Abstract
The C2′-oxidized abasic lesion (C2-AP) is produced in DNA that is subjected to oxidative stress. The lesion disrupts replication and gives rise to mutations that are dependent upon the identity of the upstream nucleotide. Ape1 incises C2-AP, but the 5′-phosphorylated fragment is not a substrate for the lyase activity of DNA polymerase β (Pol β). Excision of the lesion is achieved by strand displacement synthesis in the presence of flap endonuclease (FEN1) during which C2-AP and the 3′-adjacent nucleotide are replaced. The oxidized abasic lesion is also a substrate for the bacterial UvrABC nucleotide excision repair system. These data suggest that the redundant nature of DNA repair systems provide a means for removing a lesion that resists excision by short patch base excision repair.
Keywords: DNA repair, DNA damage, DNA oxidation, free radicals
Introduction
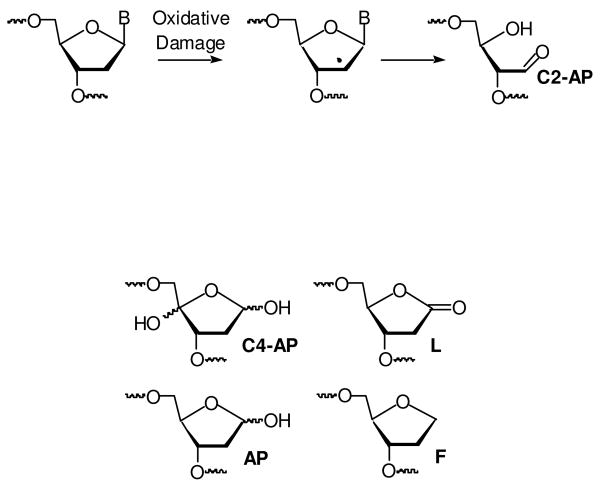
The carbon-hydrogen bonds at the C2′-position are the strongest of their type in DNA (1). Consequently, the C2′-oxidized abasic site (C2-AP), which results from hydrogen atom abstraction from this position, is most frequently associated with DNA that is subjected to strong oxidants, such as hydroxyl radical, which is generated by γ-radiolysis (2). C2-AP is also produced from 2′-deoxyuridin-5-yl radical, the σ-radical that is generated from irradiation of the 5-halopyrimidine radiosensitizing agents (5-bromo-2′-deoxyuridine, 5-iodo-2′-deoxyuridine) (3-6). Although the mechanism is uncertain, the radical resulting from C2′-hydrogen atom abstraction is likely a common precursor to C2-AP in these processes (Scheme 1). Despite the absence of a nucleobase, C2-AP exerts a distinctive effect on DNA replication in Escherichia coli (7). Mutations are introduced into DNA upon bypass of the oxidized abasic site via a mechanism that is dependent upon the identity of the 3′-adjacent (upstream) nucleotide. The high mutagenic potential of this lesion indicates that its faithful repair is necessary to protect cells exposed to oxidative stress. Bacterial base excision repair (BER) enzymes, such as Xth and Nfo, efficiently incise DNA containing the C2-AP lesion (8). However, unlike the AP and C4-AP lesions, C2-AP lacks a leaving group (phosphate) at the β-position relative to the aldehyde carbon (9-12). Consequently, C2-AP does not undergo the typical lyase reaction catalyzed by BER enzymes required to complete its excision (13). The resistance of C2-AP to short patch BER led us to investigate other excision processes. Herein, we present two alternative pathways by which C2-AP is excised from DNA.
Scheme 1.
Note: AP, F, C4-AP, L structures
The importance of maintaining the integrity of DNA is reflected by the redundancy of DNA repair pathways (14-17). In addition to short patch BER, some lesions are removed along with one or more adjacent nucleotides, which may not be damaged. DNA polymerase β (Pol β) carries out long patch BER in conjunction with flap endonuclease (FEN1) following 5′-incision of the lesion by Ape1 (18-20). Pol β extends the newly formed 3′-terminus of the incised strand in a dNTP dependent manner, while simultaneously displacing the 5′-fragment that contains the lesion. Strand displacement synthesis is facilitated by FEN1, which cleaves the 5′-fragment that is displaced as a result of polymerase extension. Repair is completed by ligation of the 3′-terminal hydroxyl and 5′-phosphate following FEN1 excision. Long patch BER has proven effective for a variety of lesions, including tandem lesions that are resistant to the related short patch method (21, 22). Longer stretches of nucleotides are removed during nucleotide excision repair. The bacterial UvrABC nucleotide excision repair (NER) system removes lesion-containing oligonucleotide fragments that are typically 12-13 nucleotides long. Incision on each side of the lesion by the nuclease component of UvrC is a coupled process (23, 24). Nucleotide excision repair is often associated with bulky lesions that distort the duplex structure (24, 25). However, smaller lesions including abasic sites are also excised (26) (10, 24-27).
