Abstract
We genotyped 13 single nucleotide polymorphisms (SNPs) in the estrogen receptor alpha gene (ESR1) region in three population-based case-control studies of epithelial ovarian cancer conducted in the United States, comprising a total of 1,128 and 1,866 non-Hispanic white invasive cases and controls, respectively. A SNP 19 kb downstream of ESR1 (rs2295190, G-to-T change) was associated with invasive ovarian cancer risk, with a per-T-allele odds ratio (OR) of 1.24 (95% confidence interval (CI), 1.06–1.44, p=0.006). rs2295190 is a non-synonymous coding SNP in a neighboring gene called spectrin repeat containing, nuclear envelope 1 (SYNE1) which is involved in nuclear organization and structural integrity, function of the Golgi apparatus, and cytokinesis. An isoform encoded by SYNE1 has been reported to be downregulated in ovarian and other cancers. rs2295190 was genotyped in an additional 12 studies through the Ovarian Cancer Association Consortium, with 5,279 invasive epithelial cases and 7,450 controls. The per-T-allele OR for this 12-study set was 1.09 (95% CI, 1.02–1.17, p=0.017). Results for the serous subtype in the 15 combined studies were similar to those overall (n=3,545; OR=1.09, 95% CI, 1.01–1.18, p=0.025), and our findings were strongest for the mucinous subtype (n=447; OR=1.32, 95% CI, 1.11–1.58, p=0.002). No association was observed for the endometrioid subtype. In an additional analysis of 1,459 borderline ovarian cancer cases and 7,370 controls, rs2295190 was not associated with risk. These data provide suggestive evidence that the rs2295190 T allele, or another allele in linkage disequilibrium with it, may be associated with increased risk of invasive ovarian cancer.
Keywords: ovarian cancer, SYNE1, ESR1, OCAC
Introduction
Ovarian cancer is hormone-related, and it has long been hypothesized that variation in steroid hormone biosynthesis, catabolism and response genes may be associated with ovarian cancer risk. To explore whether genetic variation in these pathways is associated with ovarian cancer risk, we genotyped tag and functional SNPs in and around (+/−20,000bp) genes in these pathways in three studies of epithelial ovarian cancer conducted in the United States. Herein we describe both an initial suggestive finding in our three-study effort for a coding SNP (rs2295190) in the estrogen receptor alpha (ESR1) gene region and a follow-up genotyping effort in an independent data set comprising 12 studies from the international Ovarian Cancer Association Consortium (OCAC). rs2295190 is a missense SNP located 19 kb downstream of ESR1 in the spectrin repeat containing, nuclear envelope 1 (SYNE1) gene. SYNE1 encodes several different isoforms, many of which are involved in a variety of cellular processes including cytokinesis, Golgi function, and nuclear organization and structural integrity (1–3). One isoform, Drop1, has been noted to be down regulated in ovarian and other cancers (4).
Methods
Details of each of the 15 studies that participated in this analysis have been published (5, 6). All studies were approved by the ethics review boards of their parent institutions and obtained written informed consent from study participants prior to interview and collection of blood samples. All but one study (MAY (Mayo Clinic Ovarian Cancer Case Control Study)) was population-based.
Initial analyses were based on three case-control studies in the United States: DOV (Diseases of the Ovary and Their Evaluation, Washington state); HOP (Hormones and Ovarian Cancer Prediction Study, Pennsylvania, New York, Ohio), and USC (University of Southern California/Los Angeles County Case-Control Studies of Ovarian Cancer, California). Genotyping was performed at the University of Southern California using a 1,536 Illumina Golden Gate custom array to assess steroid hormone-related polymorphisms and others. A total of 693 tag and functional SNPs were selected for 42 hormone-related candidate genes, as well as regions spanning 20,000 bp up- and downstream of the genes, in an attempt to genotype SNPs that might influence their expression. Thirteen SNPs were genotyped in the ESR1 gene region. A total of 4,195 samples were assayed and 3,943 had genotyping success of >90%. Because the minor allele frequency of rs2295190 is considerably lower in non-Whites, we restricted analyses to the 3,315 non-Hispanic white women who were successfully genotyped for rs2295190, including 1,128 women with invasive epithelial ovarian cancer, 321 women with borderline epithelial ovarian tumors, and 1,866 controls.
