Abstract
Noncompetitive immunoassays are advantageous over competitive assays for the detection of small molecular weight compounds. We recently demonstrated that phage peptide libraries can be an excellent source of immunoreagents that facilitate the development of sandwich-type noncompetitive immunoassays for the detection of small analytes, avoiding the technical challenges of producing anti-immunocomplex antibody. In this work we explore a new format that may help to optimize the performance of the phage anti-immunocomplex assay (PHAIA) technology. As a model system we used a polyclonal antibody to 3-phenoxybenzoic acid (3-PBA) and an anti-immunocomplex phage clone bearing the cyclic peptide CFNGKDWLYC. The assay setup with the biotinylated antibody immobilized onto streptavidin-coated magnetic beads significantly reduced the amount of coating antibody giving identical sensitivity (50% saturation of the signal (SC50) = 0.2–0.4 ng/ml) to the best result obtained with direct coating of the antibody on ELISA plates. The bead-based assay tolerated up to 10 and 5% of methanol and urine matrix, respectively. This assay system accurately determined the level of spiked 3-PBA in different urine samples prepared by direct dilution or clean-up with solid-phase extraction after acidic hydrolysis with overall recovery of 80–120%.
Keywords: Phage anti-immunocomplex assay, Phage peptide display, Phage ELISA, Noncompetitive immunoassay, 3-Phenoxybenzoic acid, Pyrethroid insecticides
Double-sandwich or noncompetitive immunoassays have been dominantly used for protein detection for many reasons including improved sensitivity, speed, and specificity. Very few noncompetitive immunoassays have been developed for small molecules because they are too small for double-sandwich approaches to work as discussed in more detail below. To address this problem we developed a phage anti-immunocomplex assay (PHAIA)1 which resulted in noncompetitive assays for several small molecules [1,2]. A limitation of the PHAIA approach was the use of relatively large amount of reagents which in this study we have addressed with the use of standard commercial magnetic beads.
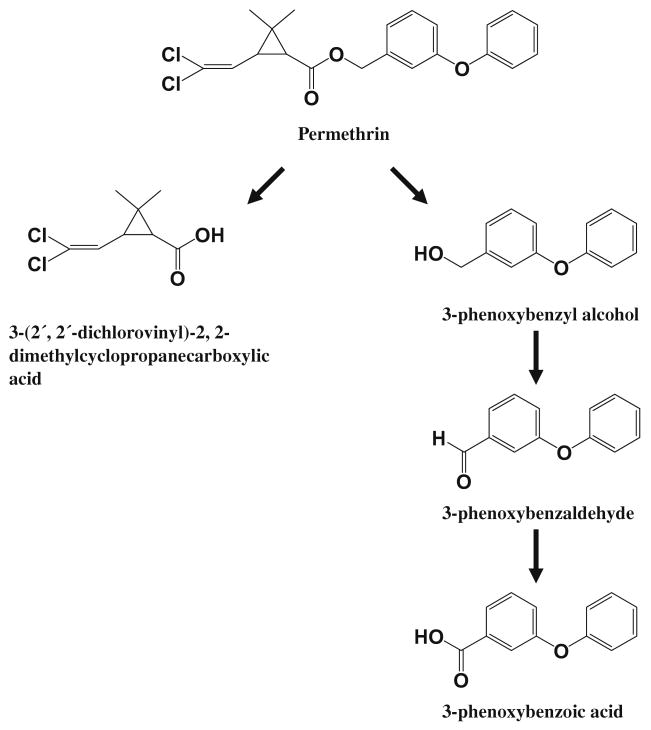
Our small molecule target to demonstrate this approach is 3-phenoxybenzoic acid (3-PBA) which is the major human metabolite of most commercial synthetic pyrethroid insecticides. The metabolism pathway of permethrin in mammals is presented in Fig. 1. Pyrethroids are hydrolyzed by esterases to 3-phenoxybenzyl alcohol or 3-phenoxybenzaldehyde which are rapidly converted to 3-PBA. Since pyrethroids were illustrated as a practical approach to insect pest control by Michael Elliott and co-workers in the 1970s they have emerged as the major agricultural insecticides in the world [3,4]. In addition treating bed nets with pyrethroids has revolutionized efforts to control malaria transmission [5,6]. Their use is being expanded by treating window curtains and other items to reduce transmission of vector-borne diseases in homes, treating recreational clothing, and treating military uniforms [7–9]. Agricultural, residential, and even personal use of these insecticides is increasing human exposure. Although the pyrethroids have a good safety record there are concerns that have arisen from animal research indicating that pyrethroid exposure may affect neurological development [10,11], induce cancer [12], suppress the immune system [13], and disrupt the endocrine system [14]. A recent study has shown that exposure in the general population is widespread [15,16]. If the pyrethroids are to be used safely in a variety of applications, techniques for rapid environmental and particularly human monitoring are needed. This is particularly true in vector control programs where human exposure is likely to be very high.
Fig. 1.
The pathway of pyrethroid metabolism in mammals.
Immunoassays have proven to be rapid, sensitive, relatively simple, and cost-effective methods. Immunoassays generally fall into two broad types, competitive or noncompetitive two-site assays. The competitive assay uses a limited amount of a specific antibody and labeled hapten (coating antigen or enzyme tracer) that competes with the analyte for the antibody binding sites. As a consequence, quantitative detection is achieved by measuring the signal produced by the unoccupied sites of the antibody, which makes it difficult to distinguish the signal generated at low concentrations of the analyte from the signal at zero concentration, limiting the assay sensitivity. On the other hand, in noncompetitive two-site assays, the presence of the analyte is directly detected by a second antibody that recognizes an independent epitope of the same molecule. Despite the many advantages of noncompetitive immunoassays, the size of a small analyte precludes detection by this method. Noncompetitive immunoassays are known to be superior to competitive ones in terms of sensitivity, precision, kinetics, and working range [17]. Furthermore, it can be easily adapted to rapid “on site” formats such as dipstick, immunochromatography, and biosensors.
