Abstract
Elevated level of homocysteine (Hcy) called hyperhomocysteinemia (HHcy) is one of the major risk factors for chronic heart failure. Although the role of Hcy in cardiac remodeling is documented, the regulatory mechanism involved therein is still nebulous. MicroRNAs (miRNAs) and dicer have been implicated in regulation of cardiovascular diseases. Dicer is the only known enzyme involved in miRNA maturation. We investigated the involvement of dicer and miRNA in Hcy-induced cardiac remodeling. HL-1 cardiomyocytes were cultured in different doses of Hcy. Total RNA was isolated and RT-PCR and real-time PCR was performed for dicer, MMP-2,-9, TIMP-1,-3, and NOX-4. MiRNA microarray was used for analyzing the differential expression of miRNAs. Individual miRNA assay was also done. Western blotting was used to assess the MMP-9 expression in HHcy cardiomyocytes. The RT-PCR results suggest that dicer expression is enhanced in HHcy cardiomyocytes suggesting its involvement in cardiac remodeling caused due to high dose of Hcy. On the other hand, high dose of Hcy increased NOX-4 expression, a marker for oxidative stress. Additionally, HHcy cardiomyocytes showed elevated levels of MMP-2,-9 and TIMP-1,-3, and reduced expression of TIMP-4, suggesting cardiac remodeling due to oxidative stress. The miRNA microarray assay revealed differential expression of 11 miRNAs and among them miR-188 show dramatic downregulation. These findings suggest that dicer and miRNAs especially miR-188 are involved in Hcy-induced cardiac remodeling.
Keywords: MicroRNA, Dicer, Hyperhomocysteinemia, Congestive heart failure, Extracellular matrix remodeling, Matrix-metalloproteinase (MMP), Tissue inhibitors of metalloproteinase (TIMP)
Introduction
Homocysteine (Hcy) is a thiol-containing, non-protein forming amino acid, which is present in blood plasma at the concentration of less than 10–12 μM/l [1]. Elevated level of Hcy is called hyperhomocysteinemia (HHcy). Depending on the plasma concentration, HHcy is classified into moderate (12–30 μM/l), intermediate (>30–100 μM/l), and severe (>100 μM/l) [1]. Although there is controversy over the fact that whether Hcy is a cause or consequence [2], there is consensus that HHcy is associated with several pathological conditions like cardiovascular diseases (CVD), diabetes, neurodegenerative diseases, osteoporosis, and birth defects [1, 3, 4]. One of the plausible reasons behind its involvement in these diseases is its relationship with oxidative stress, a very basic biological system central to the variety of pathologies [5]. The moderate HHcy is nowadays considered as an independent risk factor for CVD, and is responsible for approximately 10% of total risks [1, 4, 6]. Although the clinical and experimental results confirmed the association of Hcy in chronic heart failure (CHF) [7, 8], the mechanism underlying it is largely unknown. Experimental studies suggest HHcy as a causative factor for adverse cardiac remodeling leading to interstitial and perivascular fibrosis, and causes myocardial stiffness that ultimately results into CHF [9–12]. Previous and recent investigations from our laboratory also shed light over the mechanisms of Hcy-induced cardiac remodeling [13–16]. Nevertheless, the direct link between Hcy and CHF is still missing and needs further investigations.
Recently, deep insight into CVD revealed that microRNAs (miRNAs or miRs) play pivotal role in regulation of genes involved in cardiac remodeling [17–23]. MiRNAs are a class of endogenous, conserved and non-coding RNA molecules of ~22 nucleotides, which negatively modulate gene expression in plants and animals, primarily through base pairing to 3′ untranslated region of target mRNAs leading to mRNA cleavage and/or translation repression [24]. The binding specificity of miRNA to its target is presumably dictated by 6–7 nt sequence from 5′ region of miRNA, called “seed” sequence [25], which nucleates binding to target mRNA, and allows more 3′ region of miRNA for subsequent zippers up with the target mRNA [26]. The complementarity of seed sequence of miRNA to the target mRNA may determine the degradation of mRNA or translational inhibition [25]. MiRNAs can be derived from individual miRNA genes, introns of protein coding genes, or polycistronic transcripts [22]. They are transcribed by RNA polymerase II or III into primary miRNA (pri-miRNA), which are several kilobases long, capped (MGpppG), and polyadenylated [27]. The pri-miRNAs are processed in nucleus by RNase III enzyme drosha and the dsRNA binding protein pasha (also known as DGCR8), into a hairpin-shaped structure of 70–100 nt called preliminary-microRNA (pre-microRNA). The pre-microRNAs are transported from nucleus to the cytoplasm by RAN-GTP and exportin 5 transport [18], where they are processed by another RNase III enzyme, dicer. Dicer process pre-microRNA into a transient ~18–24 nt duplex, which is loaded into the miRNA-associated multi-protein RNA-induced silencing complex that includes the Argonoute proteins [18, 28]. Out of the two strands, one strand of miRNA duplex is preferentially retained in the complex that finally becomes the mature miRNA, whereas the other strand also called passenger strand or miRNA* is degraded and eliminated from the complex [18, 28]. Some intronic miRNA precursors however bypass drosha processing to produce miRNA by dicer, possibly representing an alternative pathway for miRNA biogenesis [29].