Material and Methods
General procedures
Oligonucleotide synthesis was carried out on an Applied Biosystems Incorporated 394 DNA synthesizer using standard protocols. Oligonucleotides containing C2-AP were synthesized as described (28). Commercially available oligonucleotide synthesis reagents were obtained from Glen Research (Sterling, VA). The 50mer containing the fluoresceinylated thymidine (Fl-dT) was obtained from Sigma-Genosys (St. Louis, MO). DNA manipulation, including enzymatic labeling, was carried out using standard procedures (29). Preparative and analytical oligonucleotide separations were carried out on 20% denaturing or native polyacrylamide gel electrophoresis (5% cross-link, 45% urea (by weight)). Ape1, T4 DNA ligase, T4 polynucleotide kinase and terminal transferase were obtained from New England Biolabs (Beverly, MA). DNA polymerase β (Pol β) and flap endonuclease (FEN1) were obtained from Trevigen (Gaithersburg, MD). UvrABC was obtained as previously described (30, 31). [γ-32P]-ATP and [α-32P]-3′-deoxyadenosine triphosphate were purchased from Perkin-Elmer (Waltham, MA). Aldehyde reactive probe (ARP) was purchased from Invitrogen. Radioactive samples were quantitated by Cerenkov counting using a Beckman LS6500 liquid scintillation counter. Quantification of radiolabeled oligonucleotides was carried out using a Molecular Dynamics Storm 840 Phosphorimager equipped with ImageQuant Version 5.1 software.
Note: The strand containing C2-AP (or derived from it) is labeled in all experiments.
Preparation of C2-AP containing oligonucleotides by oxidation of 2
The single stranded oligonucleotide containing 2, 5′ -or 3′-32P-3 (2.5 μM), was oxidized using 25 mM NaIO4 in a buffer containing 100 mM NaOAc pH 6.0 at 1 h in room temperature. The reaction mixture was filtered through a G-25 Sephadex column (equilibrated using H2O), and 32P-4 (1.25 μM) was hybridized to its complement (1.8 μM) in 10 mM sodium phosphate (pH 7.2) and 100 mM NaCl to yield 10. The formation of C2-AP was verified by reacting 5′ -or 3′-32P-10 (60 nM) with 10 mM ARP (from Invitrogen) at 37 °C for 1 h.
Ape1 Kinetics on 4
The radiolabeled duplex 5′-32P-10 (10 nM, 20 nM, 40 nM, 60 nM, 100 nM, 250 nM), was treated with Ape1 (500 pM) in a total of 10 μL at room temperature for 2 min in 20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate and 1 mM dithiothreitol. The reactions were quenched with an equal volume of 90% formamide loading buffer and analyzed by 20% denaturing PAGE.
Strand Displacement Synthesis by Pol β
The radiolabeled duplex, 5′-32P-10 (300 nM), was treated with Ape1 (15.5 nM) at 37 °C for 1 h in 20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate and 1 mM dithiothreitol, pH 7.9 in a total reaction volume of 20 μL. The reaction mixture was filtered through a G-25 Sephadex column (equilibrated using a buffer containing 50 mM Tris-HCl pH 8.0, 10 mM MnCl2, and 1 mM DTT). Pol β (0.1 nM) was added to the resulting 5′-32P-11 (200 nM) in the absence or presence of FEN1 (1 nM) and incubated in 50 mM Tris-HCl pH 8.0, 10 mM MnCl2, 1 mM DTT, 0.1 mg/ml BSA, 50 μM dGTP and/or 50 μM dTTP at 37 °C for 1 h in a total reaction volume of 30 μL. Aliquots (3 μL) were removed at 2, 5, 10, 20, 30, 40, 50 and 60 min and quenched with an equal volume of 90% formamide loading buffer containing EDTA (10 mM). The reactions were analyzed by 20% denaturing PAGE.