To approach a noncompetitive assay for small molecules, efforts have been made to derive antibodies that can bind the analyte–antibody immunocomplex, without binding the antibody when the analyte is not present. However, due to the technical difficulty of producing secondary antibodies that specifically react with the analyte–antibody immunocomplex this approach has rarely been successful. Nevertheless, for the last decade, various attempts have been made to construct two-site assays including: (1) the anti-metatype antibody-based assay, in which the anti-metatype antibody binds to the analyte–antibody immunocomplex [18–22], and (2) the open-sandwich assay, in which separately expressed engineered recombinant variable heavy and light chains are fused to signal producing molecules to form an analyte-dependent stable association [23–25]. In order to overcome these limitations we recently reported a general method for the substitution of anti-metatype antibodies by phage-borne peptides selected from phage peptide libraries [1,2]. Selected phage peptides were capable of recognizing the conformational change of the antibody binding pocket caused upon binding to its analyte with monoclonal and polyclonal antibodies. In addition, the phage particle presents a large surface formed by more than 2700 copies of the major coat protein pVIII. This serves as a highly multivalent scaffold for binding of the anti-phage antibody-horseradish peroxidase (HRP), resulting in significant signal amplification.
The PHAIAS showed considerably improved assay sensitivity compared to those of chemically synthesized competing hapten-based ELISAs [1,2]. However, when protein A-purified polyclonal antibodies (PAbs) were used to develop the phage-based assay on the 96-well plate, rather high concentrations (5–10 μg/ml) of coating PAbs were needed to obtain the lowest assay sensitivity, which meant that approximately 50–100 μg of PAb per plate was necessary. This may present a practical limitation when large-scale use or commercial fabrication of the assay is required. In order to minimize the amount of coating antibody, we explored the use of magnetic beads as a solid-phase support for the 3-PBA PAb and the phage bearing the cyclic peptide CFNGKDWLYC as a model system.
M13 bacteriophages are large filamentous structures, about 1 μm long and 9 nm wide, and thus their binding to the immunocomplex immobilized on the surface of microtiter plates is strongly limited by diffusion or hindrance by coimmobilized nonspecific subpopulations of antibody. The microsize (3 μm in diameter) and dispersed nature of the magnetic beads will allow for a nearly “in solution” reaction optimizing the access of the phage bearing peptide to the immobilized immunocomplex. Magnetic beads are also particularly advantageous for various analytical applications due to their easy manipulation in fluids and common use in automatic detection systems. In this study we present the development and validation of a PHAIA for 3-PBA based on the use of magnetic beads.
Materials and methods
Chemicals and buffers
All reagents were of analytical grade unless otherwise specified. Mouse anti-M13 phage monoclonal antibody-HRP conjugate and desalting column (PD-10) were purchased from GE Health Care (Piscataway, NJ). Helper phage M13KO7 was purchased from New England Biolabs (Ipswich, MA). Bovine serum albumin (BSA), chicken ovalbumin (OVA), polyethylene glycol (PEG) 8000, Tween 20, 3,3′,5,5′-tetramethylbenzidine (TMB), and 3-PBA standard compound were obtained from Sigma (St. Louis, MO). Streptavidin-coated magnetic beads and magnetic extractor were purchased from Invitrogen (Carlsbad, CA). EZ-Link sulfo-NHS-LC-Biotin was purchased from Pierce (Rockford, IL). Assays were preformed with 96-well microtiter plates (Nunc-Immuno plate. MaxiSorp surface, Roskilde, Denmark). Normal strength phosphate-buffered saline (PBS) (1 × PBS; 8 g/L of NaCl, 0.2 g/L of Na2HPO4, and 0.2 g/L of KCl, pH 7.5), PBST (PBS containing 0.05% Tween 20), SOP medium (Luria broth (LB) medium containing 0.25% K2HPO4, 0.1% MgSO4, 0.1% glucose, and 100 μg/ml ampicillin), and 0.05 M citrate-acetate buffer (14.71 g/L Na3C6H5O7·2H2O, pH 5.5) were used for immunoassay. HRP substrate buffer was prepared by adding 400 μl of 0.6% TMB in dimethyl sulfoxide (DMSO) and 100 μl of 1% hydrogen peroxide (H2O2) into 25 ml of citrate-acetate buffer.
Phage preparation
ARI 292 cells with the peptide encoding vector that expressed the coat protein pIII-cyclic 8 amino acids (CFNGKDWLYC) fusion was cultured in 5 ml of LB overnight at 37 °C. Three milliliters of the overnight culture was added into a 1 L culture flask containing 500 ml SOP medium and the flask was incubated with vigorous shaking until OD600 = 0.4. After adding 1 ml of helper phage (1 × 1012 transducing units (t.u.)/ml) and 30 min of incubation at 37 °C without shaking, 5 ml of 2% arabinose and 0.5 ml of kanamycin (20 mg/ml) were added. The cells were then cultured with vigorous shaking overnight at 37 °C. The overnight culture was divided into the three 250-ml centrifuge bottles followed by centrifugation at 12,000g for 20 min. The clear supernatants were transferred into new centrifuge bottles. A 0.2 vol of 20% PEG in 2.5 M NaCl (PEG–NaCl) was added to the bottles and incubated on ice for 1 h. The bottles were centrifuged again and the phage pellets were resuspended with 150 ml of PBS. After another addition of 0.2 vol of PEG–NaCl, incubation on ice for 1 h, and centrifugation, the phage pellets were resuspended with 5 ml of suspension buffer (PBS containing 0.02% NaN3 and 1× protease inhibitor cocktail of Roche Diagnostics). This phage stock solution was filtered through a 0.22-μm membrane, aliquoted into Eppendorf tubes, and stored at −70 °C.