In human genome nearly thousands of miRNAs are encoded. They are estimated to fine tune the expression of nearly 30% of mRNA transcripts [30], and thereby regulating almost all cellular functions. Recent investigations revealed its role in several pathological conditions [18], indicating its great potential for therapeutics. Investigations on miRNAs thus opened a new era of genomics called microRNomics [31].
The fact that dicer is the only known enzyme involved in maturation of all miRNAs has been established in various cell types including embryonic stem cells [32], Germline cells [33] as well as specialized cell types, such as pancreatic islet cells [34], immune cells [35], neural cells [36], and endothelial cells [37]. Dicer plays key roles in cardiomyopathy [38], angiogenesis [23], and endothelial cell function [37, 39] in a spatio-temporal manner. It has also been implicated in dilated cardiomyopathy and post-natal lethality [39–41].
In heart, matrix-metalloproteinases (MMPs) maintain the balance of elastin and collagen ratio of extracellular matrix. In healthy condition, they remain in latent state. However, they are activated in stress condition causing misbalance of elastin/collagen ratio leading to fibrosis that culminates into heart failure [39, 42, 43]. The major role of tissue inhibitors of metalloproteinase (TIMPs) is to ameliorate the effect of MMPs in cardiac remodeling [44]. NADP oxidase (NOX) expression level is an indicator of oxidative stress; upregulation reflects higher oxidative stress and vice versa.
This study addresses two questions: (1) whether dicer and miRNA are involved in cardiac remodeling in HHcy cardiomyocytes and (2) implications of HHcy on miRNA biogenesis that is illustrated by differential expression of dicer.
Materials and Methods
Cell Culture and Treatment Groups
All the experiments were performed on HL-1 cell line, which were produced from murine cardiomyocytes and show all the properties of cardiomyocytes including contractility [45]. HL-1 cells were cultured in a special medium called Claycomb medium (JRH Biosciences, catalog #51800C, Lenexa, KS, USA), which is supplemented with 10% fetal bovine serum (Sigma-Aldrich, catalog #F2442, Saint Louis, MO, USA), 1% L-glutamine (Life Technologies, catalog #25030-081, Foster City, CA, USA), 1% norepineprine (Sigma, catalog # A-0937, Saint Louis, MO, USA), and 1% Penicillin-Streptomycin (Life Technologies, catalog #15140-122). For experimental purpose, cells were cultured with three different doses (control-zero, moderate-30 μM and hyper-100 μM) of Hcy in plain Claycomb medium and incubated at 37°C with 5% carbon dioxide; and RNA and protein was extracted after 24 h of incubation.
RNA Isolation, Reverse Transcription (RT), and Real-Time PCR
Total RNA including small RNAs was isolated from HL-1 cardiomyocytes following the protocol of mirVana RNA isolation kit (Ambion, Part number #AM1560, Foster City, CA, USA). Promega kit was used for RT-PCR and syber-green method was applied for real-time PCR. The primers used for RT-PCR are shown in Table 1. The PCR reaction programs used are as follows.
Table 1.