While analyses of the steroid hormone pathway genes and gene-environment interactions within our three-study collaboration are still ongoing, the OCAC provides an early opportunity for independent fast-track evaluation of initial positive findings of participating studies, and rs2295190 was nominated because it was the SNP most strongly associated with invasive ovarian cancer risk among the hormone-related genes (additive model p=0.006). Thus, this SNP was examined in 12 additional case-control studies that are members of the OCAC (six from the US [HAW (Hawaiian Ovarian Cancer Study), MAY, NCO (North Carolina Ovarian Cancer Study), NEC (New England-based Case–control Study), STA (Genetic Epidemiology of Ovarian Cancer Study, Stanford University), UCI (UC Irvine Ovarian Cancer Study)], one from Australia [AUS (Australian Cancer Study/Australian Ovarian Cancer Study)] and five from Europe [GER (German Ovarian Cancer Case-control Study), MAL (MALOVA Ovarian Cancer Case-control Study, Danish Cancer Society, Denmark), POL (Polish Ovarian Cancer Study), SEA (Studies of Epidemiology and Risk Factors in Cancer Heredity, UK), UKO (UK Ovarian Cancer Population Study)]). Genotyping for these latter studies was done using the TaqMan allelic discrimination assay (TaqMan; Applied Biosystems, Foster City, CA) with the exception of AUS which used the Sequenom iPlex gold genotyping platform (Sequenom Inc., San Diego, CA). After excluding 65 women with missing tumor behavior, these 12 studies provided genotype data for 5,279 non-Hispanic white women with invasive and 1,138 women with borderline epithelial ovarian cancer, and 7,450 controls. Information regarding Hispanic ethnicity was not collected in some studies conducted in populations with few Hispanics; in those instances, all white women were considered to be non-Hispanic.
The OCAC has developed a set of genotyping quality control procedures, described in Ramus et al. (7). All laboratories providing data for these analyses genotyped a common set of DNA samples; the concordance across laboratories was 100% and genotyping success was >90%. All studies had concordance for duplicate samples of ≥98%, with ≥95% sample call success overall, and call rates for every 384-well plate 90% or above.
Statistical analyses
Data were compiled centrally at the OCAC data coordinating centers at Duke University and the University of Southern California. Genotype frequencies in (non-Hispanic white) controls were assessed for departure from Hardy-Weinberg Equilibrium (HWE) using the Fisher’s Exact test. Per-allele odds ratios (ORs) and their 95% confidence intervals (CIs) were estimated using unconditional logistic regression. Our analyses examined risk of invasive and borderline epithelial tumors separately, and we assessed risk within histologic subgroups. All analyses were adjusted for age and study site and were performed using Stata v. 10 (STATA Corporation, College Station, TX). The “meta” command, which implements inverse-variance weighting, was used to calculate fixed and random effects ORs and 95% CIs, as well as to generate a forest plot. Heterogeneity across studies was examined by Cochran’s Q test.
Results
The distribution of the rs2295190 GG, GT and TT genotypes among women with invasive ovarian cancer and controls by study site is presented in Table 1. We found no evidence of deviation from HWE (p>0.05 for each study), and the frequency of the T allele among controls ranged from 12–17% (Table 1). The minor allele was associated with an increased risk of invasive epithelial ovarian cancer both in the initial three-study collaboration and in the 12-study data (per-T-allele ORs and 95% CIs were, respectively: 1.24 (1.06–1.44, p=0.006), and 1.09 (1.02–1.17, p=0.017); Table 2). Results from unconditional logistic regression models are presented in Table 2; results were essentially identical for the random- and fixed-effects models. No heterogeneity between studies was observed (pheterogeneity for 15 studies=0.73; Figure 1). Among invasive cancers, the association was present for serous tumors (n=3,545, OR in 15 studies=1.09, 95% CI 1.01–1.18, p=0.025) and strongest for the mucinous subtype (n=447, OR in 15 studies=1.32, 95% CI 1.11–1.58, p=0.002). No association was observed for endometrioid tumors (Table 2). The SNP was not associated with risk of borderline tumors overall (Table 2), or with histologic subtypes of serous (n=802, OR in 15 studies=0.94, 95% CI, 0.80–1.10, p=0.415) or mucinous borderline tumors (n=580, OR in 15 studies=1.06, 95% CI, 0.89–1.26, p=0.510; data not shown).