Strand Excision by FEN1
The radiolabeled duplex, 3′-32P-10 (300 nM), was treated with Ape1 (15.5 nM) at 37 °C for 1 hr in 20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate and 1 mM dithiothreitol, pH 7.9 in a total reaction volume of 20 μL. The reaction mixture was filtered through a G-25 Sephadex column (equilibrated using a buffer containing 50 mM Tris-HCl pH 8.0, 10 mM MnCl2, and 1 mM DTT). Pol β (0.1 nM) and FEN1 (1 nM) were added to the resulting 3′-32P-11 (200 nM) and incubated in 50 mM Tris-HCl pH 8.0, 10 mM MnCl2, 1 mM DTT, 0.1 mg/ml BSA, 50 μM dGTP and/or 50 μM dTTP at 37 °C for 1 h in a total reaction volume of 30 μL. Aliquots (3 μL) were removed at 2, 5, 10, 20, 30, 40, 50 and 60 min and were quenched with an equal volume of 90% formamide loading buffer containing EDTA (10 mM). The reactions were analyzed by 20% denaturing PAGE.
Preparation of 3′- and 5′-32P-6 by enzyme ligation
The 3′-terminal oligonucleotide component 8 (25 nmol) was 5′-phosphorylated with 1 mM ATP and T4 polynucleotide kinase (20 units) in kinase buffer (70 mM Tris-HCl, 10 mM MgCl2, 5 mM dithiothreitol) at 37 °C for 45 min. The reaction mixture was filtered through a G-25 Sephadex column (equilibrated with H2O), and the 5′-phosphorylated-8 was 3′-32P-labeled with [α-32P]-3′-deoxyadenosine 5′-triphosphate and terminal transferase (40 units) in TdT buffer (20 mM Tris-acetate, 50 mM potassium-acetate, 10 mM magnesium acetate, pH 7.9) and 0.25 mM CoCl2 at 37 °C for 45 min. After filtering through a G-25 Sephadex column (equilibrated with H2O), the 5′-phosphorylated-3′-32P-8 (25 nmol) was added to 5 (37.5 nmol) and template 9 (37.5 nmol) in ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg/mL BSA). The oligonucleotides were hybridized at 80 °C for 5 min and slowly cooled to room temperature. After annealing, T4 DNA ligase (4000 units) was added along with 10 nmol of ATP, and the mixture was incubated at 16 °C for 1 h. The reaction was quenched by adding an equal volume of 90% formamide loading buffer (without dyes). The product (3′-32P-6) was purified by 20% denaturing PAGE. 5′-32P-6 was prepared as described above, except that 5′-32P-5 was employed and 5′-phosphorylated-8 was not labeled at its 3′-terminus.
Preparation of 12
The radiolabeled 50mer, 5′ -or 3′-32P-6 (250 nM) was oxidized using 25 mM NaIO4 in a buffer containing 100 mM NaOAc pH 6.0 for 1 h at room temperature. The reaction mixture was filtered through a G-25 Sephadex column (equilibrated using H2O), and 5′ -or 3′-32P-7 (200 nM) was hybridized to its complement (250 nM) in 10 mM sodium phosphate pH 7.2 and 100 mM NaCl by heating to 80 °C for 5 min and slowly cooling to room temperature. The formation of C2-AP was verified by reacting 32P-7 (20 nM) with 10 mM ARP at 37 °C for 1 h.
Incision of 5′-12 with UvrABC
UvrA, UvrB and UvrC were activated separately before application by incubating at 65 °C for 20 min in 1 × NER buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 50 mM KCl and 1 mM ATP). Duplex 32P-12 (10 nM) was reacted with 40 nM UvrA, 100 nM UvrB and 100 nM UvrC in 1 × NER buffer at 55 °C for 3 h. Aliquots (2 μL) were removed at 15, 30, 60, 90, 120, 150 and 180 min and quenched with 8 μL of a solution containing 0.5 M NH4OAc and 0.1 mg/mL calf-thymus DNA (the 0.5 M NH4OAc and 0.1 mg/mL calf-thymus DNA are final concentrations). The samples were immediately precipitated with 2.5 volumes of ethanol and concentrated. The dried DNA was suspended in 90% formamide loading buffer and resolved using 20% denaturing PAGE. The duplex containing fluoresceinylated thymidine (5′-32P-13) was subjected to comparable treatment and analysis, except that aliquots were removed over a shorter time period.