Primers used for RT-PCR and real-time PCR
| Primer | Sequence 5′–3′ | Sequence 5′–3′ |
|---|---|---|
| Dicer forward | TGG AGC GAA TTC TCA GGA GT | |
| Dicer reverse | CAC AGG GCG TGT ATT TTC CT | |
| Timp-1 forward | GCA GTG AAG AGT TTC TCA TC | |
| Timp-1 reverse | TCA TCG GGC CCC AAG GGA TCT | |
| Timp-3 forward | GCCCTCCCATATGTATACCC | |
| Timp-3 reverse | TAGGCCTCACCTCA AGTCTG | |
| Timp-4 forward | CTT ATC TGC CTT GCC CTC AG | |
| Timp-4 reverse | TGT TCA GCT CCC ACT GTG TC | |
| MMP-9 forward | CATGTCACTTTCCCTTCACC | |
| MMP-9 reverse | TTGCCGTCCTTATCGTAGTC | |
| MMP-2 forward | TGTGGGTGGAAATTCAGAAGGT | |
| MMP-2 reverse | TTGTTGCCCAGGAAAGTGAAG | |
| NOX-4 forward | TGGAACTTGGGTTCTTCCAG | |
| NOX-4 reverse | CCAGAATGAGGATCCCAGAA |
Reverse Transcription
Incubation of RNA with oligo dT at 70°C for 6.00 min. The RT cycle was 25°C for 2.00 min, 42°C for 50.00 min, 75°C for 5.00 min, 4°C for ever.
PCR program for amplification of cDNA was:

Real-time PCR for amplification of cDNA using syber-green dye was:

Western Blotting
For Western blot analyses, cells were washed with phosphate buffered saline and lysed in Ripa buffer (Boston BioProducts, Worcester, MA, USA) supplemented with protease inhibitor cocktail (Sigma, catalog #P8340, Saint Louis, MO, USA). Protein concentration was determined by Bradford assay using Bio-Rad protein assay dye (Bio-Rad Laboratories, Hercules, CA, USA; catalog #500-0006) using the softmax software and Molecular devices (CA, USA) machine. Equal amounts of protein (15 μg) were subjected to 10% SDS-PAGE and transferred to PVDF membrane (Bio-Rad laboratory). Immunoblotting was performed using antibodies of MMP-9 (Santa Cruz Bio-technology, Santa Cruz, CA, USA). Secondary antibody was applied and blots were developed using ECL plus substrate (GE Healthcare, Piscataway, NJ, USA). Densitometry analyses were carried out on Western blots using UMAX Power Lock II program (Taiwan, ROC).
MicroRNA Assay
Total RNA was isolated by miRVana method in the same manner as applied for the RT-PCR. Purity of RNA was assessed by Nano-Drop, and only highly pure RNAs (ratio of 260/280 > 1.8 and 260/230 > 1.8) were used for downstream assay. RT-PCR was performed by Megaplex without pre-amplification of miRNA method (Applied Biosystems, Foster City, CA, USA). The microarray assay was performed on 384 miRNA probe using 20 times diluted cDNA (Megaplex RT-PCR product), and following the protocol of manufacturer (Applied Biosystems). The real-time data were analyzed by using SDS RQ manager software provided by Applied Biosystems. Individual miRNA expression was analyzed by using individual miRNA primers specific for RT and real-time reactions following the protocol of Taqman miRNA assays (Applied Biosystems).
Megaplex RT program:

Results
RT-PCR for Dicer Expression in HHcy Cardiomyocytes
The RT-PCR of dicer was performed on control (without Hcy treatment), 30 μM Hcy treated (mild dose), and 100 μM Hcy-treated (high dose) cardiomyocytes. There was an increase in dicer expression in a dose-dependent manner of Hcy. The expression of GAPDH, a loading control, among the three groups was same (Fig. 1a, b).
Fig. 1.
a RT-PCR product of dicer. GAPDH is loading control. b Densitometry analysis. AU arbitrary unit
Real-Time PCR for Dicer Expression in HHcy Cardiomyocytes
To understand the quantitative value, real-time PCR was performed on the same group of cardiomyocytes. For endogenous control, GAPDH was used. It is evident from Fig. 2 that dicer expression is increased (because “ct” value is low) in HHcy cardiomyocytes. The ‘ct’ value of control and HHcy cardiomyocytes were normalized with that of GAPDH.
Fig. 2.
