Abstract
Deep pyrosequencing of a CD8+TL epitope from the Tat protein of simian immunodeficiency virus (SIV) from four infected rhesus macaques carrying the restricting MHC allele (Mamu-A*01) for that epitope, revealed that natural selection favoring escape mutations led to an increase in the frequency of haplotypes in the epitope region that differed from the inoculum. After 20 weeks of infection, a new sequence haplotype in the epitope region had increased to a frequency greater than 50% in each of the four monkeys (range 57.9%–98.9%); but the predominant haplotype was not the same in all four monkeys. Thus, even under strong selection favoring escape from CD8+TL recognition, the random nature of mutation itself is the primary factor affecting which escape mutation is likely to become predominant within an individual host. The relationship between the frequency of the inoculum haplotype in the epitope region and time post infection approximated a simple hyperbola. On this assumption, the expected ratio of the frequencies at the inoculum at two times t1 and t2, fi (t2)/fi (t1), will be given by t1 / t2. Because standard phylogenetic methods for reconstructing ancestral sequences failed to predict the inoculum sequence correctly, we used this relationship to predict the inoculum sequence with 100% accuracy, given data on haplotype frequencies at different time periods.
Keywords: cytotoxic T-lymphocyte, escape mutation, positive selection, pyrosequencing, simian immunodeficiency virus
Introduction
Both human immunodeficiency virus 1 (HIV-1), the primary worldwide cause of AIDS and the related simian immunodeficiency virus (SIV), are known to have very high mutation rates, on the order of 10−2 per site per year (Li et al. 1988). Some nonsynonymous (amino acid-altering) mutations may be positively selected because they confer escape from the host’s immune recognition (Evans et al. 1999; Allen et al. 2000; Hughes et al. 2001). Indeed, positively selected nonsynonymous mutations in genomic regions encoding epitopes bound by host class I major histocompatibility complex (MHC) molecules and recognized by CD8+ T-cells (CD8+TL) have been shown to represent a substantial fraction of the variation observed during infection of rhesus macaques (Macaca mulatta) with SIVmac239 (O’Connor et al. 2002, 2004; Hughes et al. 2005). Conventional sequencing studies have revealed substantial nucleotide sequence polymorphism within the population of SIV inhabiting a single host, including both apparently selected and presumably neutral polymorphism (Hughes et al. 2005). However, such techniques were limited in the number of sequences that could be examined; as a consequence, the evolutionary dynamics of escape from CD8+TL recognition could not be examined in detail.
Deep pyrosequencing technology has the potential to provide a much more detailed understanding of the sequence diversity of immunodeficiency virus genomes subject to selection favoring CD8+TL escape mutants (Bimber et al. 2009). Here we use results from this technology to examine the dynamics of sequence evolution in a region of the Tat gene including a well-characterized CD8+TL epitope (SL8) presented by the Mamu-A*01 positive rhesus macaques (Allen et al. 2000). This epitope is subject to strong positive selection favoring escape mutations early in infection (Allen et al. 2000; Hughes et al. 2001). However, this epitope is also subject to constraint on viable escape mutations because it is encoded by a portion of the tat reading frame that overlaps a region of the vpr reading frame, and the latter is subject to purifying selection (Hughes et al. 2001). Comparing the trajectories taken by the evolution of escape in different infected monkeys provides evidence regarding the extent to which such constraints shape the evolution of escape strategies in SIV populations infecting different monkeys.
In natural infections, it may be difficult to reconstruct ancestral sequences in genomic regions where escape mutations occur. In the present case, because all monkeys were infected with an identical known inoculum, we test whether knowledge of the dynamics of allele frequency change can be used to identify the ancestral sequence, even when that sequence is at low frequency in the viral population infecting a given host. We apply a simple method for reconstructing the ancestral sequence that depends on the dynamics of haplotype frequency change rather than on phylogenetic reconstruction. Phylogenetic reconstruction of transmitted/founder virus has been used in HIV-1 infections for sequences of the complete env gene (Keele et al. 2008) or the complete genome (Salazar-Gonzalez et al. 2009). However, phylogenetic analysis is unlikely to be able to provide adequate resolution in the case of the short sequences provided by pyrosequencing technology, especially under positive selection which promotes parallel amino acid changes (Hughes et al. 2001). Thus, our method may be useful in the analysis of pyrosequencing data from SIV and other viruses in which escape from CD8+TL recognition occurs regularly.