Results
Oligonucleotide substrate preparation
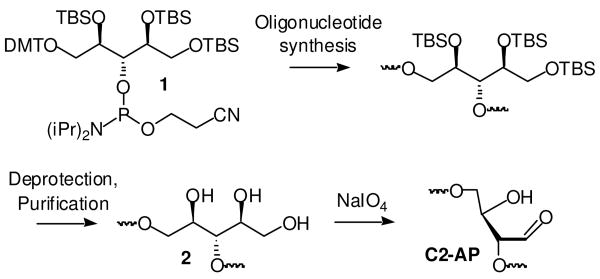
Oligonucleotides containing C2-AP were prepared by NaIO4 oxidation of those (3, 6) containing the triol (2, Scheme 2) (28). Due to the modest coupling yield of the latent C2-AP phosphoramidite (1), the 50mer (6) used in NER experiments was prepared via ligation of chemically synthesized 32mer (5) with 8 in which 9 was used as a template (Table 1). The gel purified 50mer (6) was isolated in ∼90% yield, and was subsequently treated with NaIO4 to produce 7. For each oligonucleotide (3, 6) the completeness of the periodate oxidation was demonstrated using aldehyde reactive probe (ARP) because the triol and C2-AP-containing oligonucleotides are inseparable from one another, whereas the adducted aldehyde containing oligonucleotides migrate more slowly than their precursors (See Supporting Information).
Scheme 2.
Table 1.
Oligonucleotides and duplexes employed in experiments.
5′-d(CGA CCG GCT CGT ATG  TG TGT GGA GCT GTG G) TG TGT GGA GCT GTG G) |
3  = 2 = 2
|
4  = C2-AP = C2-AP |
5′-d(GCA GAT CTG GCC TGA TTG CGG TAG  GA TGG AG) GA TGG AG) |
5  = 2 = 2
|
5′-d(GCA GAT CTG GCC TGA TTG CGG TAG  GA TGG AGC CGT AAC AGT ACG TAG TC) GA TGG AGC CGT AAC AGT ACG TAG TC) |
6  = 2 = 2
|
7  = C2-AP = C2-AP |
| 5′-d(CCG TAA CAG TAC GTA GTC) |
| 8 |
| 5′-d(ACT GTT ACG GCT CCA TC) |
| 9 |
5′-d(CGA CCG GCT CGT ATG  TG TGT GGA GCT GTG G) TG TGT GGA GCT GTG G) |
| 3′-d(GCT GGC CGA GCA TAC AAC ACA CCT CGA CAC C) |
10  = C2-AP = C2-AP |
| HO OPO3 -2 |
5′-d(CGA CCG GCT CGT ATG  TG TGT GGA GCT GTG G) TG TGT GGA GCT GTG G) |
| 3′-d(GCT GGC CGA GCA TAC AAC ACA CCT CGA CAC C) |
11  = C2-AP = C2-AP |
5′-d(GCA GAT CTG GCC TGA TTG18 CGG TAG  GA TGG30 AGC CGT AAC AGT ACG TAG TC) GA TGG30 AGC CGT AAC AGT ACG TAG TC) |
| 3′-d(CGT CTA GAC CGG ACT AA C GCC ATC ACT AC C TCG GCA TTG TCA TGC ATC AG) |
12  = C2-AP = C2-AP |
5′-d(GAC TAC GTA CTG TTA CGG CTC CAT C C TAC CGC AAT CAG GCC AGA TCT GC) C TAC CGC AAT CAG GCC AGA TCT GC) |
| 3′-d(CTG ATG CAT GAC AAT GCC GAG GTA GAG ATG GCG TTA GTC CGG TCT AGA CG) |
13  = Fl-dT = Fl-dT |
The C2-AP lesion is sufficiently stable to heat so that duplexes containing it were prepared by standard hybridization involving brief heating at 80 °C and slow cooling to room temperature. The substrate for strand displacement studies (11) was prepared from the appropriate form of 5′-32P-10 using Ape1. Analysis of 32P-11 by nondenaturing polyacrylamide gel electrophoresis showed that the ternary complex accounted for >80% of the material (data not shown). Ape1, the enzyme mainly responsible for the first step in BER of abasic lesions efficiently incises C2-AP (Km = 48.7 ± 7.6 nM, Vmax = 3.7 ± 0.3 × 10-2 pmol/min-1 at 500 pM Ape1, kcat = 7.6 ± 0.6 min-1). The reactions were carried out for 2 min and the velocities were determined by dividing the amount of product by the reaction time. Independent experiments showed the product formation was linear over this reaction period at the low and high concentrations of 5′-32P-10 (See Supporting Information). A representative plot of the incision reaction on 5′-32P-10 is shown in Figure 1.
Figure 1.