Real-time PCR of dicer in HHcy (100 μM/l)
RT-PCR for MMP-2,-9, TIMP-1,-3,-4, and NOX-4 Expression in HHcy Cardiomyocytes
The semi-quantitative RT-PCR among the three groups (control, mild Hcy, and high Hcy) of cardiomyocytes revealed that MMP-2 as well as MMP-9 expression was increased with treatment of Hcy in a dose-dependent manner (Fig. 3a–d). There was no change in GAPDH (the loading control) expression among the three groups. The TIMP-1 and TIMP-3 expression showed the similar pattern of expression as MMP-2,-9 (Fig. 4a–d). However, TIMP-4 decreased in expression with higher dose of Hcy (Fig. 5a, c). The expression of NOX-4 was increased with high dose of Hcy (Fig. 5b, d). The loading control was GAPDH in all the RT-PCR amplification.
Fig. 3.
a, b RT-PCR product of MMP-9 and MMP-2, respectively. GAPDH is loading control. c, d Densitometry analyses. AU arbitrary unit. * P < 0.05
Fig. 4.
a, b RT-PCR product of TIMP-1 and TIMP-3, respectively. GAPDH is loading control. c, d Densitometry analyses. AU arbitrary unit
Fig. 5.
a, b RT-PCR product of TIMP-4 and NOX-4, respectively. GAPDH is loading control. c, d Densitometry analyses. AU arbitrary unit. * P < 0.05
Western Blotting for MMP-9 Expression in HHcy Cardiomyocytes
The protein expression of MMP-9 was upregulated at higher dose of Hcy in cardiomyocytes. The constitutive protein GAPDH was used as a loading control, which is equally expressed among control, mild dose (30 μM) and high dose (100 μM) of Hcy-treated cardiomyocytes (Fig. 6a, b).
Fig. 6.
a Western blotting of MMP-9. GAPDH is loading control. b Densitometry analyses. AU arbitrary unit
MiRNA Microarray Expression Profile
The microarray analyses of miRNAs revealed differential expression of nearly 11 miRNAs (Fig. 7) of which 4 miRNAs were downregulated (higher and positive ‘ct’ value) and 6 miRNAs were upregulated (lower and negative ‘ct’ value). Interestingly, miRNA-188 shows significant decrease in expression in HHcy cardiomyocytes (Fig. 7). The snoRNA 202 was used as endogenous control and the ‘ct’ value of all miRNAs in the array was normalized with snoRNA 202.
Fig. 7.
MiRNA microarray showing differential expression of 11 miRNAs, snoRNA 202 was used as endogenous control. * P < 0.05
Individual miRNA Assay
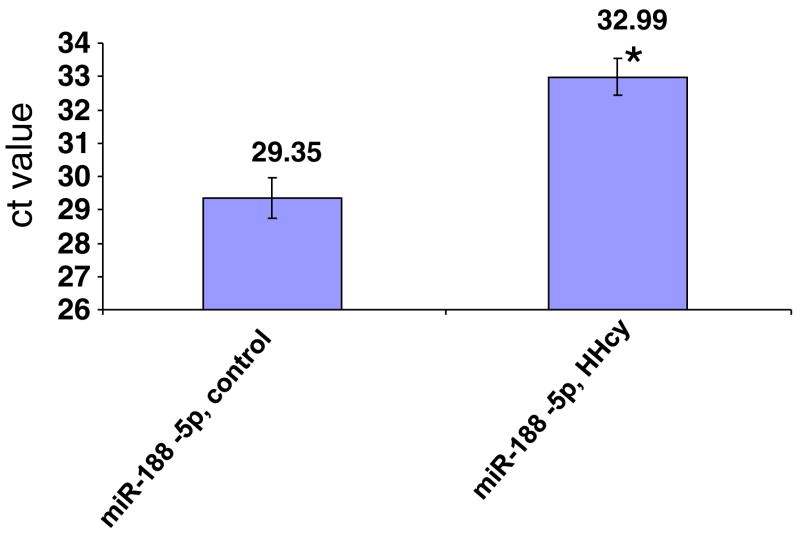
The real-time PCR of miR-188-5p showed decreased expression (higher ‘ct’ value) in HHcy cardiomyocytes. The ‘ct’ value of both control and HHcy cardiomyocytes were shown in the figure (Fig. 8). The snoRNA 202 was used as endogenous control and its ‘ct’ value for both control and HHcy was almost equal to 22.
Fig. 8.
