Summary
Bacteria perform chemotaxis utilizing core two-component signaling systems to which have been added enhanced features of signal amplification, sensory adaptation, molecular memory and high sensitivity over wide dynamic range. Chemoreceptors are central to the enhancements. These transmembrane homodimers associate in trimers and in clusters of signaling complexes containing from a few to thousands of receptors. Receptor homodimers couple ligand occupancy and adaptational modification to transmembrane signaling. Trimers activate and control the histidine kinase. Clusters enable signal amplification, high sensitivity and adaptational assistance. Homodimer signaling initiates with helical piston sliding that is converted to modulation of competing packing modes of adjacent segments of an extended helical coiled coil. In trimers, signaling and coupling may involve switching between compact and expanded forms.
Introduction
Bacterial chemotaxis is mediated by two-component systems for which the capabilities of histidine kinase-response regulator signaling have been enhanced by additional components and protein modifications. With these, chemotaxis sensory systems exhibit signal amplification and sensory adaptation, detect temporal gradients through a molecular memory and respond with high sensitivity over a wide dynamic range [1]. Chemoreceptors are central to these expanded capabilities. They carry sites of covalent modification that enable molecular memory, sensory adaptation and wide dynamic range. Receptors are homodimers that form trimers and higher order clusters of signaling complexes, structures in which interactions create signal amplification, cooperative sensing and cross-receptor adaptational assistance [1]. This review summarizes recent progress in understanding chemoreceptors, focusing on the extensively characterized receptors of Escherichia coli and Salmonella enterica and on results published since 2008. A recent review contains additional background [1] and other reviews provide complementary information [2-5].
The strategy of the chemotaxis sensory system
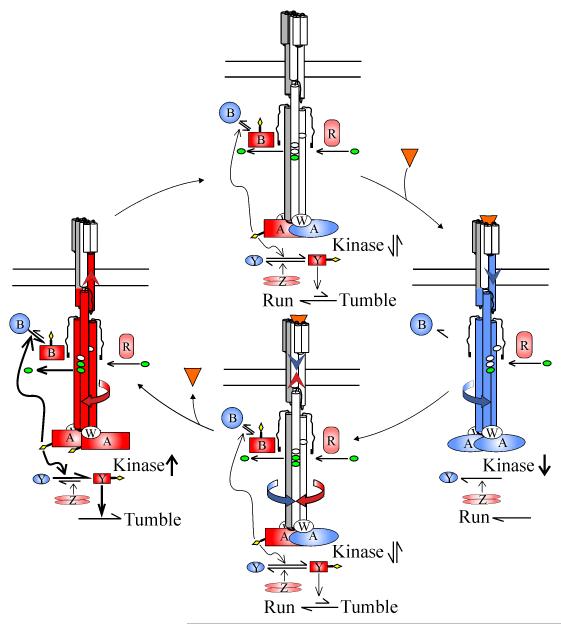
The chemotaxis system directs motile cells to favorable environments by controlling phosphorylation of histidine kinase CheA and its cognate response regulator CheY (Fig. 1). Chemoreceptors, CheA and coupling protein CheW form non-covalent but notably stable [6] signaling complexes, multi-component equivalents of single-protein sensor kinases, that couple ligand occupancy to kinase activity. Attractant binding to a chemoreceptor inhibits the kinase, reducing the concentration of short-lived phospho-CheY (CheY-P) and altering motility (Fig. 1). The system adapts by covalent modification of chemoreceptors, methylation of specific receptor glutamates by methyltransferase CheR and demethylation by methylesterase CheB (Fig. 1).
Fig. 1. The strategy of the chemotaxis sensory system of E. coli.