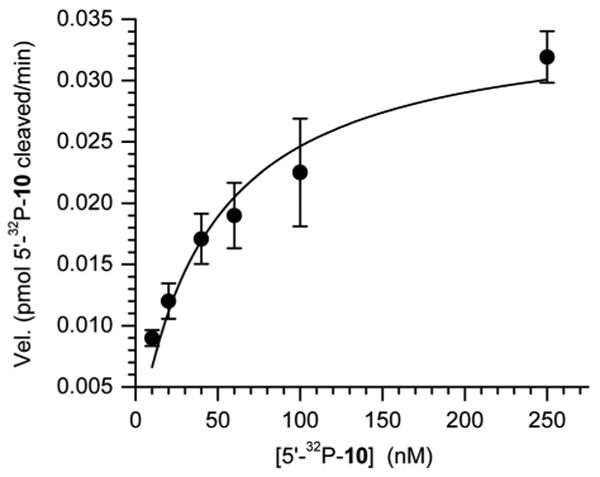
Representative plot of Ape1 (500 pM) incision of C2-AP in 5′-32P-10.
C2-AP removal by long patch base excision repairp
No reaction, especially excision, is observed when 5′ -or 3′-32P-11 is incubated with Pol β in the absence of dNTPs. (Please note that in each instance in the experiments that follow, the respective strand derived from that containing C2-AP is labeled.) Evidence for a Schiff base intermediate is obtained if the reaction mixture is reacted with NaBH4 (See Supporting Information). The protein-DNA cross-link yield is proportional to the concentration of 5′-32P-11 in the presence of a constant excess of Pol β. In contrast, extension of the labeled fragment in 5′-32P-11 (200 nM) by Pol β (0.1 nM) is observed in the presence of dTTP (50 μM) even without FEN1 (Figure 2A). Reaction proceeds to ∼60% conversion within 30 min, but only to ∼65% after 1 h. Addition of a fresh aliquot of Pol β at 30 min rapidly increases the conversion to ∼90% (data not shown). This suggests that incomplete extension is attributable to Pol β inactivation. However, we cannot determine from these observations whether inactivation is due to Schiff base formation or protein denaturation. In the absence of FEN1 at 0.1 nM Pol B, the majority of the extension product consists of a single nucleotide, and the amount of +2 nucleotide product reaches less than 12% after 1 h. In contrast, the +2 nucleotide product reaches almost 70% after 1 h in the presence of Pol β (0.1 nM) and FEN1 (1 nM) (Figure 2B). Under these conditions, the single nucleotide addition product grows very rapidly within the first 10 min of reaction, but is supplanted over time by the growth of the +2 nucleotide product. The overall conversion of 5′-32P-11 also increases to more than 80% in 30 min, and almost 85% in 1 h. Small amounts of +3 (<3 %) and + 4 (<1%) nucleotide products are observed, but only if dGTP is present in addition to dTTP (See Supporting Information).
Figure 2.
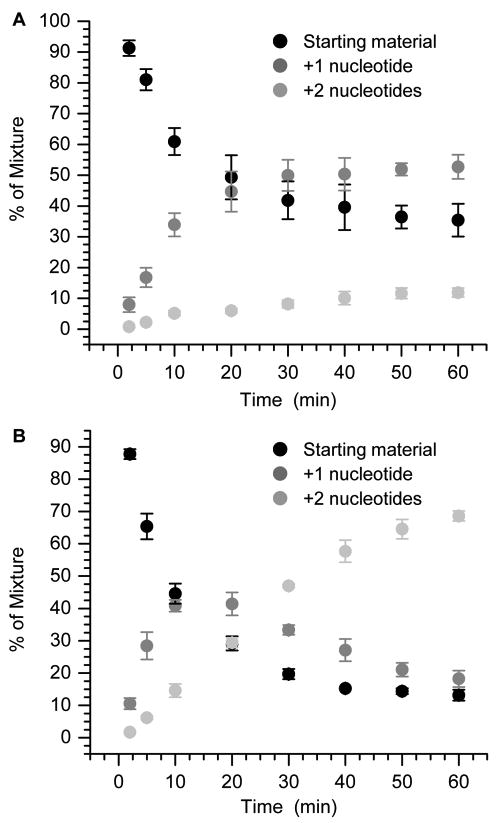
Pol β (0.1 nM) and dTTP (50 μM) mediated extension of Ape1 incised C2-AP (5′-32P-11, starting material) in the (A) absence or (B) presence of FEN1 (1 nM). Each data point is the average of 3 independent measurements ± standard deviation.
These observations are complimented by experiments using 3′-32P-11 in which we measure removal of the nucleotides displaced by Pol β mediated extension (Figure 3). The FEN1 incision reaction lags slightly behind extension observed using 5′-32P-11 (Figure 3B). In addition, the product resulting from loss of 2 nucleotides is the only major one observed when analyzing 3′-32P-11. Product consisting of loss of a single nucleotide is observed only at negligible amounts. The product contains a 5′-phosphate group and migrates slightly more rapidly in the gel than the corresponding marker whose terminus is a 5′-hydroxyl (Figure 3A). The 2-nucleotide deletion product is the sole one regardless of whether dGTP is present in addition to dTTP (See Supporting Information).