Real-time PCR of miR-188-5p individual assay. SnoRNA 202 was used as endogenous control. * P < 0.05
Statistical Analyses
The data were expressed as mean and standard error from experiments performed four times using different RNA/protein samples. Student’s t-test was used to analyze the significant difference in mean values of the samples. The difference of mean between two samples was considered significant if the P-value was <0.05.
Discussion
Although the role of miRNA in CVDs has been well documented [18] and Hcy has been established as independent risk factor for CVDs [41], the implications of miRNAs in HHcy is still unexplored. We have made an attempt to investigate the involvement of miRNAs and dicer in HHcy cardiac remodeling leading to heart failure. Increase in dicer (enzyme required for maturation of miRNAs) in high dose of Hcy suggests high demand of miRNAs required for maintaining the regulation of genes involved in cardiac remodeling.
Extracellular Matrix Remodeling in HHcy Cardiomyocytes
The elevated level of Hcy causes extracellular matrix remodeling through alteration in elastin/collagen ratio [46]. The matrix-metalloproteinases (MMP-2,-9) and their tissue inhibitors (TIMP-1,-3) contribute in this remodeling process. The RT-PCR results show increase in expression of MMP-2,-9 and TIMP-1,-3, indicating that high dose of Hcy causes extracellular matrix remodeling leading toward heart failure (Figs. 3a, c and b, d and 4a, c and b, d).
TIMP-4 and NOX-4 Expression in HHcy Cardiomyocytes
The fact that Hcy causes oxidative stress has been assessed through expression of NOX-4 (a signature of stress) and TIMP-4 (a signature of anti-stress in heart). Interestingly, there was upregulation of NOX-4 and downregulation of TIMP-4 expression in HHcy cardiomyocytes (Fig. 5a and c, b and d). These results extend concrete support to the fact that high dose of Hcy generates oxidative stress.
Western Blotting for MMP-9
To confirm whether RNA expression is also translated at protein level, Western blotting was performed for MMP-9. As expected, the expression pattern of MMP-9 at protein level is consistent with that at RNA level, and is enhanced in HHcy cardiomyocytes in a dose-dependent manner (Fig. 6a, b). MiRNAs act both at mRNA level using RNAi mechanism as well as at protein level by interfering with the translational pathways. Interestingly, the results from Figs. 3 and 6 suggest that miRNAs regulate MMP-9 at mRNA and not at protein level through post-transcriptional modification. The alteration at protein level due to change in the expression of mRNA extends further support to the fact that MMP-9 expression increases at high dose of Hcy. Further, Figs. 3 and 6 demonstrate that there is a drastic increase in the MMP-9 mRNA level with minimal changes at the protein level. These data revealed the differential role of transcription versus post-translational modifications of the protein in pathogenesis of CVDs. However, there are several regulatory steps between transcription and translation. One reason for decrease in the expression of MMP-9 at protein level while comparing to the RNA level may be the contribution of the regulatory steps. Additionally, there were biological variations among the samples. One of the plausible causes in variation at the level of mRNA and protein might be the biological variations in the individual HL1 cardiomyocytes as we used separate treatment samples for extracting proteins and RNAs.
MiRNA Expression Profile
After confirming the hyperhomocysteinemic condition of cardiomyocytes through the signatures of oxidative stress and cardiac remodeling, high dose of Hcy was applied on HL-1 cardiomyocytes and miRNAs expression was assessed by using 384 miRNA microarray probes. The cDNA samples from control (without Hcy) and HHcy (100 μM/ml Hcy) cardiomyocytes were used for the real-time PCR. The analyses of results revealed differential expression of 11 miRNAs where 7 miRNAs were upregulated and 4 were downregulated (Fig. 7). The negative ‘ct’ value represents downregulation, whereas the positive ‘ct’ value represents upregulation of miRNAs. Interestingly, miR-188-5P shows dramatic downregulation in HHcy cardiomyocytes (Fig. 7), suggesting its putative involvement in HHcy. Here, it is germane to mention that “ct” value is inversely proportional to expression level that is high “ct” value means low expression and vice versa. To confirm the microarray data, individual miR-188 Taqman assay was performed. The results from individual assay using snoRNA202 as endogenous control for real-time PCR amplification of miR-188-5P showed the downregulation of miR-188 in HHcy cardiomyocytes (Fig. 8). Therefore, miR-188 might be a candidate for HHcy cardiac remodeling. However, it is premature to conclude the direct implication of miR-188 in HHcy cardiac remodeling.