Methods
Sequencing Methods
Deep pyrosequencing (Bimber et al. 2009) was applied to a region from the tat gene of SIV from seven infected rhesus macaques (rh2122, rh2124, rh2126, rh2127, r01008, r00014, and r97113). The inoculum sequence of this region encoded the Mamu-A*01-encoded epitope SL8 (STPESANL), and all seven monkeys were A*01-positive. Sequencing was applied to samples taken 1, 2, 3, 4, 8, and 20 weeks post-infection from four animals (rh2122, rh2124, rh2126, and rh2127) and at 88 weeks (r00014), 201 weeks (r91113), and 221 weeks post-infection from individual animals. In sequence analyses a region of 144 nucleotides was analyzed; a small proportion of sequence reads that did not cover this entire region, and were excluded from statistical analyses.
Statistical Methods
We used Nei and Gojobori’s (1986) method to estimate dN and the number of synonymous substitutions per synonymous site (dS). In preliminary analyses, the methods of Li (1993) and Yang and Nielsen (2000) yielded essentially identical results, as expected because the number of substitutions per site was low in this case (Nei and Kumar 2000). We use the term “haplotype” to designate a specific combination of amino acid residues at the eight amino acid positions corresponding to the SL8 epitope. The distance between two populations of sequences with respect to the distribution of haplotype frequencies at the SL8 epitope was computed by the formula:
where n is the number of haplotypes and x1 and y1 are the frequencies of the ith haplotype in the two populations, respectively (Nei 1987, p. 216).
We used allometric regression to construct a model predicting the frequency of the inoculum sequence fi(t) as a function of time in weeks (t) post infection. This method, long a standard technique in biostatistics, involves linear regression the natural logarithm of a dependent variable (Y) on the natural logarithm of the independent variable (X). The resulting linear regression equation has the form ln(Y) = a + b ln(X). This same equation can then be re-expressed in exponential form as Y = eaXb, where e is the base of the natural logarithms (Sokal and Rohlf 1981).
Results
Amino Acid Changes
Deep pyrosequencing was used to monitor sequence evolution in the region of the SL8 epitope in four infected rhesus macaques at 1, 2, 3, 4, 8, and 20 weeks post-infection. Viral load data were collected from each animal at each of these times. The overall mean viral load was 1.55 × 106 copies/ml ± 0.45 × 106 S.E., with a range from 2.25 × 103 to 3.39 × 106 copies/ml; and the median was 3.71 × 105 copies/ml. There was not a significant difference in mean (ANOVA) or median (Kruskal-Wallis test) viral load among time points.
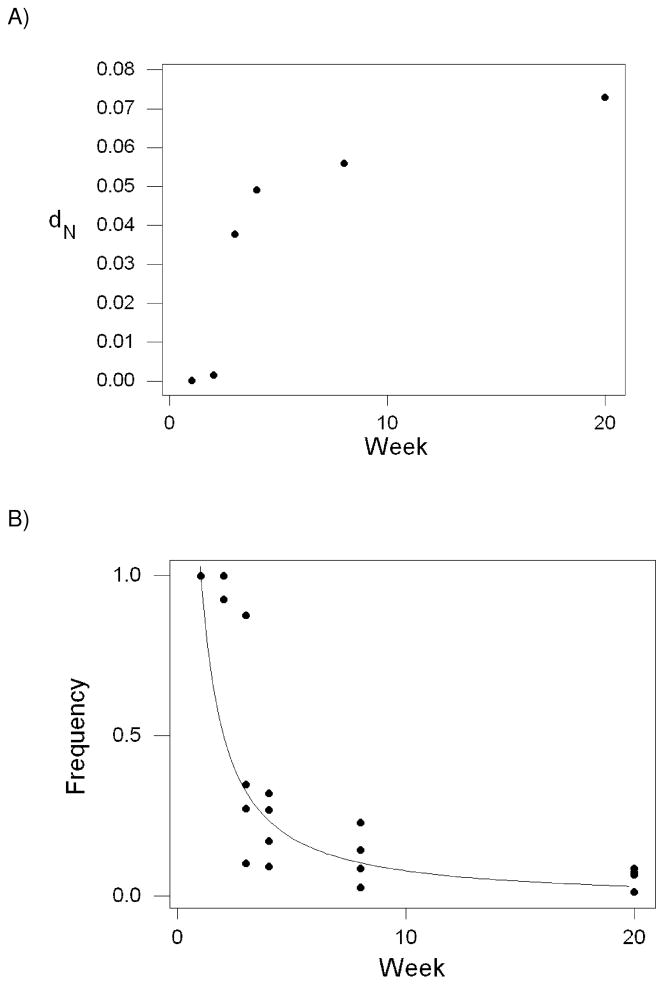
Few nucleotide sequence differences from the inoculum sequence, whether synonymous or nonsynonymous, were observed in the remainder of the region analyzed, exclusive of the SL8 epitope. And within the SL8 epitope, no synonymous differences from the inoculum were observed before week 20; and these occurred in only two of the monkeys (rh2126 and rh2127). On the other hand, in the codons encoding the SL8 epitope itself, there was a steady increase in the mean number of nonsynonymous substitutions per nonsynonymous site (dN) in comparison to the inoculum sequence (Figure 1A). The most dramatic increase in mean dN occurred between weeks 2 (0.0015) and 3 (0.0377), a 25-fold increase (Figure 1A).