The figure shows the processes of stimulation and adaptation for a positive stimulus (increase in attractant concentration; right and bottom panel) and a negative stimulus (decrease in attractant concentration; left and top panel) using cartoons of the signaling complex representing the two receptor conformations (left = “kinase-on”, right = “kinase-off”). See text for discussion of the equilibrium between these two conformations. For clarity, only one of the three receptor dimers in the signaling complex is shown. Che proteins are labeled by their respective letters. Methyl-accepting sites are marked by ovals on the chemoreceptor, methyl groups are green and phosphoryl groups are yellow diamonds. Red symbolizes activated or activating forms, blue inactive or inactivating forms and pink constitutive activity. Top panel: Methyltransferase CheR adds methyl groups to the receptor and methylesterase CheB, activated by CheA-mediated phosphorylation, removes them. Phospho-CheY (CheY-P) binds to the flagellar rotary motor, switching its default state of counter-clockwise to clockwise and thus motility from straight-line runs to swimming-direction-reorienting tumbles. A balance between creation of CheY-P by CheA and destruction by phosphatase CheZ results in a steady state concentration of CheY-P that produces tumble episodes every few seconds, causing the cell to reorient and thus trace a three-dimensional random walk. Right panel: Attractant (orange triangles) binding to a periplasmic site generates a piston movement in the transmembrane sensing module (blue helix and arrowhead) which induces a cascade of alternating stabilizing and destabilizing changes in the helical packing of the signal conversion and kinase control modules (blue helixes and curving arrow) to shift the receptor conformational equilibrium toward the kinase-off, methylation-on, demethylation-off receptor conformation. Kinase inhibition reduces the cellular content of short-lived CheY-P and CheB-P, reducing the probability of tumbles and demethylation, respectively. Bottom panel: Altered receptor propensities for methylation and demethylation plus reduction of CheB-P result in an increase in receptor methylation that counteracts (opposing red arrows) the effects of ligand binding and thus returns the conformational equilibrium, propensities for adaptational modification, kinase activity, CheY-P, CheB-P and motile behavior to their null states. Left panel: Loss of ligand eliminates the changes generated by receptor occupancy but the compensatory effects of increased methylation are still present (red arrows), driving the receptor toward a kinase-on, methylation-off, demethylation-on conformation (red arrows and helices) that results in heightened kinase activity and thus higher levels of CheY-P and CheB-P, generating increased tumbling and demethylation, respectively. Effects on receptor propensities for modification and methylesters activity result in receptor methylation dropping to the original value in the absence of stimulation, thus reestablishing the adapted state (top panel).
Chemoreceptors and signaling complexes can be viewed as allosteric proteins in equilibrium between two conformations. One, favored by attractant occupancy, is kinase-off, methylation-on, demethylation-off and ligand-affinity-high. The other, favored by methylation, is kinase-on, methylation-off, demethylation-on and ligand-affinity-low. These two states are represented in Fig. 1 by the right and left cartoons, respectively. In vivo, a steady-state extent of methylation resulting from the relative rates of methylation and demethylation creates a mixed receptor population of the two conformations, illustrated in top cartoon of Fig. 1 as a single form representing a weighted average of the two conformations and their activities. This conformational distribution generates an intermediate level of kinase activity and thus alternating periods of forward swimming (“runs”) and abrupt turns (“tumbles”) that trace a random walk. Attractant binding, aided by a 35-fold signal amplification [7], shifts the equilibrium toward the kinase-off state, greatly reducing kinase activity and thus CheY-P concentration, which in turn biases the random walk to runs (Fig. 1, right). The shifted propensities for methylation and demethylation plus reduced production of the active form of the methylesterase by CheA-mediated phosphorylation of CheB return the conformational equilibrium and thus kinase activity, CheY-P, CheB-P and motile behavior to their pre-stimulus states by balancing ligand occupancy and methylation across the receptor population (Fig. 1, bottom cartoon, symbolizes this balancing as occurring within a single receptor).
Functional roles of dimers and trimers of dimers
Some chemoreceptor functions are performed by individual dimers; others require trimers of dimers. Isolated individual dimers in Nanodiscs bind ligand, undergo adaptational modification and signal across the membrane [8]. Coupling of ligand occupancy and methylation to conformational changes and transmembrane signaling is as effective for Nanodisc-isolated dimers as for interacting dimers in native membranes [9•]. In contrast, efficient activation of kinase requires more than one receptor dimer [8]. Nanodiscs averaging ~5 dimers per disc, i.e. each contain ≥ 3 parallel, potentially trimer-forming dimers; activate kinase as effectively as receptors in native membrane vesicles [M. Li and G.L. Hazelbauer, unpublished observations]. Thus coupling of ligand occupancy and adaptational modification to receptor conformation occurs in dimers but effective kinase activation requires a trimer of dimers.