HHcy engenders reactive oxygen species that causes oxidative stress, which is indicated by increase in NOX-4 expression (Fig. 5b, d). The stress condition induces extracellular matrix remodeling indicated by increase in expression of MMP-2,-9 (Figs. 3a–d and 6a, b) as well as TIMP-1,-3 (Fig. 4a–d). The decrease in expression of TIMP-4 (Fig. 5a, b) further supports remodeling due to stress generated by high dose of Hcy. To retain the normal expression of genes and for reversal of remodeling, miRNAs (the regulatory molecule) synthesis increases, which is reflected from increased expression of dicer in HHcy (Fig. 1a, c). Therefore, increase in dicer expression in cardiomyocytes is an indication of attempt of miRNAs for reversal of cardiac remodeling to maintain the healthy condition of heart. This result supports the finding that dicer expression increases in heart failure patients treated with left ventricular assistance device, where heart tries to revert to normal condition [40].
Dicer being the terminal enzyme of miRNA biogenesis, upregulation in its expression in HHcy cardiomyocytes indicates global increase in miRNAs synthesis, suggesting involvement of miRNAs in regulation of cardiac remodeling caused due to high dose of Hcy. The results of differential expression of miRNAs from control (without Hcy) and experimental (high dose of Hcy) cardiomyocytes further extend support to the fact that miRNAs are implicated in Hcy-mediated heart failure (Fig. 7). The putative candidate is miR-188. There are two types of miR-188: miR-188-3p and miR-188-5p. The “3p” indicates the 3 prime end, whereas “5p” points to 5 prime end of the same miRNA. The individual miR-188-5P assay pin points the putative implication of miR-188 in HHcy cardiomyocytes.
In normal homeostatic condition, there is a differential expression of miRNAs because in one hand decrease in expression of certain miRNAs is essential for maintaining healthy condition, whereas on the other hand increase in certain miRNAs is required for homeostasis. The increase/decrease in dicer expression globally increases or decreases the expression of all miRNAs by increasing/decreasing their expression in the same pattern as they were in healthy condition. Dicer cannot alter the expression of a specific/individual miRNA. In Hcy-treated cardiomyocytes, the increase in dicer expression suggests that maturation of all miRNAs was induced as more regulatory molecules (miRNAs) were required to take over the remodeling process. However, the differential expression of miRNA (especially miR-188) represents the effect of Hcy on miRNA expression, which is independent of the effect caused by dicer.
Limitations
Although there is changes in the expression of dicer and miRNA-188 in hyperhomocysteinemic cardiomyocytes suggesting their involvement in cardiac remodeling at high dose of Hcy, there is no data supporting direct cause and effect relationship. Additionally, we do not know the exact mechanism of regulation of dicer and miRNAs in HHcy. The gain-of-function and loss-of-function experiments are under investigation, where miR-188 mimic will be used to enhance the expression level of miR-188 in hyperhomocysteinemic cardiomyocytes, and the amelioration in the cardiac remodeling will be assessed. Similarly dicer knock down using siRNA of dicer will be used to assess the direct role of dicer in hyperhomocysteinemic cardiomyocytes. Additionally, functional data are required to conclude the role of dicer and miR-188 in Hcy-mediated heart failure both at in vitro and in vivo levels (Fig. 9).
Fig. 9.
A model showing high dose of Hcy induces oxidative stress, which in turn causes cardiac remodeling leading to heart failure. Although high dose of Hcy downregulated microRNA-188, it is yet to confirm how miRNA-188 is associated with hyperhomocysteinemic heart failure
Conclusion
HHcy is an independent risk factor for CVDs [41] and the regulatory mechanism of Hcy-mediated heart failure is not well understood. To our knowledge, it is the first report suggesting the involvement of miRNAs and dicer in Hcy-mediated heart failure. The functional assay using miR-188 mimic (the synthetic oligos) for ameliorating Hcy-induced cardiac remodeling will provide concrete support to the present results. Considering the tremendous therapeutic potential of miRNAs [18], the present findings will provide a new platform to understand the microRNomics of HHcy.
Acknowledgments
A part of the study was supported by NIH grants HL 71010, HL-74185, and HL-88012.
