS substantially enhances signal-to-background for imaging (Iyer et al., 2001).
Figure 2. Gal4 promoter firefly luciferase reporter mouse.
A. Schematic diagram of the transgenic construct, consisting of 5 repeated Gal4 DNA binding motifs linked to a minimal promoter element (TATA box) to drive firefly luciferase expression. Polyadenylation motifs (PolyA) and SV40 intron were included to reduce baseline expression and increase Gal4 regulated transcription. B. Expression of firefly luciferase was activated in response to infection in the liver (1.4 × 107 pfu), brain (2 × 106 pfu), or skeletal muscle (2.3 × 105 pfu) with a recombinant adenovirus constitutively expressing the Gal4 DNA transcription factor.
We recently developed a transgenic reporter mouse for bioluminescence imaging of the Gal4/UAS system (Pichler et al., 2008). This mouse has a multimerized Gal4 DNA binding domain (UAS) driving expression of firefly luciferase. Under basal conditions, these mice had very low bioluminescence. Reporter mice infected with an adenovirus engineered to express the Gal4 transcription factor showed up to 4 logs of induction of firefly luciferase activity as quantified by bioluminescence imaging. Adenoviral infection activated expression of luciferase in multiple organs and tissues, including liver, brain, and skeletal muscle, showing that the imaging reporter gene could be activated throughout the entire mouse (Fig 2). To date, this mouse has been used only to study infection with recombinant adenoviruses. However, this reporter mouse could be used to investigate infection with any virus engineered to express the Gal4 transcription factor. The Gal4-VP16 hybrid transcription factor used in this system is composed of only 227 amino acids, which is notably smaller than firefly luciferase (550 amino acids). This size difference may permit insertion of Gal4-VP16 in smaller viruses. Based on experience with adenoviral infection, increases in expression of luciferase following infection equal or exceed those observed with recombinant viruses directly expressing FL, making these mice a highly sensitive, general model for studies of pathogenesis.
Genetically-engineered reporter mice for host immunity
In response to viral infection, cells activate a large number of signaling pathways to control viral infection at the cellular level and initiate inflammatory responses that recruit immune cells. Many of these host responses are regulated at the level of transcription. To visualize transcriptional regulation of immune responses, investigators have begun to develop reporter mice with the promoter for a gene of interest controlling expression of a luciferase reporter gene. Studies using these types of reporter mice have the potential to reveal dynamics of host immune responses in ways not possible with conventional assays of excised tissues.
NF-κB reporter mouse
Toll-like receptors (TLRs) are primary sensors of molecular patterns present in viruses and other pathogens. Following ligand binding, TLR signaling converges on NF-κB, a transcription factor that controls expression of multiple pro-inflammatory cytokines essential for ant-viral immunity (Kawai and Akira, 2007).
Blackwell and colleagues have developed a transgenic reporter mouse for bioluminescence imaging of NF-κB-dependent transcription (Yull et al., 2003). This mouse uses the NF-κB-dependent portion of the human immunodeficiency virus-1 (HIV-1) long terminal repeat (HIV-1 LTR) to control firefly luciferase, allowing gene expression to be detected in tissues throughout the intact mouse and in excised tissues. Following intraperitoneal injection of lipopolysaccharide (LPS) as a model of infection with Gram-negative bacteria, NF-κB-dependent expression of firefly luciferase occurred in liver, spleen, lungs, kidneys, and the forebrain. By comparison, activation of the luciferase was restricted to the lungs in response to aerosolized LPS. A separate study with these mice showed activation of NF-κB-dependent transcription in the lung following inhalation of silica particles but not a non-fibrogenic particle composed of titanium dioxide (Hubbard et al., 2002). In addition, research with these mice demonstrated that the duration of activation of NF-κB signaling determines the degree of lung damage in response to an inflammatory stimulus (Everhart et al., 2006). Overall, these results highlight how imaging studies of NF-κB-dependent gene expression can be used to identify spatial and temporal activation of inflammation and its effects on tissue pathology. We note that these mice have not yet been used for studies of the NF-κB pathway in the context of viral infection, although activation of the reporter likely will be less than observed following maximal stimulation with LPS. Since the reporter mouse integrates all pathways that regulate expression of NF-κB-dependent genes, distinguishing protective from pathologic activation of NF-κB during viral infection will be difficult based solely on non-invasive bioluminescence imaging.