Figure 1.
(A) Mean number of nonsynonymous substitutions per nonsynonymous site (dN) versus inoculum sequence in the Tat SL8 epitope of SIV samples from four rhesus macaques as a function of weeks post-infection. (B) Frequency of the inoculum sequence, fi (t),as a function of weeks post-infection. The curve is the exponential regression fi (t) = 1.20 t −1.14 (R2 = 71.7%; P < 0.001).
As a consequence of nonsynonymous substitutions in the SL8 epitope region, the population frequency of the original inoculum sequence (STPESANL) decreased rapidly over the same period, dropping from an initial frequency of 100% in all four monkeys to a mean frequency of 5.88 % (± 1.64 % S.E.; Figure 1B). Numerous alternative haplotypes were present by the second week in one monkey (rh2122) and by the third week in all other monkeys (Table 1). Eventually, a new predominant haplotype replaced the inoculum haplotype as the most frequent, reaching over 50% frequency in each monkey by week 20; but this new predominant haplotype was not the same in all four monkeys (Table 1). The pattern of increase in frequency of the new predominant haplotype differed markedly between monkeys. In rh2126, for example, the PTPSANL haplotype was already present at 60% frequency by week 3 and had reached almost 99% frequency by week 20 (Table 1). By contrast, in rh2127, the STPESAKP haplotype did not appear in detectable frequency until week 8 but had reached 72% frequency by week 20 (Table 1).
Table 1.
Frequencies of commonly occurring haplotypes1 in the Tat SL8 epitope of SIV infecting four Mamu-A*01-positive rhesus monkeys.
| Monkey | Haplotype | Week |
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 8 | 20 | ||
| rh2122 | STPESANL | 100.0% | 92.7% | 27.2% | 17.1% | 14.2% | 6.6% |
| - - - - L - - - | 0.0% | 1.9% | 17.9% | 15.9% | 14.6% | 0.9% | |
| -I - - - - - - | 0.0% | 0.0% | 32.5% | 39.5% | 22.4% | 10.6% | |
| P - - - - - - - | 0.0% | 0.0% | 14.8% | 23.7% | 41.0% | 80.7% | |
| Others (N = 22) | 0.0% | 5.4% | 7.6% | 3.8% | 7.8% | 1.0% | |
| rh2124 | STPESANL | 100.0% | 100.0% | 87.6% | 32.1% | 22.9% | 8.6% |
| -I - - - - - - | 0.0% | 0.0% | 5.5% | 23.4% | 33.4% | 57.9% | |
| P - - - - - - - | 0.0% | 0.0% | 3.3% | 28.6% | 21.7% | 17.3% | |
| Others (N = 27) | 0.0% | 0.0% | 3.6% | 15.9% | 22.0% | 16.2% | |
| rh2126 | STPESANL | 100.0% | 100.0% | 10.2% | 9.2% | 2.6% | 1.1% |
| P - - - - - - - | 0.0% | 0.0% | 60.1% | 82.8% | 97.4% | 98.9% | |
| Others (N = 28) | 0.0% | 0.0% | 29.7% | 8.0% | 0.0% | 0.0% | |
| rh2127 | STPESANL | 100.0% | 100.0% | 34.8% | 26.8% | 8.4% | 7.3% |
| -I - - - - - - | 0.0% | 0.0% | 11.3% | 14.4% | 1.1% | 0.0% | |
| - - - - - - -P | 0.0% | 0.0% | 3.7% | 14.4% | 31.6% | 1.5% | |
| - - - - - - -R | 0.0% | 0.0% | 3.5% | 7.6% | 17.7% | 0.4% | |
| - - - - - - -KP | 0.0% | 0.0% | 0.0% | 0.0% | 1.2% | 72.0% | |
| P- - - - - - - | 0.0% | 0.0% | 24.1% | 19.8% | 22.5% | 5.6% | |
| Others (N = 54) | 0.0% | 0.0% | 22.6% | 17.0% | 17.5% | 13.2% | |
Haplotypes included for a given monkey are those that were present at a frequency of 10% or greater in at least one week’s sample from that monkey.