Table 1.
Distribution of rs2295190 genotypes by case-control status, and Hardy-Weinberg equilibrium p-values for controls, by study population*
| Case† /Control genotype | |||||
|---|---|---|---|---|---|
| Study | GG | GT | TT | T allele freq., controls | pHWE |
| Initial three studies | |||||
| DOV | 329/483 | 141/162 | 16/12 | 0.142 | 0.87 |
| HOP | 214/497 | 77/166 | 3/8 | 0.136 | 0.19 |
| USC | 258/417 | 80/117 | 10/4 | 0.116 | 0.21 |
| Additional twelve studies | |||||
| AUS | 617/789 | 236/295 | 23/16 | 0.149 | 0.06 |
| GER | 148/302 | 52/111 | 3/14 | 0.163 | 0.37 |
| HAW | 45/110 | 23/46 | 1/2 | 0.158 | 0.37 |
| MAL | 281/546 | 143/223 | 15/25 | 0.172 | 0.71 |
| MAY | 193/262 | 74/99 | 7/5 | 0.149 | 0.30 |
| NCO | 446/559 | 155/169 | 9/10 | 0.128 | 0.62 |
| NEC | 449/682 | 145/200 | 11/19 | 0.132 | 0.31 |
| POL | 160/408 | 87/164 | 7/17 | 0.168 | 0.88 |
| SEA | 675/893 | 244/291 | 18/29 | 0.144 | 0.35 |
| STA | 191/129 | 72/48 | 3/4 | 0.155 | 1.00 |
| UCI | 203/331 | 64/79 | 11/10 | 0.118 | 0.06 |
| UKO | 336/411 | 122/133 | 10/19 | 0.152 | 0.05 |
Restricted to non-Hispanic white women
Invasive cases only
Table 2.
Per allele odds ratios and 95% confidence intervals for borderline and invasive ovarian cancer by histologic subtype
| Initial three studies† |
Additional twelve studies‡ |
All |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR* | 95% CI | p | Cases | Controls | OR* 95% CI | p | Cases | Controls | OR* | 95% CI | p | |
| All invasive | 1,128 | 1,866 | 1.24 | (1.06–1.44) | 0.006 | 5,279 | 7,450 | 1.09 (1.02–1.17) | 0.017 | 6,407 | 9,316 | 1.11 | (1.04–1.19) | 0.001 |
| Serous | 644 | 1,866 | 1.31 | (1.09–1.57) | 0.004 | 2,901 | 7,450 | 1.05 (0.97–1.15) | 0.248 | 3,545 | 9,316 | 1.09 | (1.01–1.18) | 0.025 |
| Mucinous | 61 | 1,866 | 1.65 | (1.01–2.68) | 0.043 | 386 | 7,450 | 1.28 (1.06–1.55) | 0.011 | 447 | 9,316 | 1.32 | (1.11–1.58) | 0.002 |
| Endometrioid | 172 | 1,866 | 0.93 | (0.66–1.31) | 0.675 | 854 | 7,450 | 1.00 (0.87–1.15) | 0.986 | 1,026 | 9,316 | 0.99 | (0.87–1.13) | 0.852 |
| Clear cell | 72 | 1,866 | 1.37 | (0.87–2.16) | 0.176 | 462 | 7,450 | 1.06 (0.88–1.28) | 0.532 | 534 | 9,316 | 1.10 | (0.93–1.31) | 0.276 |
| All other histologies | 179 | 1,866 | 1.21 | (0.89–1.64) | 0.229 | 676 | 7,450 | 1.23 (1.06–1.43) | 0.006 | 855 | 9,316 | 1.23 | (1.08–1.41) | 0.002 |
| All borderline§ | 321 | 1,866 | 1.01 | (0.78–1.32) | 0.913 | 1,138 | 5,504 | 0.99 (0.86–1.13) | 0.852 | 1,459 | 7,370 | 1.00 | (0.88–1.12) | 0.936 |
Adjusted for age and study site
DOV, HOP and USC.