Interferon β reporter mouse
Type I interferons (α and β) are well-established as essential mediators of host defense against most viral infections, and type I interferons and their downstream signaling pathways are targets of numerous viral immune evasion genes (Stark et al., 1998; Malmgaard, 2004). Of the type I interferons, interferon β is regarded as the master regulator of this signaling cascade, positioning this molecule as a central determinant of antiviral defense (Taniguchi and Takaoka, 2001). To enable bioluminescence imaging of interferon β expression during viral infection, Lienenklaus et al generated mice in which firefly luciferase was recombined into the genome under control of the endogenous promoter for interferon β (Lienenklaus et al., 2009). Tissue-specific deletion of one copy of the interferon β gene using Cre-lox technology allowed expression of firefly luciferase, reproducing the regulation of magnitude of interferon β gene expression in vivo.
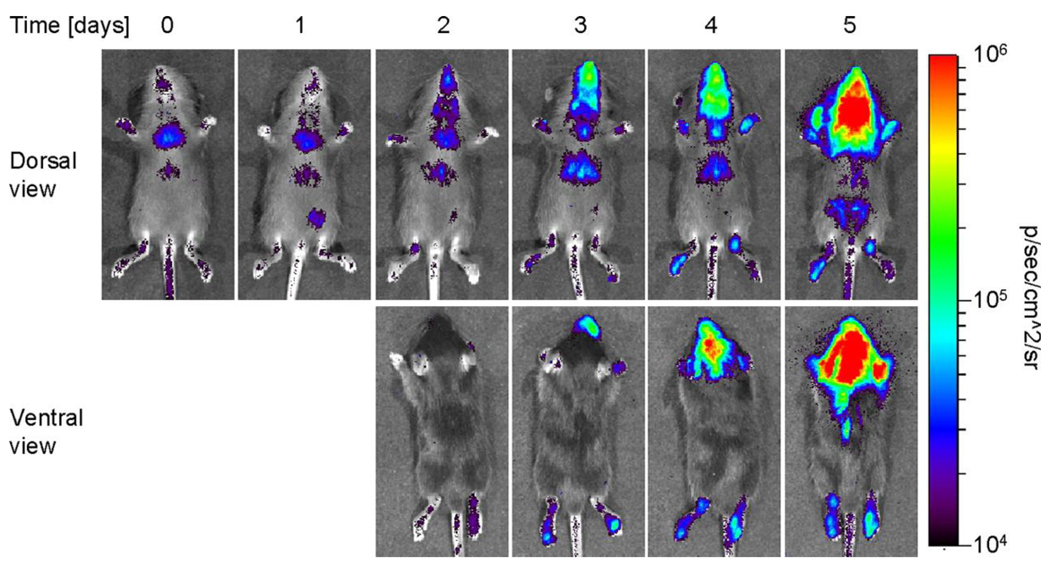
Bioluminescence imaging showed tissue-specific, high levels of interferon β gene expression in mice infected with a mutant influenza A virus lacking the viral non-structural protein NS1. The kinetics of bioluminescence paralleled the course of viral infection and production of mRNA for interferon β assayed in excised tissues. Activation of the interferon β -FL reporter was nearly completely abolished in mice infected with wild-type influenza A, demonstrating the extent to which NS1 counters interferon signaling pathways in vivo (Kochs et al., 2007). Similarly, bioluminescence imaging showed robust activation of the interferon β promoter following infection with a mutant LaCrosse virus lacking the interferon antagonist protein NSs (Fig 3) (Blakqori et al., 2007). These studies using two different viral pathogens clearly demonstrate the power of bioluminescence imaging to detect activation of host immune response pathways and effects of viral proteins to counter the response.
Figure 3. Activation of Interferon-β promoter in response to infection with LaCrosse virus.