In order to examine the rate of decline in frequency of the inoculum haplotype, we fitted a allometric (logarithmic) regression to the relationship between the frequency of the inoculum sequence at time t, fi (t), and time (t) in weeks. We obtained the following relationship: fi (t) = 1.20 t −1.14 (R2 = 71.7%; t-test of the hypothesis that the exponent equals zero: t = −7.79; 22 d.f.; P < 0.001). The exponent (−1.14) was not significantly different from −1.0 (t = −0.95; 22 d.f.; N.S.); thus, the fitted function is close to a simple hyperbola.
As a qualitative test of the predictive value of this relationship, we examined sequences from three additional A*01-positive monkeys at 88, 201, and 221 weeks post-infection, respectively. In the monkey examined at 88 weeks, no inoculum sequences were observed (predicted 0.0073). In the monkey examined at 201 weeks, the observed frequency of the inoculum sequence was 0.0303 (predicted 0.0028); and at 221 weeks, the observed frequency was 0.0407 (predicted 0.0026). Thus, even outside the range of the original observations on which the regression was based, the observed values were not very far from the predicted values.
Differences among Monkeys
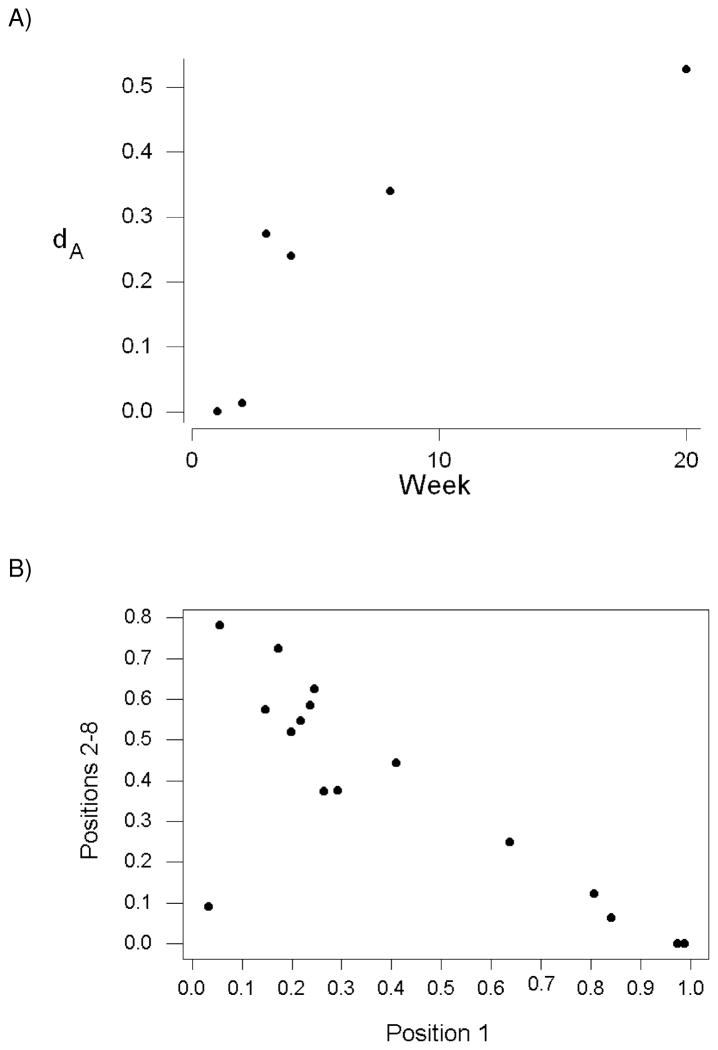
We computed the pairwise genetic distance (dA) with respect to the haplotype frequencies in the SL8 region among monkeys for each week of collection. The mean dA was positively correlated with weeks post-infection (Figure 2A; r = 0.872; P = 0.024). Thus, although the sequences of SL8 region in the SIV population within each monkey were diverging from that of the inoculum, they were also diverging from the sequence populations in the other monkeys.