Checkerboard titration of phage peptide
Each of four different concentrations of biotinylated 3-PBA PAb (1, 0.5, 0.25, and 0.125 μg) was added to separate Eppendorf tubes containing 0.1 mg of beads (6–7 × 106 beads) in 100 μl of PBS. The tubes were incubated at room temperature for 30 min with gentle shaking. After washing with PBST, 300 μl of 3% skim milk in PBST was added for blocking followed by 1 h incubation at room temperature. After another washing, all of the beads in each tube were placed in an individual well of a ELISA plate. The mixture of 50 μl of each of various phage dilutions with the same volume of 20 or 0 ng/ml of 3-PBA diluted in PBST was added to the wells and the plate was incubated for 2 h at room temperature with gentle shaking. Bound phage peptides were detected as described below.
Selection of the amount of beads and phage concentration
The saturated immobilization of 3-PBA PAb onto the magnetic beads was carried out by incubating 5 μg of biotin conjugated 3-PBA PAb with 1 mg streptavidin-coated magnetic beads (6–7 × 107 beads, average diameter of 3 μm) for 30 min at room temperature with gentle shaking as suggested by the supplier's instruction manual. The beads were then washed three times with PBST and finally resuspended with 500 μl PBS containing 0.1% BSA and 0.02% NaN3. Bead preparations (5 or 10 μg) were placed in the wells of microtiter plates and then serial dilutions of phage stock solution were added with 1 ng/ml of 3-PBA for maximal signals, or without 3-PBA for the background signals.
Magnetic bead-based PHAIA
Five microliters of the bead preparation (10 μg beads) was placed in each well of the 96-well plate. One hundred microliters of the mixture of phage solution (2.5 × 108 phage particles, 1:4000 dilution in PBST) with various concentrations of 3-PBA standard was added and the plate was incubated for 2 h with shaking on a plate orbital shaker. The plate was held on a magnetic separator while the wells were washed three times with PBST. Anti-phage antibody MAb-HRP conjugate (1:40,000 dilution in PBST) was added and the plate was incubated for 1 h with shaking. The plate was again held on a magnetic separator and washed with PBST three times. TMB solution was added and the plate was incubated for 15 min and the beads were drawn to the corner of the wells by the aid of the 96-well plate magnet. Fifty microliters of 4 N sulfuric acid was added and the plate was read at 450 nm in a plate reader (Molecular Devices, Sunnyvale, CA).
Effect of blocking reagents
Three common blocking reagents (BSA, nonfat dried skim milk, and OVA, 3% in PBST) were tested. Five hundred microliters of each blocking solution was added to the Eppendorf tubes containing an appropriate amount of antibody-coated beads and then incubated for 1 h by gentle shaking. The 96-well plate was also blocked with 300 μl of the same blocking buffer covering each well. Ten micrograms of beads was placed in the wells of the plate and then the phage (2.5 × 108 phage particles, 1:4000 dilution in PBST) with 0.5 ng/ml of 3-PBA or without 3-PBA was added. The rest of assay procedures were as described above.
Effect of methanol (MeOH) and urine matrix
The effects of MeOH and urine matrix on assay performance were estimated by observing the signal trend in a sample containing the same concentration of 3-PBA with increasing concentrations of MeOH or urine in PBST buffer. MeOH was added to the PBST buffer to final concentrations of 0, 10, 20, and 40%. 3-PBA standard was spiked to each preparation at 0, 0.5, and 1 ng/ml. For evaluation of its potential interference, urine was collected from donors that had no known exposure to pyrethroid insecticides. Prior to use, urine samples were filtered through 0.22-μm membranes and then diluted with PBST to 0, 10, 20, or 40%. For each urine dilution, 3-PBA standard was added to a final concentration of 0, 0.4, and 1 ng/ml. Fifty microliters of these solutions was mixed with 50 μl of phage solution and then added to the well containing the antibody-immobilized beads. Assay was performed as described above.
Determination of inter and intraassay variation
Urine sample was spiked with 3-PBA at the concentrations of 5, 10, and 20 ng/ml and each sample was diluted 25-fold with PBST. Assay procedures were as described above. Interassay variation was determined by performing assays on seven different days with four replicates of each concentration. Intraassay variation was determined by a single assay using 16 replicates for each concentration.
Validation of PHAIA
Three urine samples were collected from donors with no known exposure and kept at −15 °C prior to analysis. For the validation, the frozen urine sample was thawed at room temperature. The thawed sample was shaken vigorously for 30 s to completely homogenize the sample. The sample was allowed to stand for at least 2 h. A 0.5-ml aliquot of the clear supernatant was added into a 4-ml tube. 3-PBA was spiked into the urine sample. For application of the 3-PBA analysis in urine, typically the urine samples are hydrolyzed prior to analysis to release conjugated 3-PBA. To mimic this procedure, the urine samples were hydrolyzed by adding 100 μl of 6 N hydrochloric acid and heating at 100 °C for 1 h. The hydrolyzed urine was neutralized by adding 100 μl of 6 N sodium hydroxide. Sodium acetate buffer (0.2 M; pH4.5; 1 ml) was added to the tube and mixed thoroughly. To reducing the matrix effect of the hydrolyzed urine, a solid-phase extraction (SPE) method was slightly modified from the method of Ahn et al. [26]. That is, the diluted urine samples were loaded onto the mixed-mode SPE column (100 mg, Strata Screen-A, Phenomenex, Torrance, CA) which was preequilibrated with 1 ml each of methanol, water, and the buffer. The column was sequentially washed with 1 ml each of water and methanol. The column was then dried under high vacuum (10″ Hg) for 5 min. The 3-PBA was finally eluted with 1.5 ml of 1% acetic acid in the mixture of ethyl acetate:hexane (30:70, v/v). The eluate was evaporated to dryness using a centrifugal vacuum concentrator (Speed Vac, Laurel, MD) and dissolved with 2.5 ml PBS containing 10% methanol.