Advances in receptor homodimer structure
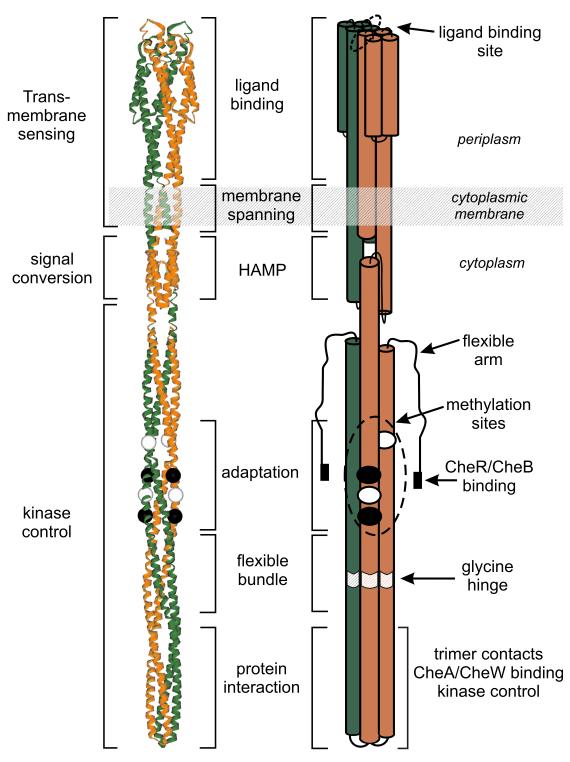
The fundamental structural unit of the chemoreceptor is a homodimer, a ~300 Å elongated cylinder of helical bundles and coiled coils comprising three functional modules: transmembrane sensing, signal conversion and kinase control (Fig. 2, [1]). Electron tomography revealed that chemoreceptors from diverse bacteria have similar elongated shapes, implying shared structure [10-14••]. For kinase control modules, the shared structure is an extended, dimeric, anti-parallel four-helix coiled coil, seen in three crystal structures [15•-17] and identified by bioinformatics as shared by all kinase control modules [18]. Models of receptor dimers commonly orient the modules of known structures with their long axes in parallel (Fig. 2). However, dimers can bend at the glycine hinge [1,16,17,19] and recent cryo-tomography observed the periplasmic portion of the transmembrane sensing module tilted relative to the receptor long axis (Fig 3A, [20••]).
Fig. 2. The chemoreceptor dimer.
A ribbon diagram (left) and a cartoon (right) of a chemoreceptor dimer illustrate structural and functional features. Modules are identified to the left of the ribbon diagram, functions or motifs are indicated between the ribbon diagram and cartoon, and specific features noted to the right of the cartoon. See text for details. The figure is based on Fig. 1, Box 3 of [1].
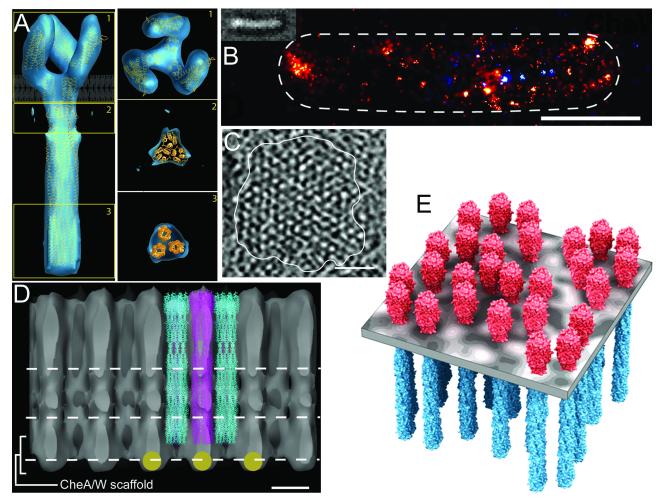
Fig. 3. Higher order interactions of chemoreceptor dimers.
A. Trimer structure. The panels show the “compact” conformation detected by cryo-electron tomography of E. coli chemoreceptors in a crystalline array, fit with three receptor dimers [20••]. The left-hand panel is a view perpendicular to the long axis of the receptor with the position of the cytoplasmic membrane indicated. The three right-hand panels are views parallel to the membrane at the three positions indicated in the left-hand image. The panels are from Fig. 4 of ref. [20••] with permission. B. Distribution of signaling complexes in an E. coli cell imaged by photoactivated localization microscopy (PALM). Chemotactically functional CheW fused to a photoactivatable fluorescent protein is visualized in a fixed cell by a procedure that can detect each individual molecule in a field of view. The image is Fig. 3D of ref. [31••] with permission. C. A tomographic slice of a face-on view of a chemoreceptor patch in C. cresentus showing partially ordered hexagonal packing. The image is Fig. 4A of ref. [11•] with permission. D. An edge-on view of the densities of an averaged packing arrangement of trimeric receptors in a C. crescentus signaling complex patch. Although beyond the scope of this review, it is tantalizing to note that the averaged densities imply that CheA and CheW associate with every second receptor trimer. The image is Fig. 3B of ref. [11•] with permission. E. Model derived from tomographic analysis of the organization of chemoreceptor trimers of dimers in patches of C. cresentus signaling complexes. The image is Fig. 4C of ref. [11•] with permission.