Figure 2.
(A) Average genetic distance (dA) based on haplotype frequencies of amino acid sequence in the Tat SL8 epitope as a function of weeks post-infection (r = 0.872; P = 0.024). (B) In samples from weeks 3–20, frequency of haplotypes with amino acid changes at position 1 of the epitope versus frequency of haplotypes with amino acid changes in positions 2–8 of the epitope (r = −0.780; P < 0.001).
An important factor in the divergence among monkeys was related to the portion of the SL8 epitope where the amino acid changes occurred. Within each monkey from weeks 3 through 20, there was a strong negative correlation between the frequency of haplotypes with amino acid changes at position 1 of the epitope and the frequency of haplotypes with amino acid changes in positions 2–8 of the epitope (Figure 2B; r = −0.780; P < 0.001). This relationship implied that in any sample in which the frequency of haplotypes with changes at position 1 of the epitope was high, the frequency of haplotypes with changes in positions 2–8 was low; and vice versa. There was one point, corresponding to week 3 from monkey rh2124, with low frequencies of both haplotypes with a change in position 1 (3.3%) and haplotypes with changes in positions 2–8 (9.1%; Figure 2B). This outlier evidently occurred because the frequency of the inoculum sequence was still high (87.6%) in monkey rh2124 at week 3. When only data from weeks 4 through 20 were included, the negative correlation between the frequency of haplotypes with amino acid changes at position 1 of the epitope and the frequency of haplotypes with amino acid changes in positions 2–8 of the epitope became even stronger (r = −0.971; P < 0.001).
By week 20, there was a predominant haplotype in each monkey which differed from the inoculum at one or two amino acid positions (Table 1). In both rh2122 and rh2126, the predominant haplotype was PTPESANL (Table 1). In rh2124, SIPESANL was the predominant haplotype, while in rh2127 STPESAKP was the predominant haplotype (Table 1). Interestingly, the PTPESANL haplotype, which became the predominant haplotype independently in two monkeys, requires a nonsynonymous mutation in the vpr reading frame (leading to I → T replacement), whereas the other two predominant involved only synonymous changes in the vpr reading frame. As a consequence of increase in frequency of these synonymous mutations, in rh2124 and rh2127, there was a strong positive correlation between dS vs. the inoculum in the vpr reading frame and week post-infection (r = 0.840; P < 0.001). By contrast, in rh2122 and rh2126, there was no correlation between dS vs. the inoculum in the vpr reading frame and week post-infection (r = −0.092; n.s.).
Inferring the Ancestral Epitope Sequence
We used standard phylogeny-based methods, using both parsimony and maximum likelihood, to infer the ancestral sequence of the sequenced region based on the sequences available from deep sequencing. None of these methods were able to reconstruct the true ancestral sequence accurately (not shown). Therefore, we implemented a method based on the relationship between time and the frequency of the inoculum sequence that might be used to infer that sequence if it were unknown. First, we note that in the fitted relationship, fi (t) = 1.20 t −1.14, the exponent was not significantly different from −1.0. Assuming an exponent of −1.0, the expected ratio of the frequencies at the inoculum at two times t1 and t2, fi (t2)/fi (t1), will be given by t1 / t2.
In order to infer the inoculum sequence, we searched for a haplotype that was decreasing monotonically over time. Suppose we are given only the samples from weeks 4, 8, and 20. In the case of monkeys rh2126 and rh2127, there was only one haplotype that showed a decrease in frequency from week 4 to 8 and from week 8 to 20; namely the inoculum haplotype (STPESANL). Therefore, in these cases, it was easy to infer the inoculum haplotype, since it was the only one to show a decrease over the three time points examined.