Results
Checkerboard titration of phage peptide
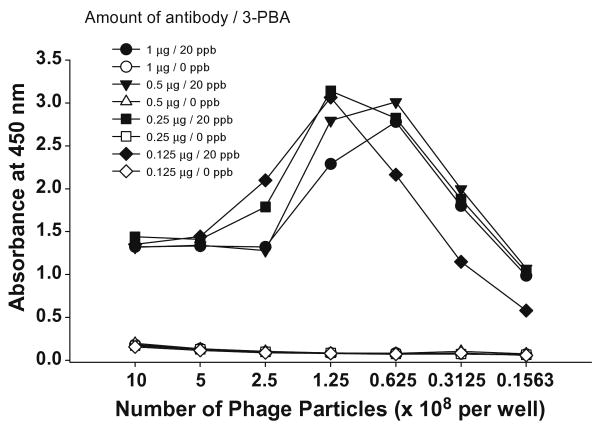
Since a major aim of the study was to reduce the amount of antibody for the practical application of this assay to a large number of samples we first studied this feature by titering the amount of phage peptide. Using a checkerboard, we used a fix amount of beads (0.1 mg) and analyte (20 or 0 ng 3-PBA/ml), and then used different concentrations of antibody in combination with different number of phage particles. As shown in Fig. 2, in the combination of 1.25 × 108 phage particles (1:8000 dilution of phage stock) with 0.125 μg antibody, the signal was similar to those in combinations with higher concentrations of antibody showing signals ranging from 2.8 to 3.1. A slightly lower signal for 1 μg of antibody resulted from the precipitation of enzymatic products. When the number of phage particles exceeded 1.25 × 108 significant interference occurred due to precipitation of enzymatic products. These results indicate that compared to the antibody concentration used for the 96-well plate-based assay, 10-fold less amount of PAb could be used for phage peptide assay development.
Fig. 2.
Checkerboard titration of phage peptide. Each of four different concentrations of protein A-purified 3-PBA PAb conjugated with biotin was immobilized on 0.1 mg of streptavidin-coated magnetic beads. Various numbers of phage peptide diluted in PBST were added with 20 or 0 ng/ml of 3-PBA standard.
Selection of the amount of beads and phage concentration
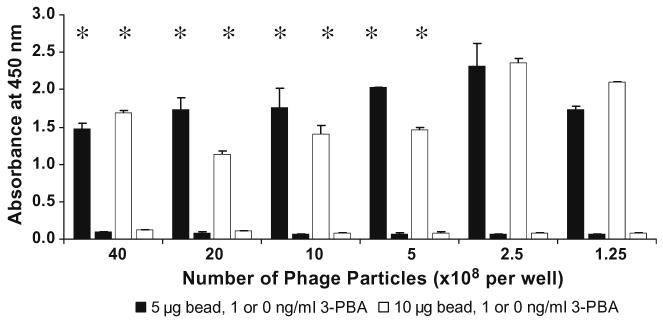
Because PHAIA is a noncompetitive two-site assay, it is necessary to determine the highest concentration of phage that can saturate the antibody immobilized on a given number of beads. Since each phage particle serves as a multiple binding scaffold (more than 2700 pVIII major coat proteins), precipitation of substrate products by excess binding of HRP-conjugated secondary antibody should be avoided. An excessive number of phage particles can also cause a high nonspecific background signal. To optimize the concentrations of phage and determine the minimal number of beads giving an appropriate difference between background and maximal signals, the assay was further optimized by titering the phage peptide at a fixed concentration of secondary antibody (1:40,000 dilution), adding serial dilutions of phage to the wells containing 5 or 10 μg of bead preparation (equivalent to 0.1 and 0.2 μg PAb, respectively) with or without 3-PBA (1.0 ng/ml). As shown in Fig. 3, when the number of phage particles was more than 5 × 108 phage per well (1:2000 dilution) the precipitation of substrate products occurred at both bead concentrations, but at the concentrations lower than 2.5 × 108 phages per well (1:4000 dilution), no precipitation was observed. Throughout this study, 10 μg of bead preparation and 1:4000 dilution of phage were used to assure the binding of coating antibody by a maximal number phage without producing precipitation of substrate.
Fig. 3.
Optimization of phage concentration. Five or 10 μg of the bead preparation was used. Serial dilutions of phage were added with or without 3-PBA. Bound phage was detected with 1:40,000 dilution of HRP-conjugated secondary antibody. Star markers indicate the precipitation of substrate product. Each bar represents the mean value of four replicates. Short bars represent the signals at no 3-PBA added.
Effect of blocking buffer
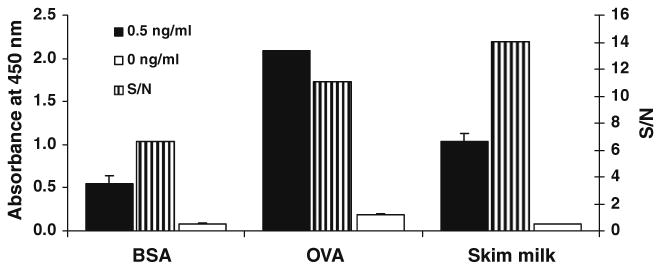
BSA, OVA, and skimmed milk were assayed as blocking agents and signal/noise (S/N) revealed that the latter reagent produced the highest signal to noise ratio (Fig. 4).
Fig. 4.
Blocking efficiency. For each blocking reagent (3% each in PBST), the left and right bars indicate the maximal signal with 0.5 ng/ml 3-PBA added and the background signal without analyte, respectively. The center bar represents the S/N value. Each value represents the mean of five replicates.