The signal conversion module is a HAMP motif, found in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins and certain phosphatases. Analyses of this module in three intact E. coli chemoreceptors [21-24•] are consistent with the parallel four-helix bundle structure determined for an isolated archaeal HAMP module [25]. In this homodimeric structure, each subunit contains two parallel, amphipathic helices, AS-1 and AS-2, connected by a linker segment packed on the surface of the bundle (Fig. 2). The available evidence argues that AS-1 is an uninterrupted extension of TM2 [26]. The AS-2 helix may extend to become the first helix of the kinase control module, but the region immediately membrane-distal to the HAMP module exhibits substantial proteolytic sensitivity and thus may not be as tightly structured. HAMP modules can also receive signals from input domains located in the cytoplasm, on the same or separate proteins [23,27].
Advances in higher order interactions
Images of clusters of E. coli signaling complexes from cryo-electron tomography had shown closely packed receptor densities extending from the cytoplasmic membrane with a layer containing CheA and CheW at their membrane-distal tips [10]. Tomography of Caulobacter crescentus produced higher resolution images that revealed partially ordered hexagonal arrays of densities in face-on views (Fig. 3C) and closely packed cylinders of transmembrane densities in edge-on views (Fig. 3D) [11•,12•]. The volume at each hexagonal vertex could be fit with three parallel receptor dimers (Fig. 3D), providing structural evidence for trimers of dimers in vivo. The parallel arrangement is different from the splayed tripod of dimers observed in the crystals of a kinase control module [16], but parallel dimers in groups of three have been observed for kinase-control modules in complex with CheA and CheW [28]. Dimers in signaling complexes might flex, for instance at the glycine hinge [1,16,17,19], to occupy an approximately cylindrical volume yet interact at their membrane distal tips as in receptor fragment crystals. Alternatively, dimer tips may make different interactions in signaling complexes, shifting dimer orientation [28]. The highest resolution images of trimers of intact dimers, obtained in the absence of CheA and CheW, suggest a third possibility, that there are alternative dimer orientations, parallel or splayed [20••].
Cryo-electron tomography of bacteria representing several major taxonomic groups revealed that all had clusters of chemotaxis signaling complexes organized like those in C. crescentus, in 12-nm hexagonal arrays of densities in which three, approximately parallel receptor dimers fit into each hexagonal vertex and the distance of the CheA/CheW layer from the membrane was a direct function of the bioinformatically deduced length of chemoreceptors in the respective species [14••]. Thus trimers of dimers and the hexagonal organization of clusters appear common and perhaps universal features of chemotaxis signaling systems.
Investigation of receptor localization at the resolution of individual dimers using photoactivated localization microscopy (PALM) revealed not only the one or two large polar clusters and several smaller, lateral clusters observed by other microscopic techniques [10-14••,29,30] but also many clusters scattered across the membrane surface in a roughly exponential size distribution, with 1/3 of the receptors in smaller lateral clusters and numerous solitary receptors (Fig. 3B, [31••]). This previously undocumented number and distribution of clusters creates the dispersion of receptors providing optimum detection sensitivity [32] and could change our view of the significance of large clusters, particularly because cooperating signaling teams appear to contain a few tens of receptors, not hundreds or thousands [33]. Heterogeneous cluster size could create a range of phosphorylation and adaptational modification rates in a cell, because these rates are influenced by receptor density [34]. Both the PALM data and observations using fluorescence argue that clusters and their approximately periodic distribution along the membrane occur via stochastic self-assembly [30,31••]. In addition the proteins associated with or part of signaling complexes are in dynamic equilibrium with a pool of individual components [35-37].