ACS, GER, HAW, MAL, MAY, NCO, NEC, POL, SEA, STA, UCI and UKO
Sites MAL, POL and UKO had no cases in this subgroup and their controls are excluded from this analysis
Figure 1.
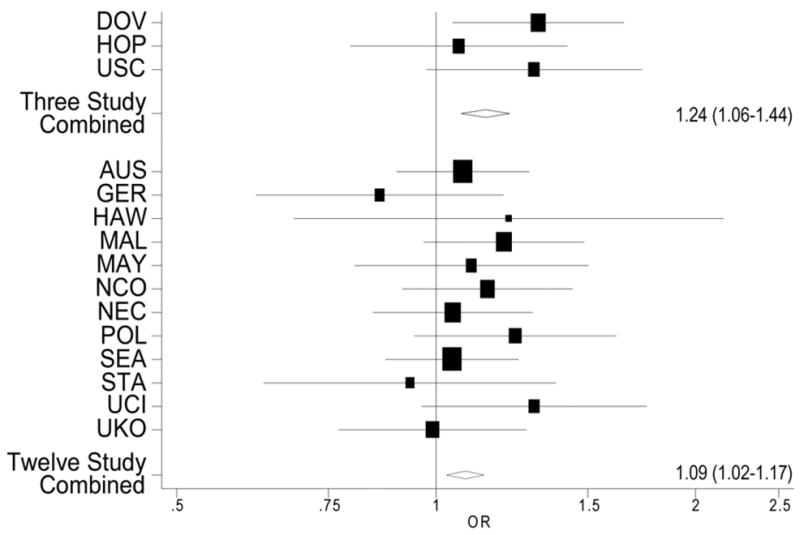
Per-allele age-adjusted odds ratios (boxes) and 95% confidence intervals (lines) for the association between rs2295190 and invasive ovarian cancer, by study (pheterogeneity =0.73), and combined odds ratios and 95% confidence intervals for the initial three-study set and the additional twelve-study set. The size of the box reflects the size of the study.
Discussion
The T allele of rs2295190 was associated with increased invasive ovarian cancer risk in our initial three-study investigation, and an association (although smaller in magnitude) subsequently was observed in an independent set of 12 additional studies. Although rs2295190 was selected because it is located in the ESR1 gene region, it is also a missense SNP in an adjacent gene, SYNE1.
A recent genome-wide association study reported that a SNP 180 bp upstream of ESR1 (rs2046210) was associated with breast cancer risk (8). This SNP is located nearly 296 kb away from rs2295190 and is not in linkage disequilibrium with it. rs2295190 is located 19 kb downstream of ESR1 in the C-terminus region of SYNE1, and is a G-to-T change that results in a conservative amino acid substitution from leucine to methionine. Based on a >1,300 kb region encompassing 250 kb upstream of ESR1 to 250 kb upstream of SYNE1 in the International Haplotype Map Project (HapMap, build 24) Caucasian data, three SNPs are in linkage disequilibrium with rs2295190 (r2>80%; rs6934016, rs13211987, and rs17082180); all are non-coding SNPs in SYNE1. The T allele of rs2295190 is predicted to be damaging to SYNE1 protein function based on the Sorting Intolerant from Tolerant (SIFT) algorithm which aligns related protein sequences and predicts the functional impact of SNPs based on the concept that an amino acid change in a highly conserved area across protein sequences would be damaging to protein function, whereas a poorly conserved region is more likely to be tolerant to such changes (9). While rs2295190 is a coding SNP in the SYNE1 gene, it is also possible that this SNP, or one in linkage disequilibrium with it, could lie in a region encoding an as-of-yet uncharacterized enhancer or repressor of SYNE1, or less plausibly, ESR1.