Magnetic bead-based PHAIA
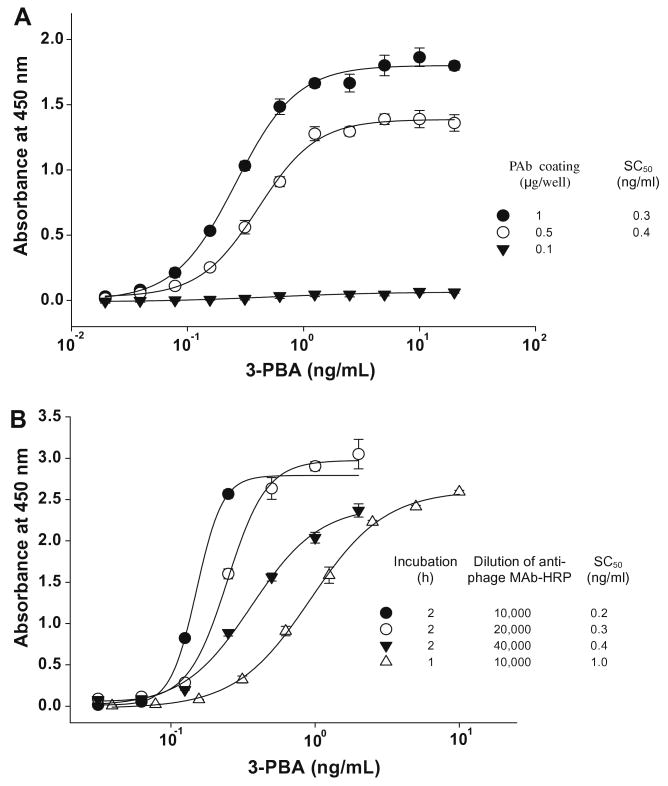
Fig. 5 shows the scheme of bead-based PHAIA. We previously selected the phage peptide used for this study from the phage display library using a well-established panning method [2]. This phage peptide is capable of distinguishing the free antibody binding pocket from the pocket–analyte complex. Thus, the phage peptide binds to the antibody predominantly in the presence of analyte. The PHAIA on the 96-well plate showed that 50% of a saturating concentration (SC50) of 0.3 ng/ml was 400-fold better in sensitivity than the chemical competing hapten-based homologous ELISA previously reported. Fig. 6A shows standard curves obtained from the 96-well plate-based PHAIA. SC50s of 0.3 and 0.4 ng/ml were observed with the plates coated with 1.0 and 0.5 μg of PAb/well, respectively. However, when the coating PAb concentration was reduced to 0.1 μg/well, concentration-dependent signals were not obtained. Although the antibody-coated plate PHAIA was very sensitive, the fact that the plate needed to be coated with relatively high concentrations of PAb could limit an application of this assay to high production. To overcome this drawback, we explored the use of magnetic beads as a solid support for antibody immobilization. The large surface area of the magnetic beads might reduce steric hindrance of the phage access to antibody binding sites requiring less amount of antibody, while still maintaining the same sensitivity. Biotinylated 3-PBA PAb was immobilized on streptavidin-coated magnetic beads. An aliquot of antibody-immobilized beads was placed in the wells of the 96-well ELISA plate. Addition of phage and analyte to the wells allows phage to bind to the antibody in the proportion to the concentration of analyte. Bound phages were detected with HRP-labeled anti-M13 phage antibody. During the incubation step, settling of the beads was prevented by shaking the plate with an orbital plate shaker. Bead washing was carried out four times with 100 μl of PBST by tapping the plate several times on the paper towel immediately after dumping washing buffer while holding the plate on the magnet. The magnetic extractor utilized for this study was strong enough to hold all bead particles during the washing and drying procedure. To avoid light scattering by suspended bead particles, beads were drawn to the corner of wells before adding the stop solution. We observed no difference in signal read-out between the plate with bead particles drawn to the corner of wells and the new plate with final solution transferred (data not shown). Fig. 6B shows the dose-dependent signal curves obtained for the magnetic bead-based assay. SC50 values and the steep slope of the linear range of the curves were slightly affected by the incubation time and the concentration of secondary antibody. With 2 h incubation, similar SC50 values (0.2–0.4 ng/ml) were observed, but the slope of the curve was gradually decreased giving a more extended dynamic range from 0.1–0.2 to 0.1–1 ng/ml as the dilution of secondary antibody increased from 1:10000 to 40000. At 1:10,000 dilution of secondary antibody, precipitation of substrate product was produced at high concentration of analyte. With 1 h incubation and 1:10,000 dilution of secondary antibody, the SC50 was increased to 1 ng/ml giving a detectable range of 0.3–2.5 ng/ml. These data indicate that the bead-based PHAIA gives an identical sensitivity to the 96-well-based PHAIA requiring 10- to 20-fold less amount of antibody. Furthermore, the sensitivity and detectable linear range could be tuned by adjusting the combination of incubation time and the concentration of secondary antibody.
Fig. 5.
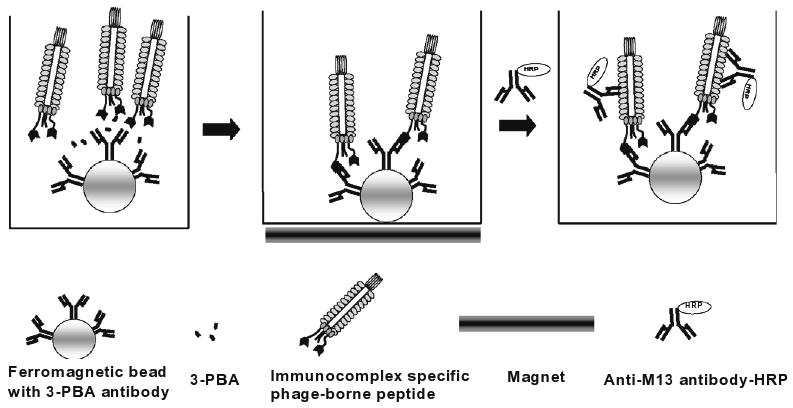
Scheme of magnetic bead-based PHAIA. Magnetic beads coated with antibody are placed in the wells of microtiter plates. A constant amount of phage solution and various concentrations of 3-PBA standard solutions are added into the wells. Unbound phage and analyte are washed away with the aid of a magnetic extractor. Bound phages are detected with anti-phage MAb-HRP.
Fig. 6.
PHAIA for 3-PBA using antibodies immobilized on ELISA plates (A) or on magnetic beads (B) as solid support. The 96-well plate directly coated with PAb was incubated for 2 h at room temperature after addition of the mixture of 3-PBA standards and phage solution.