Figure 3.
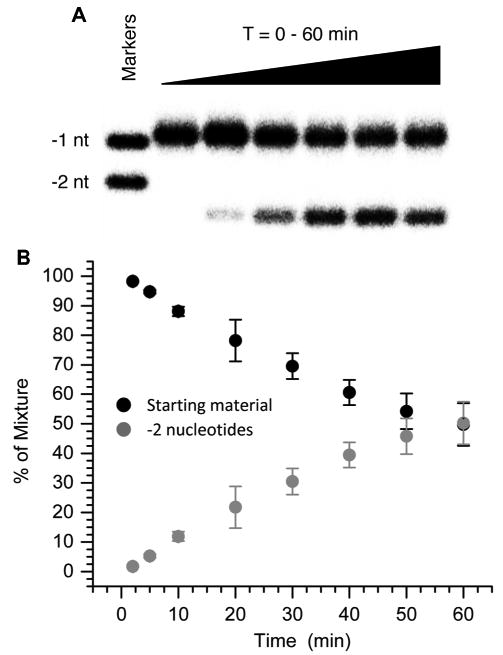
FEN1 (1 nM) excision of flap created by Pol β (0.1 nM) extension of Ape1 incised C2-AP (3′-32P-11) in the presence of dTTP (50 μM) (A) Sample autoradiogram. (B) 3′-32P-11 (starting material) and -2 nucleotide incision product as a function of time. Each data point is the average of 3 independent measurements ± standard deviation.
C2-AP removal by nucleotide excision repair
The incision sites in 12 generated by reaction with UvrABC were determined using 5′- and 3′-32P-labeled material in separate experiments. Incision occurred exclusively at G18 and G30 in 12 (See Table 1 for positions relative to lesion). These positions correspond to cleavage at the 8th phosphate diester to the 5′-side of C2-AP and 5th phosphate diester to the 3′-side of C2-AP respectively (See Table 1 for positions relative to lesion) (See Supporting Information). Coupled incision by UvrABC results in the loss of a 12-nucleotide fragment that includes the C2-AP lesion (32). The rate of incision was followed using 5′-32P-12 and compared to an analogous 50-nucleotide duplex containing a C5-fluoresceinylated thymidine (Figure 4). The latter modification is often used as a standard when determining the susceptibility of lesions to UvrABC. In side-by-side reactions, ∼95% of the fluoresceinylated thymidine standard is incised within 30 min. However, only 26% of 12 is incised in this period, and the incision only reaches 71% even after 3 h of incubation with the enzyme. No cleavage of 5′-32P-12 is observed under these conditions in the absence of UvrABC.
Figure 4.
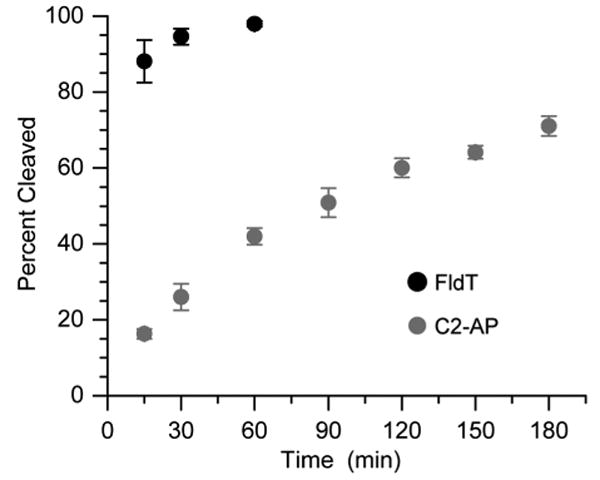
UvrABC incision of C2-AP (5′-32P-12) as a function of time in comparison with Fl-dT (5′-32P-13). Data are the average of 3 independent experiments ± standard deviation.