SYNE1 is an unusually large gene (0.5 Mb) on chromosome 6q25.1–25.2 (10) that encodes several protein isoforms through alternative transcriptional initiation, termination and splicing. Many of the isoforms localize to the nuclear envelope, Golgi apparatus and cytoskeleton and are involved in a variety of cellular processes including cytokinesis and Golgi function as well as organization, structural integrity and positioning of the nucleus (1–3). Mutations towards the 3′ end of this gene are associated with cerebellar ataxia and Emery-Dreifuss muscular dystrophy (10, 11).
One of the shorter transcripts, Drop1, is encoded in the 5′ end of the SYNE1 gene and has been observed to be markedly downregulated in ovarian tumors compared to normal ovarian surface epithelium from the same individual. These results were supported by expression profiling, in situ hybridization, and immunostaining for Drop1 protein. Estrogen receptor alpha expression was not correlated with Drop1 expression in tumor samples, though their expression was correlated in ovarian carcinoma cell lines (4). In the same study, Drop1 mRNA was also observed to be down regulated in carcinomas of the uterus, cervix, kidney, thyroid, pancreas, and lung (4). SYNE1 is frequently methylated in lung cancer cell lines, lung adenocarcinoma (12), and colorectal cancer (13), and has been observed to be mutated in colorectal cancer (14).
Among invasive ovarian cancers, the association with rs2295190 was observed for the serous subtype and was strongest for the mucinous subtype, with no association observed for endometrioid tumors. These results should be interpreted cautiously as differences may exist between studies in defining histology. Although some evidence suggests that mucinous invasive and borderline ovarian tumors may share a common origin (15), we noted no clear association of rs2295190 with risk of mucinous borderline tumors.
Even with this relatively large sample set, because the frequency of the T allele is low (12–17%), the statistical power for some subgroup analyses is limited, and it is possible that our findings are due to chance. It is also possible that residual confounding due to population stratification may influence our results, though it is unlikely because analyses were restricted to non-Hispanic white women and adjusted for study site, and results were similar across studies with different populations.
In conclusion, our study based in the international OCAC provides suggestive evidence that the T allele of rs2295190, or another allele in linkage disequilibrium with it, may be associated with an increased risk of invasive ovarian cancer. Efforts to further characterize this region around ESR1/SYNE1 are warranted.
Acknowledgments
The OCAC is supported by the Ovarian Cancer Research Fund provided by the family and friends of Kathryn Sladek Smith.
The DOV study (MAR, JD, KLC-H, CC) was supported by the US National Cancer Institute grants CA87538 and CA112523.
Financial support for the AOCS study was provided by U.S. Army Medical Research and Materiel Command under DAMD17-01-1-0729, and the Cancer Council Tasmania and Cancer Foundation of Western Australia. Funding for the ACS study was provided by the National Health and Medical Research Council (NHMRC) of Australia (199600). GCT and PW are supported by the NHMRC.
The GER study (JC-C and SW-G) was supported by the German Federal Ministry of Education and Research of Germany, Programme of Clinical Biomedical Research grant 01 GB 9401, the genotyping in part by the state of Baden-Württemberg through Medical Faculty of the University of Ulm (P.685) and data management by the German Cancer Research Center.
The HAW study (MTG, GL, PJT, MEC) was supported in part by Public Health Service grant R01-CA-58598 and by contract N01-PC-35137 from the Department of Health and Human Services, National Institutes of Health.
The HOPE study (RBN, KM) was supported by grants from the Department of Defense DAMD 17-02-1-0669 and National Cancer Institute RO1CA095023.
The MALOVA study (EH, SKK, CH) was supported by grants from Mermaid 1, The Danish Cancer Society and The National Cancer Institute, Bethesda, MD, USA (RO1 CA61107).
The MAY study (ELG, JMC, BLF, RAV) was supported in part by the US National Cancer Institute grant CA122443.
The NCO study (AB, PGM, JMS) was supported by the US National Cancer Institute Grant R01-CA76016. JMS and RP were supported by the US National Cancer Institute Grant R01-HL090559.