Effect of MeOH and urine matrix
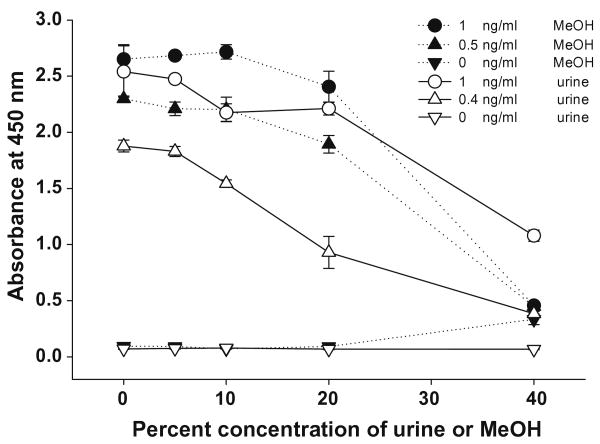
An estimation of the assay tolerance to solvent or matrix is necessary for an accurate detection of unknown samples when samples are prepared by solvent extraction or dilution of crude samples. The effects of MeOH and urine matrix were determined by reading signals at a given concentration of 3-PBA spiked into samples with increasing concentration of urine or MeOH (Fig. 7). The signals were reduced by approximately 15% at 20% MeOH at both concentrations of 3-PBA. However, negligible changes in signals were observed up to 10% MeOH, indicating that unknown samples containing 10% MeOH can be used by the assay with no matrix effects. For urine, there was a 20% decrease in signal at 10% urine with 0.4 ng/ml 3-PBA, indicating that 10% urine could affect the linear range. Therefore, unknown urine samples should be diluted by more than 10-fold or if prepared by SPE, by dissolving the eluate with buffer to <20% MeOH.
Fig. 7.
The effect of MeOH and urine matrix in assay buffer. Three different concentrations of 3-PBA standard in each concentration of MeOH or urine solution were mixed with the same volume of phage solution (2.55 × 108 phage particles, 1:4000 dilution in PBST) followed by 2 h incubation on the orbital plate shaker. One hundred microliters of anti-phage MAb-HRP solution in PBST (1:40,000 dilution) was added and incubated for 1 h. Each value represents the mean value of four replicates.
Inter and intraassay variations and assay validation
Urine samples collected from people who had no known exposure to pyrethroid insecticides were used for the experiments. For the determination of inter and intraassay variations, one urine sample spiked with three different concentrations was diluted 25-fold with PBST to minimize potential matrix effects. The results are shown in Table 1. The recoveries are generally within 91–104% of the spiked concentrations with coefficients of variation of 16% or less. The samples for assay validation were spiked with 3-PBA, then prepared by simple dilution of crude urine with PBST or hydrolysis followed by SPE clean-up. Simple dilution of samples is suitable for detection in urine containing high concentrations of 3-PBA. Since 3-PBA is known to exist as different conjugates such as glycine or glucuronide, hydrolysis is necessary to form the free 3-PBA. The utilized SPE method for urine clean-up was established and validated by LC-MS analysis in this laboratory (unpublished data). As shown in Table 2, for both methods of sample preparation, general recoveries ranged from 80 to 120%. This result demonstrated that the bead-based PHAIA could be a useful assay to accurately detect 3-PBA in urine.
Table 1.
Inter and intraassay variations of PHAIA.
| Fortification level (ng/ml) | aInterassay variation | bIntraassay variation | ||||||
|---|---|---|---|---|---|---|---|---|
| N | Measured (ng/ml) | Average recovery (%) | CV (%) | N | Measured (ng/ml) | Average recovery (%) | CV (%) | |
| 5 | 7 | 4.9 ± 0.4 | 98 | 8.2 | 16 | 4.9 ± 0.3 | 98 | 6.1 |
| 10 | 7 | 10.3 ± 1.6 | 103 | 15.5 | 16 | 10.4 ± 0.6 | 104 | 5.8 |
| 20 | 7 | 18.1 ± 1.7 | 91 | 9.4 | 16 | 20.2 ± 1.5 | 101 | 7.4 |
Interassay variation was determined by four replicates on seven different days.
Intraassay variation was determined by 16 replicates on a single day.
Table 2.
Assay validation: 6 different concentrations of 3-PBA were spiked to 3 different urine samples diluted with PBST and two samples prepared by SPE clean-up.
| Spiked (ng/ml) | Urine A | Urine B | Urine C | Average recovery (%) |
|---|---|---|---|---|
| Sample prepared by dilution | ||||
| 0 | <0.1 | <0.1 | <0.08 | |
| 5 | 5.9 | 4.2 | 3.1 | 101 |
| 10 | 12.2 | 8.1 | 10.0 | 92 |
| 20 | 21.4 | 18.0 | 15.8 | 97 |
| 50 | 56.2 | 40.3 | 49.0 | 97 |
| 100 | 96.5 | 90.2 | 105.7 | 88 |
| Sample prepared by SPE | ||||
| 0 | N.D | <0.2 | aND | |
| 3 | 2.5 | 3.2 | ND | 95 |
| 5 | 4.7 | 4 | ND | 87 |
| 10 | 10 | 11.7 | ND | 109 |
| 50 | 52.6 | 54.3 | ND | 107 |
Not determined.