IFN-β reporter mice were infected i.p. with 1 × 104 pfu of mutant LaCrosse virus lacking the interferon antagonist protein NSs. Dorsal and ventral views of an infected mouse show distribution and kinetics of gene expression in response to viral infection (Copyright 2009. The American Association of Immunologists, Inc.).
In addition to activation during the course of viral infection, imaging studies showed basal activity of the interferon β promoter throughout the mouse with particularly elevated expression in the thymus. Based on imaging data and ex vivo analysis, the authors determined that thymic epithelium was the predominant source of interferon β gene expression in the absence of infection. Notably, ex vivo studies showed absence of interferon α in this organ in uninfected mice. By crossing reporter mice to animals lacking Aire, a gene required for ectopic expression of antigens by thymic epithelium and establishing central T cell tolerance (Derbinski et al., 2001), this study determined that Aire is required for basal expression of interferon β in the thymus. Based on these data, the authors proposed a function for interferon β in establishment and/or maintenance of T cell tolerance. These data from uninfected mice highlight the potential for in vivo imaging studies to reveal unexpected aspects of in vivo physiology and pathophysiology.
Reporter mice for damage to host tissues
Morbidity and mortality from viruses are caused by damage to host organs and tissues due to direct result of infection and/or associated inflammation. To assess overall effects of anti-viral therapies, it is essential to determine that an agent blocks viral replication and also reduces damage to infected tissues. Ideally, an imaging strategy would account for both aspects of viral disease. Towards this goal, investigators have developed reporter mice for imaging various aspects of changes in gene expression due to tissue damage.
Glial fibrillary acid protein (GFAP)-firefly luciferase reporter mice
GFAP is the major intermediate filament present in central nervous system astrocytes. Levels of GFAP protein increase substantially following injury, mediated at least in part by changes in gene transcription. To image neuronal injury in vivo, imaging reporter mice have been developed using the promoter for GFAP to regulate expression of firefly luciferase. Cho et al generated a novel dual bioluminescent reporter mouse for imaging activation of the GFAP promoter (Cho et al., 2009). In this mouse, the GFAP promoter controlled firefly luciferase, while Renilla luciferase was expressed constitutively by the promoter for the housekeeping gene glyceraldehyde 3 phosphate dehydrogenase (GAPDH). The dual reporter strategy allowed changes in GFAP-firefly luciferase expression to be normalized to the basal expression of GAPDH-Renilla luciferase, reducing mouse-to-mouse variations in bioluminescence from the GFAP reporter alone. Following experimentally induced seizures, these mice had elevated levels of firefly luciferase activity due to neuronal injury and activation of the GFAP promoter.
Kadurugamuwa et al also used a dual bioluminescent reporter strategy to image glial injury in response to Streptococcus pneumoniae (Kadurugamuwa et al., 2005). These authors also generated transgenic mice with the GFAP promoter controlling expression of firefly luciferase. The reporter mice were infected with Streptococcus pneumoniae engineered with the lux operon, which constitutively produces bioluminescence with an emission wavelength distinct from firefly luciferase. Using optical filters to distinguish bacterial bioluminescence from GFAP-firefly luciferase, this study correlated progression of bacterial meningitis with progressive injury to the nervous system. Importantly, bioluminescence imaging showed that early intervention with antibiotics limited the extent of neuronal injury as quantified by expression of GFAP-firefly luciferase and validated ex vivo. These imaging studies are notable both for the ability to image effects of tissue damage and the innovative use of dual bioluminescence imaging reporters to improve the quality of imaging data and monitor two separate biologic processes in the same animal. This type of imaging strategy will allow investigators to more accurately assess viral replication, associated damage to tissues, and response to therapy in over time in a wide variety of experimental model systems.
Inducible nitric oxide synthase (iNOS) reporter mice
Inducible nitric oxide synthase (iNOS) contributes to host defense against both viral and bacterial pathogens, including Listeria monocytogenes, reovirus, and influenza A (Remer et al., 2001) (Goody et al., 2005; Aldridge JR et al., 2009). Levels of iNOS protein are very low under basal conditions, but the enzyme is upregulated in response to infection and inflammation. Expression of iNOS also increases in response to tissue injury, increasing oxidative damage to organs and tissues (Soejima et al., 2000). iNOS expression in tissues is regulated predominantly by transcription (Kinugawa et al., 1997), making it an opportune target for monitoring with a gene promoter-imaging reporter construct.