Advances in signaling and adaptation
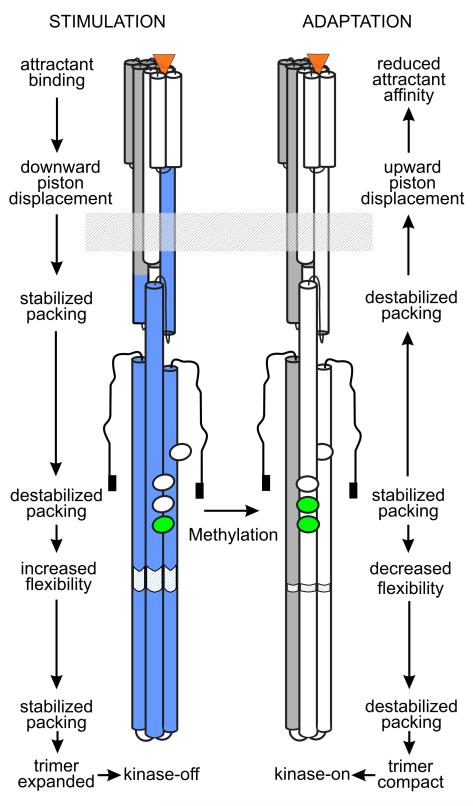
Conformational signaling links attractant occupancy at one end of a chemoreceptor to kinase activity of the non-covalently associated CheA at the other. For the transmembrane signaling module of E. coli chemoreceptors, a large body of structural, biochemical and mutational evidence indicates that the ligand-induced conformational change is a piston sliding of a few Ångströms of a helix that begins at the attractant-binding site and extends across the membrane to the HAMP module [38]. Furthermore, the conformational change of methylation-mediated adaptation is sliding of the same helix in the opposite direction [39]. Recent work suggests that signaling by piston sliding is common in receptors linked to two-component signaling [40-43].
The parallel four-helix bundle of the HAMP motif appears well designed to convert one kind of conformational signal into another. An initial model suggested it could function as a “gearbox” in which the four-helix bundle would ratchet by helical rotation between x-da (knob-on-knob) packing observed in the crystal structure and the more common a-d (knob-in-hole) packing [25]. However, extensive mutational analysis of the HAMP module in an intact chemoreceptor implies that only x-da packing, not a-d packing, is functionally important and argues against a functional cycling between two packing modes [24•]. Instead, the authors propose that HAMP signaling involves changes in stability of the x-da bundle generated by helical sliding of TM2 and that these changes modulate destabilizing phase clashes between AS-2 and the helix of the kinase control module into which it extends [24•].
Ideas about helical stability and phase clash also emerged from study of the kinase control module. That module can be divided into three regions, adaptation, flexible bundle and protein interaction (Fig. 2, [1,18]). Mutational and biochemical analyses suggest that signals are conveyed between regions by changes in helix-helix packing and that packing in the adaptation and protein interactions regions are coupled tightly and anti-symmetrically [44•]. Strong packing in the adaptation region stabilizes the kinase-on state and strong packing in the protein interaction region stabilizes the kinase-off state [44•]. Thus neutralizing negatively charged glutamyl side chains in the adaptation region by methylation would reduce charge density, favor closer helical packing and the kinase-on state. In a complementary set of observations, the crystal structure of the kinase-control fragment of a soluble T. maritima chemoreceptor exhibited a bulge on one helix in what would be the adaptation region [15•]. This perturbation generates changes in local helical packing in the extended four-helix coiled-coil, resulting in a > 25 Å displacement of the kinase-interacting tip. Taken together the data suggest that conformational signals impinging on the HAMP module as a piston sliding of AS-1 are conveyed through the kinase control module by opposing influences of out-of-register or alternatively packed helices along the length of the signal conversion and kinase control modules (Fig. 4).
Fig. 4. Chemoreceptor signaling.
Cartoons of a stimulated and adapted receptor dimer illustrate our understanding, explained in the text, of the conformational changes that couple stimulation by attractant binding to kinase inhibition (left-hand image and labels) and adaptation by methylation to re-establish pre-stimulation kinase activity (right-hand image and labels) . See Fig. 2 for labels of the receptor features shown in the cartoon and Fig. 1 for the symbolism of the colors. The figure is based on Fig. 3 of [1].