Conclusion
In the current study, we described the development of a sensitive magnetic bead-based PHAIA for the detection of a human urinary biomarker of pyrethroid exposure, 3-PBA. This method used significantly less PAb than required for the 96-well plate-based assay. PHAIA is a sandwich-type two-site immunoassay initially developed for the detection of small molecular weight compounds in which the phage-borne peptide binds to the analyte–antibody complex. In order to circumvent the drawback of the 96-well-based PHAIA which requires a large amount of PAb for coating, we used micrometer-sized magnetic beads as a solid support with the assumption that the large surface area of beads likely decreased steric hindrance of mega Dalton-sized phage particles to the 3-PBA-specific subpopulation of PAb. The checkerboard titration test showed that a 10-fold lower amount of PAb compared to the microplate assay was acceptable because approximately 0.1 μg of antibody immobilized on 0.1 mg of beads showed a significant difference between in the presence and in the absence of analyte. Further experiments indicated that the amount of beads could also be significantly reduced showing that the use of 5 or 10 μg of beads gave a similar difference in signals observed from the checkerboard titration. The final concentrations of antibody and beads were 0.1 and 5 μg, respectively. The standard curve generated using 5 μg of beads immobilized with biotinylated 3-PBA PAb not only significantly reduced the amount of coating antibody but also gave identical sensitivity in terms of SC50 value (0.2–0.3 ng/ml) to that of the assay with PAb directly coated on the wells of the plate at 1.0 μg per well. In estimation of blocking efficiency with three common blocking agents (3% of BSA, nonfat dried skim milk, OVA), 3% nonfat dried skim milk showed the most effective blocking giving the highest S/N value. For an application of this assay to unknown urine samples, crude urine samples need to be diluted at least the 10 times with assay buffer because the assay was tolerant to 5% final concentrations of urine matrix. Since the assay tolerance to MeOH was up to 10% final concentration, the concentration of MeOH is allowable up to 20% during sample preparation. The estimation of inter and intraassay variation with 25-fold diluted urine showed no significant variations with general recoveries of 90–105 and 16% of coefficients of variation or less when assays were performed over seven different days. When this assay was used for the detection of 3-PBA in urine samples, we observed a good recovery of spiked 3-PBA with overall recoveries of 80–120% for two different types of urine samples prepared by direct dilution of urine with PBST or urine clean-up with a SPE method. In addition, because of the demand in developing the sensing technologies for multiplexed, reliable, in-field measurements beads have gained popularity as material for a variety of formats of assays [27–29]. This bead-based PHAIA could be easily extended to various high throughputs or multiplexing assay systems. Such systems could utilize the versatility of the phage particle such as PCR-amplified detection of phage gene [30] or the direct multiple binding of fluorophores for the nanoparticle-based fluoroimmunoassay or the microchannel-based immunoassays which reduce the sample volume and speed assay procedures [31–33].
Acknowledgments
This work was supported in part by the following: The NIEHS Superfund Basic Research Program, P42 ES004699; the National Institutes of Environmental Health Sciences R37 ES002710. The support of the Western Center for Agricultural Health and Safety at the University of California Davis, PHS OH07550 is also acknowledged.
Footnotes
Abbreviations used: HRP, horseradish peroxidase; MeOH, methanol; OVA, ovalbumin; PAbs, polyclonal antibodies; PBS, phosphate-buffered saline; PEG, polyethylene glycol; PHAIA, phage anti-immunocomplex assay; 3-PBA, 3-phenoxybenzoic acid; PBST, PBS containing 0.05% Tween 20; TMB, 3,3′,5,5′-tetramethylbenzidine.
References
- 1.González-Techera A, Vanrell L, Last JA, Hammock BD, González-Sapienza G. Phage anti-immune complex assay: general strategy for noncompetitive immunodetection of small molecules. Anal Chem. 2007;79:7799–7806. doi: 10.1021/ac071323h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González-Techera A, Kim HJ, Gee SJ, Last JA, Hammock BD, González-Sapienza G. Polyclonal antibody-based noncompetitive immunoassay for small analytes developed with short peptide loops isolated from phage libraries. Anal Chem. 2007;79:9191–9196. doi: 10.1021/ac7016713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott M, Farnham AW, Janes NF, Needham PH, Pulman DA. Potent pyrethroid insecticides from modified cyclopropane acids. Nature. 1973;244:456–457. doi: 10.1038/244456a0. [DOI] [PubMed] [Google Scholar]
- 4.Elliott M. Properties and applications of pyrethroids. Environ Health Perspect. 1976;14:3–13. doi: 10.1289/ehp.76141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyimo EO, Msuya FHM, Rwegoshora RT, Nicholson EA, Onzava AEP, Lines JD, Curtis CF. Trial of pyrethroid impregnated bednets in an area of Tanzania holoendemic for malaria. Part 3. Effects on the prevalence of malaria parasitaemia and fever. Acta Trop. 1991;49:157–163. doi: 10.1016/0001-706x(91)90034-h. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay SW, Adiamah JH, Miller JE, Armstrong JRM. Pyrethroid-treated bednet effects on mosquitoes of the Anopheles gambiae complex in The Gambia. Med Vet Entomol. 1991;5:477–483. doi: 10.1111/j.1365-2915.1991.tb00576.x. [DOI] [PubMed] [Google Scholar]
- 7.Elnaiem DA, Aboud MA, El Mubarek SG, Hassan HK, Ward RD. Impact of pyrethroid-impregnated curtains on Phlebotomus paratosi sandflies indoors at Khartoum, Sudan. Med Vet Entomol. 1999;13:191–197. doi: 10.1046/j.1365-2915.1999.00183.x. [DOI] [PubMed] [Google Scholar]
- 8.Demirel N, Cranshaw W. Permethrin treatment of pollination bags as a protecting from false chinch bug, Nysius raphanus (Howard) (Hemiptera: Lygaeidae), Injury to canola in Colorado. Crop Prot. 2006;25:1062–1064. [Google Scholar]
- 9.Abdel-mohdy FA, Moustafa MG, Fouda MF, Aly Rehan AS. Repellency of controlled-release treated cotton fabrics based on cypermethrin and prallethrin. Carbohydr Polym. 2008;73:92–97. [Google Scholar]
- 10.Kolaczinski JH, Curtis CF. Chronic illness as a result of low-level exposure to synthetic pyrethroid insecticides: a review of the debate. Food Chem Toxicol. 2004;42:697–706. doi: 10.1016/j.fct.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Shafer TJ, Meyer DA, Croffon KM. Developmental neurotoxicity of pyrethroid insecticides: critical review and future research needs. Environ Health Perspect. 2005;113:123–136. doi: 10.1289/ehp.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.U.S.EPA. Permethrin Facts. U.S. Environmental Protection Agency. [July 10, 2006];2006 Available from: < http://www.epa.gov/oppsrrd1/REDs/factsheets/permethrin-fs.htm>.