Zhang et al generated a transgenic mouse with firefly luciferase under transcriptional control of the murine iNOS promoter (Zhang et al., 2003). Using an experimental model of sepsis elicited by interferon γ and LPS, bioluminescence imaging revealed significant increases in luciferase activity in liver. Changes in bioluminescence correlated directly with upregulation of iNOS mRNA and protein in excised organs, validating the reporter mouse for quantifying iNOS in vivo. iNOS-firefly luciferase also was activated in selectively in inflamed joints in an experimental model of arthritis. To establish that the reporter mouse could be used to quantify effects of therapeutic agents, these investigators tested effects of various compounds designed to block induction of iNOS. Treatment with agents including dexamethasone and epigallocatechin gallate suppressed activation of the iNOS promoter, showing the power of this imaging strategy to test pharmacodynamics and dosing schedules for potential drugs targeting the iNOS pathway.
Constitutive expression of luciferase in multiple tissues for adoptive transfer studies
Trafficking of various types of innate and adaptive immune cells to sites of infection is a critical part of the host response to infection (Salazar-Mather and Hokeness, 2003). Kinetics and/or numbers of responding cells are key determinants that distinguish control of infection versus progressive disease. Bioluminescence imaging provides a non-invasive method to detect and quantify trafficking of immune cells to sites of viral infection.
To enable bioluminescence imaging studies of immune cell trafficking in living mice, investigators have developed transgenic mice that express firefly luciferase in selected cell types or in all hematopoietic cells. For example, Azadniv et al generated transgenic mice in which the human CD2 promoter directed expression of firefly luciferase to CD8 T cells (Azadniv et al., 2007). CD8 T cells in these mice were specific to the immunodominant epitope from the model antigen ovalbumin. Mice were infected with an HSV-1 amplicon vector expressing the ovalbumin antigen, followed by adoptive transfer of transgenic CD8 T lymphocytes. In this model system, as few as 103 transferred CD8 cells could be localized in vivo with bioluminescence imaging. This imaging strategy could be used to monitor trafficking of CD8 T lymphocytes in response to other existing viruses engineered to express ovalbumin peptides as model antigens (Norbury et al., 2001; El-Gogo et al., 2007; Albrecht et al., 2008). It should also be possible to study infection directly in the CD8 T cell transgenic mice, although sensitivity might be reduced by CD8 lymphocytes already present in lymphoid tissues and other organs prior to infection.
As a more general source of donor hematopoietic cells for trafficking studies, other investigators have developed genetically-engineered mice in which firefly luciferase is expressed constitutively in all tissues, including hematopoietic stem cells (Cao et al., 2004). Using these mice, investigators were able to monitor engraftment of a single hematopoietic stem cell adoptively transferred to a recipient mouse and analyze foci of bioluminescent hematopoietic cells that varied in anatomic distribution among animals. These transgenic reporter mice also have been used as donors of purified populations of immune cells, such as regulatory T cells, for bioluminescence imaging studies of cell trafficking and biologic effects in graft versus host disease (Nguyen et al., 2007). For studies of viral infection and immune responses, similar adoptive transfer strategies could be used to image kinetics of cell recruitment to sites of infection and effects of specific chemokines or viral immune evasion proteins to regulate cell trafficking. In combination with intravital microscopy or other imaging techniques, adoptive transfer studies of bioluminescent immune cells would allow host immune responses to viral infection to be imaged and analyzed at both microscopic and macroscopic levels in vivo .