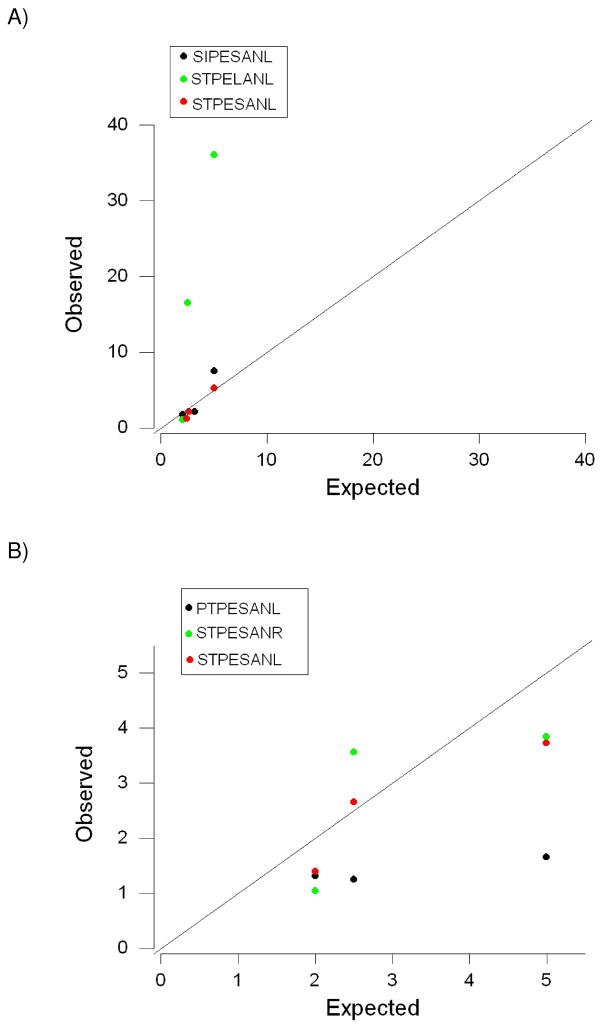
In the case of monkey rh2122, there were three haplotypes that showed a decrease in frequency from week 4 to week 8 and from week 8 to week 20: SIPESAL, STPELANL, and STPESANL (Figure 3A). In order to decide which of these was the inoculum, we computed the sum of the squared deviations of observed frequency ratios from expected frequency ratios (Figure 3A). The sum of squared deviations was much lower for STPESANL (0.788) than for STPELANL (1167.600) or for SIPESANL (6.398; Figure 3A), identifying STPESANL as the inoculum. In the case of monkey rh2124, there were again three haplotypes that showed a decrease in frequency from week 4 to 8 and from week 8 to week 20: PTPESANL, STPESANL, and STPESANR (Figure 3B). The sum of squared deviations of observed frequency ratios from expected frequency ratios was lower for STPESANL (1.973) than for STPESANR (3.669) or PTPESANL (13.195; Figure 3B), again identifying STPESANL as the inoculum. Therefore, this simple method was able to identify the inoculum sequence with perfect accuracy.
Figure 3.
Observed versus expected ratios of haplotype frequencies at weeks 4, 8, and 20 post-infection in monkeys (A) rh2122 and (B) rh2124.
Discussion
Deep pyrosequencing of a CD8+TL epitope from the Tat protein of simian immunodeficiency virus (SIV) from four infected monkeys, each of which carried Mamu-A*01, revealed that natural selection favoring escape mutations led to an increase in the frequency of haplotypes in the epitope region that differed from the inoculum. After 20 weeks of infection, a new sequence haplotype in the epitope region had increased to a frequency greater than 50% in each of the four monkeys (range 57.9%–98.9%); but the predominant haplotype was not the same in all four monkeys. In fact, three different predominant epitopes were seen in the four monkeys; thus the same haplotype (PTPESANL) became predominant in two of the monkeys but not in the others.