The cooperative and interactive nature of the chemotactic response means that conformational signals generated in one receptor dimer must affect neighboring dimers [1]. Trimers of receptor dimers provide the first level of dimer-dimer interaction. Measurement of fluorescence anisotropy of fluorescently labeled receptors in signaling complexes in vivo [45,46] and cryo-electron tomography of intact, membrane-embedded receptors produced without CheA or CheW [20••] each indicate that separation between dimers in a trimer is variable. Both approaches find that ligand occupancy favors an expanded separation among dimers whereas methylation favors a compact geometry. The arrangements could be the kinase-on and kinase-off conformations of trimers that alter enzyme activity and allow one dimer to influence its neighbor.
Like signaling, adaptation occurs in groups of receptors, “assistance neighborhoods”, in which sub-stoichiometric [47] modification enzymes interacting at the carboxyl terminus of one receptor dimer can modify sites equivalent to those on ~6 neighboring dimers [48]. Assistance is necessary for precise adaptation [49,50]. Molecular modeling of the ~30-residue flexible arm between the receptor body and a carboxyl-terminal tether demonstrated CheR could physically reach some but not all modification sites on 8 neighbors, totaling the same number of accessible sites observed experimentally but indicating a larger assistance neighborhood in terms of receptors served [51].
Conclusions
As key contributors to the enhanced two-component signaling systems of chemotaxis, chemoreceptors provide the modification sites and accompanying conformational coupling central to sensory adaptation, gradient sensing and wide dynamic range. They also provide a hierarchy of oligomerization from dimers to trimers of dimers to clusters. In this hierarchy, dimers mediate transmembrane signaling and conformational coupling, trimers mediate kinase activation and control, and clusters enable signal amplification, interacting signaling teams and adaptational assistance. The result is a high-performance signaling system that illustrates how sophisticated complexity can be built from a few simple components and reactions.
Box 1. Key unanswered questions about chemoreceptors.
What is the structure of an intact dimer, what are its conformational states and how are they influenced by ligand occupancy and adaptational modification?
What is the stoichiometry and structure of the core signaling complex, what are its conformational states and how are they influenced by ligand occupancy and adaptational modification?
- What are the mechanisms by which conformational signals are passed
- between modules within a receptor dimer?
- among dimers in a trimer?
- among trimers?
- between receptor(s) and kinase?
- For clusters of signaling complexes:
- What are the structural and mechanistic bases of formation and maintenance?
- Are there functional differences between different sized clusters?
- Is there a functional role for large clusters?
- For adaptational modifications:
- What mechanisms balance rates of methylation and demethylation with only one methyltransferase for every 60 receptor dimers and one methylesterase for every 40?
- What is the structural basis for making a site of modification available for methylation but not for demethylation, or vice versa?
What are the variations in organization and mechanisms among chemoreceptors and signaling complexes across bacterial diversity?
Acknowledgements
Our laboratory is supported by GM29963 from the National Institute of Health. We thank Sriram Subramaniam and Jan Liphardt for permissions and images, as well as our many colleagues who responded to requests for information. We ask their understanding that limitations on article length and references kept us from discussing all recent publications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 4.Rao CV, Glekas GD, Ordal GW. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 2008;16:480–487. doi: 10.1016/j.tim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vladimirov N, Sourjik V. Chemotaxis: how bacteria use memory. Biol Chem. 2009;390:1097–1104. doi: 10.1515/BC.2009.130. [DOI] [PubMed] [Google Scholar]
- 6.Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •9.Amin DN, Hazelbauer GL. The chemoreceptor dimer is the unit of conformational coupling and transmembrane signaling. J Bacteriol. 2010 doi: 10.1128/JB.01391-09. In press. This work shows fully efficient coupling of receptor conformation and transmembrane signaling to ligand binding and adaptational modification in isolated receptor dimers, indicating that neither trimers nor higher order interactions are required for these intial steps of signaling.
- 10.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •11.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. This study and ref. [12•] provide the currently highest resolution images of native signaling complexes in vivo, revealing partially ordered hexagonal arrays of densities and providing important structural evidence that in vivo receptors are organized as trimers of dimers.
- •12.Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borgnia MJ, Subramaniam S, Milne JLS. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J Bacteriol. 2008;190:2588–2596. doi: 10.1128/JB.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••14.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. This cryo-electron tomography study documents wide-spread if not universal hexagonal organization of clusters of trimers of chemoreceptor dimers in signaling complexes.