Conclusion
Mice described in this review highlight the growing number of genetically-engineered reporter animals developed for bioluminescence imaging studies (Table 1). These mice substantially expand potential applications of bioluminescence imaging for studies of viral infection in small animal models of disease. Using these and similar transgenic models, it now is possible to image wild-type viruses with virus-specific reporter mice, which will enable investigators to perform imaging studies with existing mutant viruses. By constructing reporter mice specific for other viruses, it will be possible to image infection with viruses having genomes too small to accommodate an imaging reporter gene, such as many RNA viruses. Transgenic reporter mice also provide general tools for imaging viruses expressing model transcription factors (Gal4) or antigens (ovalbumin), allowing in vivo studies of viral replication and host immune trafficking. Transgenic imaging reporter mice also are available for imaging temporal patterns of expression for key host response genes in anti-viral defense, such as interferons and NF-KB. These mice provide nearly real-time analysis of host immunity, opening the possibility for longitudinal studies of immune responses to wild-type viruses and viruses with mutations in specific immune evasion genes (Goody et al., 2005).
Table 1.
Summary of reporter mice with potential applications for imaging viral infection
| Mouse (promoter and luciferase enzyme) |
Application | Reference |
|---|---|---|
| HSV-1 thymidine kinase (TK) promoter-firefly luciferase |
HSV-1 infection | {Luker, 2006 #1232} |
| Gal4-UAS-firefly luciferase | Recombinant viruses expressing Gal4 transcription factor |
{Pichler, 2008 #1233} |
| NF-κB-firefly luciferase | Protective and pathologic host immune responses mediated by NF-κB transcription factor |
{Yull, 2003 #1539} |
| IFN-β-firefly luciferase | Activation of type I interferon response to infection |
{Lienenklaus, 2009 #1527} |
| Glial fibrillary acidic protein (GFAP)-firefly luciferase |
Central nervous system response to infection/injury |
{Cho, 2009 #1541} {Kadurugamuwa, 2005 #1173} |
| CD2-firefly luciferase | Trafficking of CD8 T cells | {Azadniv, 2007 #1525} |
| β-actin-firefly luciferase | Ubiquitous expression of firefly luciferase in cells for adoptive transfer experiments |
{Cao, 2004 #585} |
In addition to studies using one type of luciferase, we envision that future studies of viral pathogenesis in small animal models will combine recombinant viruses and transgenic reporter mice with different types of luciferase. For example, using a virus expressing Renilla luciferase to infect a reporter mouse with the promoter for an immune response gene of interest driving firefly luciferase would allow real-time monitoring of viral infection and dissemination relative to activation of host immunity. This strategy will provide an integrated assessment of both viral and host factors that determine pathogenesis without the need to euthanize multiple animals at separate time points to collect tissue samples. This type of combined bioluminescence imaging study is eminently feasible, as evidenced by studies cited above. We also anticipate that future transgenic mice for imaging studies will use reporters that extend beyond promoter-reporter constructs with unmodified luciferase enzymes. For example, destabilized luciferase enzymes can be used to improve temporal resolution for kinetics of infection, although such constructs may have lower total amounts of bioluminescence (Luker et al., 2003; Banaszynski et al., 2008). Imaging reporter genes also could be activated or regulated in a tissue-specific manner, using mice with inducible promoters. These types of studies will continue to expand the power of bioluminescence imaging to study viral infection, identify viral and host genes that determine outcomes of infection, and analyze effects of anti-viral therapies.