Abbreviations
- Hcy
Homocysteine
- HHcy
Hyperhomocysteinemia
- MMP
Matrix metalloproteinase
- TIMP
Tissue inhibitor of metalloproteinase
- NOX
Nicotinamide adenine diphosphate oxidase
- CHF
Congestive heart failure
- CVD
cardiovascular diseases
- miR/miRNA
microRNA
- RT-PCR
Reverse transcription polymerase chain reaction
References
- 1.Herrmann W, Herrmann M, Obeid R. Hyperhomocysteinaemia: A critical review of old and new aspects. Current Drug Metabolism. 2007;8(1):17–31. doi: 10.2174/138920007779315008. [DOI] [PubMed] [Google Scholar]
- 2.Martin-Herrero F, Jimenez-Candil J, Martin-Moreiras J, Pabon P, Cruz-Gonzalez I, Martin-Garcia A, et al. Homocysteine, cause or consequence? International Journal of Cardiology. 2008;129(2):276–277. [Google Scholar]
- 3.Refsum H, Smith AD, Ueland PM, Nexo E, Clarke R, McPartlin J, et al. Facts and recommendations about total homocysteine determinations: An expert opinion. Clinical Chemistry. 2004;50(1):3–32. doi: 10.1373/clinchem.2003.021634. [DOI] [PubMed] [Google Scholar]
- 4.Wierzbicki AS. Homocysteine and cardiovascular disease: A review of the evidence. Diabetes & Vascular Disease Research. 2007;4(2):143–150. doi: 10.3132/dvdr.2007.033. [DOI] [PubMed] [Google Scholar]
- 5.Jhee KH, Kruger WD. The role of cystathionine beta-synthase in homocysteine metabolism. Antioxidants Redox Signaling. 2005;7(5–6):813–822. doi: 10.1089/ars.2005.7.813. [DOI] [PubMed] [Google Scholar]
- 6.Graham IM, Daly LE, Refsum HM, Robinson K, Brattstrom LE, Ueland PM, et al. Plasma homocysteine as a risk factor for vascular disease. The European Concerted Action Project. JAMA. 1997;277(22):1775–1781. doi: 10.1001/jama.1997.03540460039030. [DOI] [PubMed] [Google Scholar]
- 7.Blacher J, Demuth K, Guerin AP, Vadez C, Moatti N, Safar ME, et al. Association between plasma homocysteine concentrations and cardiac hypertrophy in end-stage renal disease. Journal of Nephrology. 1999;12(4):248–255. [PubMed] [Google Scholar]
- 8.Blacher J, Safar ME. Homocysteine, folic acid, B vitamins and cardiovascular risk. Journal of Nutrition, Health & Aging. 2001;5(3):196–199. [PubMed] [Google Scholar]
- 9.Lee RT. Matrix metalloproteinase inhibition and the prevention of heart failure. Trends in Cardiovascular Medicine. 2001;11(5):202–205. doi: 10.1016/s1050-1738(01)00113-x. [DOI] [PubMed] [Google Scholar]
- 10.Sakata Y, Yamamoto K, Mano T, Nishikawa N, Yoshida J, Hori M, et al. Activation of matrix metalloproteinases precedes left ventricular remodeling in hypertensive heart failure rats: Its inhibition as a primary effect of angiotensin-converting enzyme inhibitor. Circulation. 2004;109(17):2143–2149. doi: 10.1161/01.CIR.0000125741.88712.77. [DOI] [PubMed] [Google Scholar]
- 11.Spinale FG, Coker ML, Bond BR, Zellner JL. Myocardial matrix degradation and metalloproteinase activation in the failing heart: A potential therapeutic target. Cardiovascular Research. 2000;46(2):225–238. doi: 10.1016/s0008-6363(99)00431-9. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi SC, Smiley LM, Mujumdar VS, Clonts B, Parker JL. Reduction-oxidation (Redox) and vascular tissue level of homocyst(e)ine in human coronary atherosclerotic lesions and role in extracellular matrix remodeling and vascular tone. Molecular and Cellular Biochemistry. 1998;181(1–2):107–116. doi: 10.1023/a:1006882014593. [DOI] [PubMed] [Google Scholar]
- 13.Lominadze D, Roberts AM, Tyagi N, Moshal KS, Tyagi SC. Homocysteine causes cerebrovascular leakage in mice. American Journal of Physiology. Heart and Circulatory Physiology. 2006;290(3):H1206–H1213. doi: 10.1152/ajpheart.00376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moshal KS, Sen U, Tyagi N, Henderson B, Steed M, Ovechkin AV, et al. Regulation of homocysteine-induced MMP-9 by ERK1/2 pathway. American Journal of Physiology. Cell Physiology. 2006;290(3):C883–C891. doi: 10.1152/ajpcell.00359.2005. [DOI] [PubMed] [Google Scholar]