- •15.Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. The results provide a potentially important hint as to how conformational signals are transmitted through the kinase-control module, i.e. by minor alterations in local helical packing that can displace the kinase-interacting tip of a chemoreceptor > 25 Å.
- 16.Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 17.Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 18.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••20.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. The study provides the currently highest resolution images of intact chemoreceptors. The images reveal two distinct conformations of chemoreceptor trimers of dimers, expanded and compact, which are correlated with the “kinase-on” and “kinase-off” state, respectively.
- 21.Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: A disulfide mapping study. Biochemistry. 2007;46:13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ames P, Zhou Q, Parkinson JS. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:6676–6685. doi: 10.1128/JB.00750-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watts KJ, Johnson MS, Taylor BL. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J Bacteriol. 2008;190:2118–2127. doi: 10.1128/JB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Molecular Microbiology. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. This work provides evidence against the “gearbox” model of HAMP signaling in chemoreceptors and proposes an alternative model based on signaling by modulating destabilizing phase clashes between helical regions.
- 25.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 26.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott KT, Zhulin IB, Stuckey JA, DiRita VJ. Conserved residues in the HAMP domain define a new family of proposed bipartite energy taxis receptors. J Bacteriol. 2009;191:375–387. doi: 10.1128/JB.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis NR, Wolanin PM, Stock JB, DeRosier DJ, Thomas DR. Three-dimensional structure and organization of a receptor/signaling complex. Proc Natl Acad Sci U S A. 2004;101:17480–17485. doi: 10.1073/pnas.0407826101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 30.Thiem S, Sourjik V. Stochastic assembly of chemoreceptor clusters in Escherichia coli. Mol Microbiol. 2008;68:1228–1236. doi: 10.1111/j.1365-2958.2008.06227.x. [DOI] [PubMed] [Google Scholar]
- ••31.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. Our view of clusters of signaling complexes is expanded by results of photoactivated localization microscopy that show many more clusters and a much wider size distribution that previously detected. The results support a stochastic mechanism of cluster assembly and have implications for our notions about cluster function.
- 32.Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endres RG, Oleksiuk O, Hansen CH, Meir Y, Sourjik V, Wingreen NS. Variable sizes of Escherichia coli chemoreceptor signaling teams. Mol Syst Biol. 2008;4:211. doi: 10.1038/msb.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besschetnova TY, Montefusco DJ, Asinas AE, Shrout AL, Antommattei FM, Weis RM. Receptor density balances signal stimulation and attenuation in membrane-assembled complexes of bacterial chemotaxis signaling proteins. Proc Natl Acad Sci U S A. 2008;105:12289–12294. doi: 10.1073/pnas.0802868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Kentner D, Sourjik V. Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai W-C, Beel BD, Hazelbauer GL. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol Microbiol. 2006;61:1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x. [DOI] [PubMed] [Google Scholar]
- 40.Sevvana M, Vijayan V, Zweckstetter M, Reinelt S, Madden DR, Herbst-Irmer R, Sheldrick GM, Bott M, Griesinger C, Becker S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore JO, Hendrickson WA. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure. 2009;17:1195–1204. doi: 10.1016/j.str.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 43.Cheung J, Hendrickson WA. Sensor domains of two-component regulatory systems. Curr Opin Microbiol. 2010;13 doi: 10.1016/j.mib.2010.01.016. ???-??? [DOI] [PMC free article] [PubMed] [Google Scholar]
- •44.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: Evidence for a Yin-Yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. The paper suggests that signaling between the adaptation and protein interaction regions of the kinase control module involves helix packing changes and a strong coupling between tight packing in one region with loose packing in the other.
- 45.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaknin A, Berg HC. Direct evidence for coupling between bacterial chemoreceptors. J Mol Biol. 2008;382:573–577. doi: 10.1016/j.jmb.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 49.Hansen CH, Endres RG, Wingreen NS. Chemotaxis in Escherichia coli: A molecular model for robust precise adaptation. PLoS Comput Biol. 2008;4:e1. doi: 10.1371/journal.pcbi.0040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc Natl Acad Sci U S A. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muppirala UK, Desensi S, Lybrand TP, Hazelbauer GL, Li Z. Molecular modeling of flexible arm-mediated interactions between bacterial chemoreceptors and their modification enzyme. Protein Sci. 2009;18:1702–1714. doi: 10.1002/pro.170. [DOI] [PMC free article] [PubMed] [Google Scholar]