Acknowledgments
The authors acknowledge grant support from NIH P50CA093990 and R24CA083099 for the University of Michigan Small Animal Imaging Resource.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albrecht M, Suezer Y, Staib C, Sutter G, Vieths S, Reese G. Vaccination with a Modified Vaccinia Virus Ankara-based vaccine protects mice from allergic sensitization. J Gene Med. 2008;10:1324–1333. doi: 10.1002/jgm.1256. [DOI] [PubMed] [Google Scholar]
- Aldridge J, JR, Moseley C, Boltz D, Nagovetich N, Reynolds C, Franks J, Brown S, Doherty P, Webster R, Thomas P. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci U S A. 2009;106:5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azadniv M, Dugger K, Bowers W, Weaver C, Crispe I. Imaging CD8+ T cell dynamics in vivo using a transgenic luciferase reporter. Int Immunol. 2007;10:1165–1173. doi: 10.1093/intimm/dxm086. [DOI] [PubMed] [Google Scholar]
- Banaszynski L, Sellmyer M, Contag C, Wandless T, Thorne S. Chemical control of protein stability and function in living mice. Nat Med. 2008;14:1123–1127. doi: 10.1038/nm.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A, West A, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Berger F, Paulmurugan R, Bhaumik S, Gambhir S. Uptake kinetics and biodistribution of 14C-D-luciferin--a radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: impact on bioluminescence based reporter gene imaging. Eur J Nucl Med Mol Imaging. 2008;35:2275–2285. doi: 10.1007/s00259-008-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakqori G, Delhaye S, Habjan M, Blair C, Sanchez-Vargas I, Olson K, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol. 2007;81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wagers A, Beilhack A, Dusich J, Bachman M, Negrin R, Weissman I, Contag C. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci USA. 2004;101:221–226. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Hagemann T, Johnson D, Johnson J, Messing A. Dual transgenic reporter mice as a tool for monitoring expression of glial fibrillary acidic protein. J Neurochem. 2009;110:343–351. doi: 10.1111/j.1471-4159.2009.06146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag C, Bachmann M. Advances in in vivo bioluminescence imaging of gene expression. Ann Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- Cook S, Griffin D. Luciferase imaging of a neurotropic viral infection in intact animals. J Virol. 2003;77:5333–5338. doi: 10.1128/JVI.77.9.5333-5338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- El-Gogo S, Staib C, Meyr M, Erfle V, Sutter G, Adler H. Recombinant murine gammaherpesvirus 68 (MHV-68) as challenge virus to test efficacy of vaccination against chronic virus infections in the mouse model. Vaccine. 2007;25:3934–3945. doi: 10.1016/j.vaccine.2007.02.054. [DOI] [PubMed] [Google Scholar]
- Everhart M, Han W, Sherrill T, Arutiunov M, Polosukhin V, Burke J, Sadikot R, Christman J, Yull F, Blackwell T. Duration and intensity of NF-kappaB activity determine the severity of endotoxin-induced acute lung injury. J Immunol. 2006;176:4995–5005. doi: 10.4049/jimmunol.176.8.4995. [DOI] [PubMed] [Google Scholar]
- Goody R, Hoyt C, Tyler K. Reovirus infection of the CNS enhances iNOS expression in areas of virus-induced injury. Exp Neurol. 2005;195:379–390. doi: 10.1016/j.expneurol.2005.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Moss B, Piwnica-Worms D. Veni, vidi, vici: in vivo molecular imaging of immune response. Immunity. 2007;27:533–538. doi: 10.1016/j.immuni.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Hubbard A, Timblin C, Shukla A, Rincon M, Mossman B. Activation of NF-kappaB-dependent gene expression by silica in lungs of luciferase reporter mice. Am J Physiol Lung Cell Mol Physiol. 2002;282:L968–L975. doi: 10.1152/ajplung.00327.2001. [DOI] [PubMed] [Google Scholar]