- 15.Moshal KS, Tipparaju SM, Vacek TP, Kumar M, Singh M, Frank IE, et al. Mitochondrial matrix metalloproteinase activation decreases myocyte contractility in hyperhomocysteinemia. American Journal of Physiology. Heart and Circulatory Physiology. 2008;295(2):H890–H897. doi: 10.1152/ajpheart.00099.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyagi SC, Rodriguez W, Patel AM, Roberts AM, Falcone JC, Passmore JC, et al. Hyperhomocysteinemic diabetic cardiomyopathy: Oxidative stress, remodeling, and endothelial-myocyte uncoupling. Journal of Cardiovascular Pharmacology and Therapeutics. 2005;10(1):1–10. doi: 10.1177/107424840501000101. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: Do they play a role in cardiac hypertrophy? American Journal of Pathology. 2007;170(6):1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra PK, Tyagi N, Kumar M, Tyagi SC. MicroRNAs as a therapeutic target for cardiovascular diseases. Journal of Cellular and Molecular Medicine. 2009;13(4):778–789. doi: 10.1111/j.1582-4934.2009.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circulation Research. 2007;100(3):416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 20.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. Journal of Molecular and Cellular Cardiology. 2007;42(6):1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thum T, Catalucci D, Bauersachs J. MicroRNAs: Novel regulators in cardiac development and disease. Cardiovascular Research. 2008;79(4):562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 22.Van Rooij E, Olson EN. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. Journal of Clinical Investigation. 2007;117(9):2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. Journal of Biological Chemistry. 2005;280(10):9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circulation Research. 2007;101(12):1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 26.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genetics. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 27.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature Structural & Molecular Biology. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C. MicroRNAs: Role in cardiovascular biology and disease. Clinical Science. 2008;114(12):699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 29.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C. MicroRNomics: A newly emerging approach for disease biology. Physiological Genomics. 2008;33(2):139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nature Genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 33.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, et al. Critical roles for Dicer in the female germline. Genes and Development. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56(12):2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 35.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 36.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. Journal of Neuroscience. 2008;28(17):4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circulation Research. 2007;101(1):59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 39.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circulation Research. 2007;100(8):1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 40.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proceedings of the National Academy of Science of the United States of America. 2008;105(6):2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Ruijter W, Westendorp RG, Assendelft WJ, den Elzen WP, de Craen AJ, le Cessie S, et al. Use of Framingham risk score, new biomarkers to predict cardiovascular mortality in older people: Population based observational cohort study. BMJ. 2009;338:a3083. doi: 10.1136/bmj.a3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kundu S, Kumar M, Sen U, Mishra PK, Tyagi N, Metreveli N, et al. Nitrotyrosinylation, remodeling and endothelial-myocyte uncoupling in iNOS, cystathionine beta synthase (CBS) knockouts and iNOS/CBS double knockout mice. Journal of Cellular Biochemistry. 2009;106(1):119–126. doi: 10.1002/jcb.21982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyagi SC, Hoit BD. Metalloproteinase in myocardial adaptation and maladaptation. Journal of Cardiovascular Pharmacology and Therapeutics. 2002;7(4):241–246. doi: 10.1177/107424840200700407. [DOI] [PubMed] [Google Scholar]
- 44.Hsu CP, Huang CY, Wang JS, Sun PC, Shih CC. Extracellular matrix remodeling attenuated after experimental postinfarct left ventricular aneurysm repair. Annals of Thoracic Surgery. 2008;86(4):1243–1249. doi: 10.1016/j.athoracsur.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 45.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proceedings of the National Academy of Science of the United States of America. 1998;95(6):2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyagi SC. Homocysteine and heart disease: Patho-physiology of extracellular matrix. Clinical and Experimental Hypertension. 1999;21(3):181–198. doi: 10.3109/10641969909068660. [DOI] [PubMed] [Google Scholar]