The epitope studied is subject to particular functional constraint because it is in a region of the tat reading frame that overlaps the vpr reading frame, and this constraint may limit the number of escape mutations that are viable and thus likely to increase in frequency (Hughes et al. 2001). In the present case, the haplotype that became predominant independently in two monkeys (PTPESANL) involved a nonsynonymous change in the vpr reading frame, while the other two only involved synonymous changes in the vpr reading frame. Thus, although the presence of overlapping reading frames may have to some extent limited the range of viable escape mutations, such constraints did not lead to occurrence of a unique escape response in all four monkeys. Rather, the results seem most consistent with the hypothesis that, even under strong selection favoring escape from CD8+TL recognition, the random nature of mutation itself is the primary factor affecting which escape mutation is likely to become predominant within an individual host. This result is consistent with results of artificial selection experiments with viruses such as the bacteriophage ϕX174, in which different replicate lineages showed distinct mutational patterns in response to selection for growth at high temperature (Wichman et al. 1999).
Because the inoculum sequence was known in the present case, we used our data to test the feasibility of reconstructing the inoculum sequence from the sequence samples uncovered by deep pyrosequencing. We found that phylogenetic methods of reconstructing ancestral sequences were of no use in this case, probably because the sequences analyzed were short (144 nucleotides) and because parallel amino acid substitutions often occur in response to selection on this epitope (Hughes et al. 2001). However, we found that a method based on the dynamics of haplotype frequency changes was able to reconstruct the ancestral sequence with perfect accuracy.
An empirically derived formula for the frequency of the inoculum haplotype at time t in weeks, fi (t), was the following: fi (t) = 1.20 t −1.14. Because the exponent (−1.14) was not significantly different from −1.0, this relationship approximated a simple hyperbola. On this assumption, the expected ratio of the frequencies at the inoculum at two times t1 and t2, fi (t2)/fi (t1), will be given by t1 / t2. This relationship can be used to predict the inoculum haplotype given data on haplotype frequencies at different time periods in a CD8+TL epitope subject to natural selection favoring escape mutations.
First, the inoculum haplotype should be expected to be decreasing monotonically over time. In two of the four monkeys, there was only one haplotype decreasing monotonically over the data from weeks 4, 8, and 20; and this haplotype was the inoculum haplotype. In the other two monkeys, we compared the ratios fi (t2)/fi (t1) with the expected ratio t1 / t2, computing the sum of squared deviations of observed from expected values. In each case, the haplotype with the lowest sum of squared deviations was the inoculum sequence. As in the present case, this method may work best with data from at least three time points, but in many cases it may provide the correct answer with only two time points.
The Tat SL8 epitope is highly immunodominant and subject to strong selection favoring escape; as a result it is very rapidly evolving in Mamu-A*01 positive hosts (Allen et al. 2000). Other CD8+TL epitopes of SIV are not so rapidly evolving (O’Connor et al. 2002). One factor that can slow the evolution of escape in CD8+TL epitopes may be the requirement for compensatory mutations in cases where the escape mutation is deleterious to the virus (Friedrich et al. 2004; Peyerl et al. 2004; Yeh et al. 2006). In HIV-1 infection in humans, Goonetilleke et al. (2009) identified three different patterns of escape from CD8+TL recognition: simple escape (where a single haplotype quickly replaces the transmitted haplotype); rapid complex escape (where numerous mutated haplotypes appear quickly, with a dominant haplotype eventually emerging); and delayed complex escape (characterized by slow increase of a dominant haplotype without the rapid appearance of numerous mutated haplotypes).
The evolution of the Tat SL8 epitope fit complex escape category, at least in three of the four monkeys (Table 1). However, in rh2126, a predominant escape mutant was already present at 60% frequency by week 3, although pyrosequencing revealed numerous other escape mutants at low frequencies (Table 1). Thus, the pattern of evolution in rh2126 seemed was more like that of simple escape (Goonetilleke et al. 2009). The occurrence of very different trajectories of escape at the same epitope in four different Mamu-A*01-positive hosts (Table 1) suggests that the pattern of escape is not a property of a given epitope alone but may vary from host to host. In addition, since we did not have data for the four monkeys past 28 weeks, we could not rule out the possibility that the haplotype predominant at 28 weeks might in some cases be replaced later by yet another haplotype, adding further complexity to the process.