Discussion
The C2′-oxidized abasic site induces mutations in DNA during replication in E. coli via an unusual mechanism that involves the upstream nucleotide (7). Hence, it is imperative that this lesion be repaired. Its structure prohibits excision via a short patch BER mechanism because the lesion cannot undergo β-elimination. The data above suggest that redundancies in DNA repair pathways enable this obstacle to be overcome. Mammalian enzymes involved in the first two steps of long patch base excision repair excise the C2-AP lesion (18, 33, 34). Ape1 incises the lesion at its 5′-phosphate. The Km for this process is comparable to those describing AP and L incision by Ape1, but the kcat is about 30-fold slower (35). In addition, Ape1 incises C2-AP approximately 10-fold less efficiently than does Xth (8). Pol β efficiently extends the 3′-terminus of the fragment at the incision site by 2 nucleotides, or one nucleotide past the lesion, and is assisted by FEN1. A very small amount of product containing 3 or even 4 additional nucleotides is formed when an appropriate nucleotide triphosphate is present. The predominant addition of 2 nucleotides is consistent with our own work in which a 2-nucleotide long tandem lesion is excised by Pol β and FEN1 (21). In that case the major extension product also results from addition of one nucleotide greater than the length of the lesion, and FEN1 removes a 3-nucleotide fragment. These observations are slightly different than studies on strand displacement synthesis of the tetrahydrofuran model (F) of an AP site (19). In those studies a comparable nicked substrate containing F was extended a single nucleotide in the presence of the appropriate dNTP. However, the sequence was such that incorporation of a second nucleotide would require formation of a mismatch. In our substrate (11), dT incorporation is correct for both nucleotides. In addition, Pol β could have correctly added up to 8 nucleotides to the 5′-fragment of 11 in the presence of dTTP and dGTP. The fact that we only observe mostly the addition of 2 nucleotides and very small amounts of 3- and 4-nucleotide extended products suggests that the C2-AP lesion destabilizes the 3′-adjacent A:T base pair, providing a thermodynamic driving force for nucleotide insertion by Pol β.
Nucleotide excision repair by the bacterial UvrABC system is similar to other lesions. For instance, UvrABC incision typically produces incised regions that are 12-14 nucleotides long, and the C2-AP incision sites are 12 nucleotides apart (23, 24, 26). Of greatest relevance is the comparison to other abasic lesions. In this regard, UvrABC incises a duplex containing C2-AP at exactly the same positions as DNA containing an AP site (36). The incision efficiency of the C2-AP lesion was lower relative to the fluoresceinylated thymidine containing duplex (Figure 4), which has been used as a benchmark. C2-AP incision is also moderately lower than that of a thymidine glycol containing tandem lesion, but the source of this difference is unknown (21). Overall, the data presented here suggest two possible mechanisms by which DNA repair systems may overcome the recalcitrance of a mutagenic lesion to undergo short patch BER, and they point out the importance of redundant repair pathways for maintaining genomic integrity.
Supplementary Material
Acknowledgments
We are grateful for support of this research from the National Institute of General Medical Sciences (GM-063028). We are grateful to Professor Ben Van Houten for generously providing the plasmids and protocols for isolating Uvr A, Uvr B, UvrC. We thank Cortney Kreller for carrying out preliminary experiments.
Abbreviations
- C2-AP
C2′-oxidized abasic site
- C4-AP
C4′-oxidized abasic site
- AP
abasic site
- L
2-deoxyribonolactone
- Xth
exonuclease III
- Nfo
endonuclease IV
- BER
base excision repair
- NER
nucleotide excision repair
- ARP
aldehyde reactive probe
- FEN1
flap endonuclease
- Ape1
apurinic endonuclease
- Pol β
DNA polymerase β
- PAGE
polyacrylamide gel electrophoresis
- BSA
bovine serum albumin
Footnotes
Supporting Information. ESI-MS of 3 and 5. Verification of C2-AP formation in 4 and 7. Time course studies for Ape1 incision of 5′-32P-10. Strand displacement synthesis and FEN1 excision in the presence of dTTP and dGTP. FEN1 excision in the presence of dTTP and dGTP. Determination of UvrABC incision sites in 12. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Li MJ, Liu L, Wei K, Fu Y, Guo QX. Significant effects of phosphorylation on relative stabilities of DNA and RNA sugar radicals: remarkably high susceptibility of H-2′ abstraction in RNA. J Phys Chem B. 2006;110:13582–13589. doi: 10.1021/jp060331j. [DOI] [PubMed] [Google Scholar]
- 2.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; London: 1987. [Google Scholar]
- 3.Sugiyama H, Fujimoto K, Saito I. Evidence for intrastrand C2′ hydrogen abstraction in photoirradiation of 5-halouracil-containing oligonucleotides by using stereospecifically C2′-deuterated deoxyadenosine. Tetrahedron Lett. 1996;37:1805–1808. [Google Scholar]