- 13.Repetto RC. In: Pesticides and the Immune System: The Public Health Risk. Repetto R, Baliga SS, editors. WRI (World Resources Institute), National Center for Food & Agricultural Policy; Washington, DC: 1996. [Google Scholar]
- 14.Go V, Garey J, Wolff MS, Pogo BG. Estrogenic potential of certain pyrethroid compounds in the MCF-7 human breast carcinoma cell line. Environ Health Perspect. 1999;107:173–177. doi: 10.1289/ehp.99107173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC. Department of Health and Human Services, Centers for Disease Control and Prevention; Atlanta, GA: 2005. Third National Report on Human Exposure to Environmental Chemicals. Available from: < http://www.cdc.gov/exposurereport/pdf/results_12.pdf>. [Google Scholar]
- 16.Fortin MC, Bouchard M, Carrier G, Dumas P. Biological monitoring of exposure to pyrethrins and pyrethroids in a metropolitan population of the Province of Quebec, Canada. Environ Res. 2008;107:343–350. doi: 10.1016/j.envres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Jackson TM, Ekins RP. Theoretical limitations on immunoassay sensitivity. Current practice and potential advantages of fluorescent Eu3+ chelates as non-radioisotopic tracers. J Immunol Methods. 1986;87:13–20. doi: 10.1016/0022-1759(86)90338-8. [DOI] [PubMed] [Google Scholar]
- 18.Voss EW., Jr Anti-metatype antibody reactivity: a model for T-cell receptor recognition. Immunol Today. 1990;11:355–357. doi: 10.1016/0167-5699(90)90140-5. [DOI] [PubMed] [Google Scholar]
- 19.Ullman EF, Milburn G, Jelesko J, Radika K, Pirio M, Kempe T, Skold C. Anti-immune complex antibodies enhance affinity and specificity of primary antibodies. Proc Natl Acad Sci USA. 1993;90:1184–1189. doi: 10.1073/pnas.90.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towbin H, Motz J, Oroszlan P, Zingel O. Sandwich immunoassay for the hapten angiotensin II. A novel assay principle based on antibodies against immune complexes. J Immunol Methods. 1995;181:167–176. doi: 10.1016/0022-1759(94)00343-u. [DOI] [PubMed] [Google Scholar]
- 21.Voss EW, JR, Mummert ME. Anti-metatype antibodies in immunoassays. Mikrochim Acta. 1997;126:193–202. [Google Scholar]
- 22.Nagata S, Tsutsumi T, Yoshida F, Ueno Y. A new type sandwich immunoassay for microcystin: production of monoclonal antibodies specific to the immune complex formed by microcystin and anti-microcystin monoclonal antibodies. Nat Toxins. 1999;7:49–55. [PubMed] [Google Scholar]
- 23.Suzuki C, Ueda H, Mahoney W, Nagamune T. Open sandwich enzyme-linked immunosorbent assay for the quantitation of small haptens. Anal Biochem. 2000;286:238–246. doi: 10.1006/abio.2000.4800. [DOI] [PubMed] [Google Scholar]
- 24.Ueda H, Yokozeki T, Arai R, Tsumoto K, Kumagai I, Nagamune T. An optimized homogeneous noncompetitive immunoassay based on the antigen-driven enzymatic complementation. J Immunol Methods. 2003;279:209–218. doi: 10.1016/s0022-1759(03)00256-4. [DOI] [PubMed] [Google Scholar]
- 25.Lim SL, Ichinose H, Shinoda T, Ueda H. Noncompetitive detection of low molecular weight peptides by open sandwich immunoassay. Anal Chem. 2007;79:6193–6200. doi: 10.1021/ac070653z. [DOI] [PubMed] [Google Scholar]
- 26.Ahn KC, Lohstroh P, Gee SJ, Gee NA, Lasley B, Hammock BD. High-throughput automated luminescent magnetic particle-based immunoassay to monitor human exposure to pyrethroid insecticides. Anal Chem. 2007;79:8883–8890. doi: 10.1021/ac070675l. [DOI] [PubMed] [Google Scholar]
- 27.Aytur T, Foley J, Anwar M, Boser B, Harris E, Beatty PR. A novel magnetic bead bioassay platform using a microchip-based sensor for infectious disease diagnosis. J Immunol Methods. 2006;314:21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Riegger L, Grumann M, Nann T, Riegler J, Ehlert O, Bessler W, Mittenbuehler K, Urban G, Pastewka L, Brenner T, Zengerle R, Ducrée J. Read-out concepts for multiplexed bead-based fluorescence immunoassays on centrifugal microfluidic platforms. Sens Actuators A. 2006;126:455–462. [Google Scholar]
- 29.Dosev D, Nichkova M, Dumas R, Gee SJ, Hammock BD, Liu K, Kennedy I. Spray pyrolysis synthesis of particles possessing magnetic and luminescent properties, application of magnetic/luminescent particles in immunoassays. Nanotechnology. 2007;18:055102. doi: 10.1088/0957-4484/18/5/055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo YC, Zhou YF, Zhang XE, Zhang ZP, Qiao YM, Bi LJ, Wen JK, Liang MF, Zhang JB. Phage display mediated immuno-PCR. Nucleic Acids Res. 2006;34:e62. doi: 10.1093/nar/gkl260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichkova M, Dosev D, Gee SJ, Hammock BD, Kennedy I. Microarray immunoassay for phenoxybenzoic acid using polymer encapsulated Eu:Gd2O3 nanoparticles as fluorescent labels. Anal Chem. 2005;77:6864–6873. doi: 10.1021/ac050826p. [DOI] [PubMed] [Google Scholar]
- 32.Kim KS, Park JK. Magnetic force-based multiplexed immunoassay using superparamagnetic nanoparticles in microfludic channel. Lab Chip. 2005;5:657–664. doi: 10.1039/b502225h. [DOI] [PubMed] [Google Scholar]
- 33.Nichkova M, Dosi D, Gee SJ, Kennedy IM, Hammock BD. Multiplexed immunoassays for proteins using magnetic luminescent nanoparticles for internal calibration. Anal Biochem. 2007;369:8471–8475. doi: 10.1016/j.ab.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]