- Hutchens M, Luker GD. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 2007;9:2315–2322. doi: 10.1111/j.1462-5822.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- Hwang S, Wu T-T, Tong L, Kim K, Martinez-Guzman D, Colantonio A, Uittenbogaart C, Sun R. Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J Virol. 2008;82:12498–12509. doi: 10.1128/JVI.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir S. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci U S A. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa J, Francis K. Bioluminescent imaging of bacterial biofilm infections in vivo. Methods Mol Biol. 2008;431:225–239. doi: 10.1007/978-1-60327-032-8_18. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa J, Modi K, Coquoz O, Rice B, Smith S, Contag P, Purchio T. Reduction of astrogliosis by early treatment of pneumococcal meningitis measured by simultaneous imaging, in vivo, of the pathogen and host response. Infect Immun. 2005;73:7836–7843. doi: 10.1128/IAI.73.12.7836-7843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kinugawa K, Shimizu T, Yao A, Kohmoto O, Serizawa T, Takahashi T. Transcriptional regulation of inducible nitric oxide synthase in cultured neonatal rat cardiac myocytes. Circ Res. 1997;81:911–921. doi: 10.1161/01.res.81.6.911. [DOI] [PubMed] [Google Scholar]
- Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007;81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament M, Brown W, Schumacher A, Harris R. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;15:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Lienenklaus S, Cornitescu M, Zietara N, Lyszkiewicz M, Gekara N, Jablonska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, Weiss S. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-β in vivo. J Immunol. 2009;283:3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- Lin M, Mckeown M, Ng H, Aguilera T, Shaner N, Campbell R, Adams S, Gross L, Ma W, Alber T, Tsien R. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening A, Wu A, Gambhir S. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- Luker G, Bardill J, Prior J, Pica C, Piwnica-Worms D, Leib D. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J Virol. 2002;76:12149–12161. doi: 10.1128/JVI.76.23.12149-12161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- Luker K, Hutchens M, Schultz T, Pekosz A, Luker G. Bioluminescence imaging of vaccinia virus: effects of interferon on viral replication and spread. Virology. 2005;341:284–300. doi: 10.1016/j.virol.2005.06.049. [DOI] [PubMed] [Google Scholar]
- Luker K, Luker G. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation. Antiviral Res. 2008;78:179–187. doi: 10.1016/j.antiviral.2008.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Schultz T, Romine J, Leib D, Luker G. Transgenic reporter mouse for bioluminescence imaging of herpes simplex virus 1 infection in living mice. Virology. 2006;347:286–295. doi: 10.1016/j.virol.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Maggi A, Ciana P. Reporter mice and drug discovery and development. Nat Rev Drug Discov. 2005;4:249–255. doi: 10.1038/nrd1661. [DOI] [PubMed] [Google Scholar]
- Mahlknecht U, Deng C, Lu M, Greenough T, Sullivan J, O'Brien W, Herbein G. Resistance to apoptosis in HIV-infected CD4+ T Lymphocytes is mediated by macrophages: role for Nef and immune activation in viral persistence. J Immunol. 2000;165:6437–6446. doi: 10.4049/jimmunol.165.11.6437. [DOI] [PubMed] [Google Scholar]
- Malmgaard L. Induction and regulation of IFNs during viral infections. J Interferon and Cytokine Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- Mandl S, Schimmelpfennig C, Edinger M, Negrin R, Contag C. Understanding immune cell trafficking patterns via in vivo bioluminescence imaging. J Cell Biochem. 2002;39:239–248. doi: 10.1002/jcb.10454. [DOI] [PubMed] [Google Scholar]
- Massoud T, Paulmurugan R, De A, Ray P, Gambhir S. Reporter gene imaging of protein-protein interactions in living subjects. Curr Opin Biotechnol. 2007;18:31–37. doi: 10.1016/j.copbio.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight R, Shamay A, Sankaran L, Wall R, Hennighausen L. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V, Zeiser R, Dasilva D, Chang D, Beilhack A, Contag C, Negrin R. In vivo dynamics of regulatory T-cell trafficking and survival predict effective strategies to control graft-versus-host disease following allogeneic transplantation. Blood. 2007;109:2649–2656. doi: 10.1182/blood-2006-08-044529. [DOI] [PubMed] [Google Scholar]
- Norbury C, Princiotta M, Bacik I, Brutkiewicz R, Wood P, Elliott T, Bennick J, Yewdell J. Multiple antigen-specific processing pathways for activating naive CD8+ T cells in vivo. J Immunol. 2001;166:4355–4362. doi: 10.4049/jimmunol.166.7.4355. [DOI] [PubMed] [Google Scholar]