- 4.Sugiyama H, Fujimoto K, Saito I. Stereospecific 1,2-hydride shift in ribonolactone formation in the photoreaction of 2′-iododeoxyuridine. J Am Chem Soc. 1995;117:2945–2946. [Google Scholar]
- 5.Sugiyama H, Tsutsumi Y, Fujimoto K, Saito I. Photoinduced deoxyribose C2′ oxidation in DNA. Alkali-dependent cleavage of erythrose-containing sites via a retroaldol reaction. J Am Chem Soc. 1993;115:4443–4448. [Google Scholar]
- 6.Xu Y, Sugiyama H. Photochemical approach to probing different DNA structures. Angew Chem Int Ed. 2006;45:1354–1362. doi: 10.1002/anie.200501962. [DOI] [PubMed] [Google Scholar]
- 7.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Replication of an oxidized abasic site in Escherichia coli by a dNTP-stabilized misalignment mechanism that reads upstream and downstream nucleotides. Biochemistry. 2006;45:5048–5056. doi: 10.1021/bi052276v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg MM, Weledji YN, Kroeger KM, Kim J. In vitro replication and repair of DNA containing a C2′-oxidized abasic site. Biochemistry. 2004;43:15217–15222. doi: 10.1021/bi048360c. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg MM, Kreller CR, Young SE, Kim J. Reactivity of the C2′-oxidized abasic lesion and its relevance to interactions with type I base excision repair enzymes. Chem Res Toxicol. 2006;19:463–468. doi: 10.1021/tx060001q. [DOI] [PubMed] [Google Scholar]
- 10.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 11.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 12.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 13.Kow YW, Wallace SS. Mechanism of action of escherichia coli endonuclease III. Biochemistry. 1987;26:8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- 14.Parsons JL, Elder RH. DNA N-glycosylase deficient mice: a tale of redundancy. Mutat Res. 2003;531:165–175. doi: 10.1016/j.mrfmmm.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Ho E, Satoh MS. Repair of single-strand DNA interruptions by redundant pathways and its implication in cellular sensitivity to DNA-damaging agents. Nucleic Acids Res. 2003;31:7032–7040. doi: 10.1093/nar/gkg892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiederholt CJ, Patro JN, Jiang YL, Haraguchi K, Greenberg MM. Excision of formamidopyrimidine lesions by endonucleases III and VIII is not a major DNA repair pathway in Escherichia coli. Nucleic Acids Res. 2005;33:3331–3338. doi: 10.1093/nar/gki655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishchenko AA, Deprez E, Maksimenko A, Brochon JC, Tauc P, Saparbaev MK. Uncoupling of the base excision and nucleotide incision repair pathways reveals their respective biological roles. Proc Natl Acad Sci USA. 2006;103:2564–2569. doi: 10.1073/pnas.0508582103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dianov GL, Sleeth KM, I DI, L AS. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- 20.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 21.Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung JS, Demott MS, Demple B. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase β. J Biol Chem. 2005;280:39095–39103. doi: 10.1074/jbc.M506480200. [DOI] [PubMed] [Google Scholar]
- 23.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. ‘Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Reardon JT, Sancar A. Nucleotide excision repair. Progr Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 26.Lin JJ, Sancar A. A new mechanism for repairing oxidative damage to DNA: (A)BC exonuclease removes AP sites and thymine glycols from DNA. Biochemistry. 1989;28:7979–7984. doi: 10.1021/bi00446a002. [DOI] [PubMed] [Google Scholar]
- 27.Kow YW, Wallace SS, Van Houten B. UvrABC nuclease complex repairs thymine glycol, an oxidative DNA base damage. Mutat Res. 1990;235:147–156. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Weledji YN, Greenberg MM. Independent generation and characterization of a C2′-oxidized abasic site in chemically synthesized oligonucleotides. J Org Chem. 2004;69:6100–6104. doi: 10.1021/jo049033d. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1982. [Google Scholar]
- 30.Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J Biol Chem. 2006;281:26370–26381. doi: 10.1074/jbc.M603093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J Biol Chem. 2006;281:15227–15237. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- 32.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 33.Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J Biol Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 34.Podlutsky AJ, Dianova II, Podust VN, Bohr VA, Dianov GL. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Yj, DeMottt MS, Hwang JT, Greenberg MM, Demple B. Action of human apurinic endonuclease (Ape1) on C1′-oxidized deoxyribose damage in DNA. DNA Repair. 2003;2:175–185. doi: 10.1016/s1568-7864(02)00194-5. [DOI] [PubMed] [Google Scholar]
- 36.Snowden A, Kow YW, Van Houten B. Damage repertoire of the escherichia coli UvrABC nuclease complex includes abasic sites, base -damage analogues, and lesions containing adjacent 5′ or 3′ nicks. Biochemistry. 1990;29:7251–7259. doi: 10.1021/bi00483a013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.