The NEC study (DWC, KLT) was supported by the US National Cancer Institute grants CA54419 and CA105009.
The POL study (MGC, HPY, SC, JL) was supported by funds from the intramural program of the National Cancer Institute, National Institutes of Health.
PDPP is a Cancer Research UK Senior Clinical Research Fellow. The SEA study (SEARCH; HS, MS, BP) is funded by Cancer Research UK.
The STA study was supported by US National Institute of Health grants U01 CA71966 (AS, VM), CA16056 (RD) and U01 CA69417 (for recruitment of controls by the Northern California Cancer Center).
The UCI OCAC study (HA-C, AZ, WB) is supported by the National Institutes of Health, National Cancer Institute USA grant CA-58860 and the Lon V. Smith Foundation grant LVS-18840.
The UKOPS study is funded by the OAK Foundation. SJR is funded by the Mermaid component of the Eve Appeal. A portion of this work was done at UCLH/UCL within the “women’s health theme” of the NIHR UCLH/UCL Comprehensive Biomedical Research Centre supported by the Department of Health.
The USC study was supported by the US National Cancer Institute grants P01-CA17054, CA14089, CA61132, CA63464, N01-PC67010, R03-CA113148; California Department of Health Services subcontract 050-E8709; California Cancer Research Program Grants 00-01389V-20170 and 2110200.
We thank all the individuals who took part in this study and the project staff of all the participating studies. The AOCS Management Group gratefully acknowledges the contribution of all the collaborators (see http://www.aocstudy.org/). The ACS Management Group comprises A. Green, P. Parsons, N. Hayward, P. Webb, D. Whiteman. The POL study would like to thank Louise Brinton from the National Cancer Institute (USA), Beata Peplonska and Neonila Szeszenia-Dabrowska of the Nofer Institute of Occupational Medicine (Lodz, Poland), Witold Zatonski of the M. Sklodowska-Curie Institute of Oncology and Cancer Center (Warsaw, Poland), and Pei Chao and Michel Stagner (IMS, Silver Spring, MD, USA) for their valuable contributions to the study. The GER study would like to thank Ursula Eilber and Tanja Koehler for their competent technical assistance. The UKOPS would like to thank all members of the research team, including research nurses, research scientists, data entry personnel and consultant gynaecological oncologists for their help in establishing the UKOPS case-control collection.
References
- 1.Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J Cell Sci. 2004;117:619–29. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q, Skepper JN, Yang F, et al. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–98. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 3.Gough LL, Fan J, Chu S, Winnick S, Beck KA. Golgi localization of Syne-1. Mol Biol Cell. 2003;14:2410–24. doi: 10.1091/mbc.E02-07-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marme A, Zimmermann HP, Moldenhauer G, et al. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int J Cancer. 2008;123:2048–56. doi: 10.1002/ijc.23763. [DOI] [PubMed] [Google Scholar]
- 5.Pearce CL, Near AM, Van Den Berg DJ, et al. Validating genetic risk associations for ovarian cancer through the international Ovarian Cancer Association Consortium. Br J Cancer. 2009;100:412–20. doi: 10.1038/sj.bjc.6604820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, Ramus SJ, Kjaer SK, et al. Association between invasive ovarian cancer susceptibility and 11 best candidate SNPs from breast cancer genome-wide association study. Hum Mol Genet. 2009;18:2297–304. doi: 10.1093/hmg/ddp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramus SJ, Vierkant RA, Johnatty SE, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. Int J Cancer. 2008;123:380–8. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gros-Louis F, Dupre N, Dion P, et al. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet. 2007;39:80–5. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Bethmann C, Worth NF, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–33. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 12.Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–14. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 13.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 15.Jordan SJ, Green AC, Whiteman DC, Webb PM. Risk factors for benign, borderline and invasive mucinous ovarian tumors: epidemiological evidence of a neoplastic continuum? Gynecol Oncol. 2007;107:223–30. doi: 10.1016/j.ygyno.2007.06.006. [DOI] [PubMed] [Google Scholar]