- Olivo P. Detection of herpes simplex virus by measurement of luciferase activity in an infected-cell lysate. J Virol Methods. 1994;47:117–128. doi: 10.1016/0166-0934(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Pichler A, Prior J, Luker G, Piwnica-Worms D. Generation of a highly inducible Gal4 - Fluc universal reporter mouse for in vivo bioluminescence imaging. Proc Natl Acad Sci U S A. 2008;105:15932–15937. doi: 10.1073/pnas.0801075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Prior J, Piwnica-Worms D. Imaging reversal of multidrug resistance in living mice with bioluminescence: MDR1 P-glycoprotein transports coelenterazine. Proc Natl Acad Sci USA. 2004;101:1702–1707. doi: 10.1073/pnas.0304326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T, Diamond M, Ahmed A, Valentine L, Davis C, Samuel M, Hanna S, Puffer B, Doms R. An infectious West Nile virus that expresses a GFP reporter gene. Virology. 2005;334:28–40. doi: 10.1016/j.virol.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Ray P, Tsien R, Gambhir S. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67:3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- Remer K, Jungi T, Fatzer R, Tauber M, Leib S. Nitric oxide is protective in listeric meningoencephalitis of rats. Infect Immun. 2001;69:4086–4093. doi: 10.1128/IAI.69.6.4086-4093.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Mather T, Hokeness K. Calling in the troops: regulation of inflammatory cell trafficking through innate cytokine/chemokine networks. Viral Immunol. 2003;16:291–306. doi: 10.1089/088282403322396109. [DOI] [PubMed] [Google Scholar]
- Soejima K, Snyder N, Uchida T, Szabo C, Salzman A, Traber L, Traber D. The effect of inducible nitric oxide synthase (iNOS) inhibition on smoke inhalation injury in sheep. Shock. 2000;13:261–266. doi: 10.1097/00024382-200004000-00002. [DOI] [PubMed] [Google Scholar]
- Stark G, Kerr I, Williams B, Silverman R, Schreiber R. How cells respond to interferons. Ann Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- Tung C, Zeng Q, Shah K, Kim D, Schellingerhout D, Weissleder R. In vivo imaging of beta-galactosidase activity using far red fluorescent switch. Cancer Res. 2004;64:1579–1583. doi: 10.1158/0008-5472.can-03-3226. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu Y, Shabahang S, Wang G, Szalay A. Renilla luciferase-Aequorea GFP (Ruc-GFP) fusion protein, a novel dual reporter for real-time imaging of gene expression in cell cultures and in live animals. Mol Genet Genomics. 2002;268:160–168. doi: 10.1007/s00438-002-0751-9. [DOI] [PubMed] [Google Scholar]
- Wehrman T, von Degenfeld G, Krutzik P, Nolan G, Blau H. Luminescent imaging of beta-galactosidase activity in living subjects using sequential reporter-enzyme luminescence. Nat Methods. 2006;3:295–301. doi: 10.1038/nmeth868. [DOI] [PubMed] [Google Scholar]
- Wilson K, Yu J, Lee A, Wu J. In vitro and in vivo Bioluminescence Reporter Gene Imaging of Human Embryonic Stem Cells. J Vis Exp. 2008 May 2;:pii 740. doi: 10.3791/740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Jiang P, Hoffman R. Whole-body subcellular multicolor imaging of tumor-host interaction and drug response in real time. Cancer Res. 2007;67:5195–5200. doi: 10.1158/0008-5472.CAN-06-4590. [DOI] [PubMed] [Google Scholar]
- Yull F, Han W, Jansen E, Everhart M, Sadikot R, Christman J, Blackwell T. Bioluminescent detection of endotoxin effects on HIV-1 LTR-driven transcription in vivo. J Histochem Cytochem. 2003;51:741–749. doi: 10.1177/002215540305100605. [DOI] [PubMed] [Google Scholar]
- Zaitseva M, Kapnick S, Scott J, King L, Manischewitz J, Sirota L, Kodihalli S, Golding H. Application of bioluminescence imaging to the prediction of lethality in vaccinia virus-infected mice. J Virol. 2009;83:10437–10447. doi: 10.1128/JVI.01296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz B, Imamota A, Fiering S, Herzenberg L, Kerr W, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Weber A, Li B, Lyons R, Contag P, Purchio A, West D. An inducible nitric oxide synthase-luciferase reporter system for in vivo testing of anti-inflammatory compounds in transgenic mice. J Immunol. 2003;170:6307–6319. doi: 10.4049/jimmunol.170.12.6307. [DOI] [PubMed] [Google Scholar]