While the current method was efficacious in identifying the inoculum in the case of the SL8 epitope, it may be also possible to adapt the same approach to other epitopes that evolve more slowly than Tat SL8. Note that, as long as the exponent (−b) of the relationship between fi(t) = a t −b is approximately equal to −1.0, the ratio fi (t2)/fi (t1) is expected to be approximately t1 / t2 no matter what the value of a. Moreover, the case where the exponent is equal to −1.0 constitutes a special case of a more general relationship. In general, fi (t2)/fi (t1) will be equal to t2b/t1b. Different rates of elimination of the inoculum haplotype will be reflected by different values of the coefficient a and/or of the exponent (−b). In cases where one does not know the exponent, a number of different values of −b, ranging from about −0.5 to −1.0, might be tried, on the assumption that, if another epitope differs in rate from Tat SL8, it is likely to evolve more slowly that Tat SL8 rather than more rapidly (O’Connor et al. 2002). Application of exponents in the range −0.5 to −1.0 to the present yielded correct identification of the inoculum haplotype (not shown), suggesting that the correct epitope is likely to be identified even if the exponent is only approximately known.
In the case of human infection with HIV-1, the sequence of the initially infecting virus is not generally known; yet a knowledge of this sequence may be useful both for epidemiological and therapeutic purposes (Bull and Wichman 2001; Kaye et al. 2008; Pistello et al. 2004; Yerly et al. 2004). The pyrosequencing technique used here showed advantages in its ability to provide a picture of viral diversity within a host, but involves such short sequences that phylogenetic methods (Keele et al. 2008; Salazar-Gonzalez et al. 2009) may not provide sufficient resolution to determine the ancestral sequence. Unlike experimental SIV infections, data as early as the first few weeks after infection are rarely available in the case of HIV-1. However, as the results of Goonetilleke et al. (2009) show, haplotype frequencies at CD8+TL epitopes of HIV-1 may continue to evolve over the first several years after infection. Given pyrosequencing data on epitopes in the process of escape, the present results suggest that the dynamics of sequence haplotype frequency change may yield insights regarding the source of infection.
Acknowledgments
This work was supported by NIH grant numbers 1 R01 AI077376-01 and 1 R21 AI082880-01 to D.H.O. and by 1 R01 GM GM43940-21 to A.L.H. This publication was made possible in part by Grant Number P51 RR000167 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), to the Wisconsin National Primate Research Center, University of Wisconsin-Madison. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01. This publication’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen TM, O’Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Leibl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- Bimber BN, Burwitz BJ, O’Connor S, Detmer A, Gostick E, Lank SN, Price D, Hughes A, O’Connor D. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J Virol. 2009;83:8247–8253. doi: 10.1128/JVI.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Wichman HA. Applied evolution. Annu Rev Ecol Syst. 2001;32:183–217. [Google Scholar]
- Evans DT, O’Connor DH, Jing P, Dzuris JL, Sidney J, Da Silva J, Allen TM, Horton H, Venham JE, Rudersdorf RA, Vogel T, Pauza CD, Bontrop RE, DeMars R, Sette A, Hughes AL, Watkins DI. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nature Medicine. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- Friedrich TC, Frye CA, Yant LJ, O’Connor DH, Kriewaldt NA, Benson M, Vojnov L, Dodds EJ, Cullen C, Rudersdorf R, Hughes AL, Wilson N, Watkins DI. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J Virol. 2004;78:2581–2585. doi: 10.1128/JVI.78.5.2581-2585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke N, Liu MK, Salazar-Gonzaler JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Pereslson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ CHAVI Clinical Core B. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Westover K, da Silva J, O’Connor DH, Watkins DI. Simultaneous positive and purifying selection on overlapping reading frames of the tat and vpr genes of simian immunodeficiency virus. J Virol. 2001;75:7666–7672. doi: 10.1128/JVI.75.17.7966-7972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Piontkivska H, Krebs KC, O’Connor DH, Watkins DI. Within-host evolution of CD8+-TL epitopes encoded by overlapping and non-overlapping reading frames of simian immunodeficiency virus. Bioinformatics. 2005;21(Suppl 3):iii39–iii44. doi: 10.1093/bioinformatics/bti1203. [DOI] [PubMed] [Google Scholar]
- Kaye M, Chibo D, Birch C. Phylogenetic investigation of transmission pathways of drug-resistant HIV-1 utilizing pol sequences derived from resistance genotyping. J Acquir Immune Defic Syndr. 2008;49:9–16. doi: 10.1097/QAI.0b013e318180c8af. [DOI] [PubMed] [Google Scholar]
- Keele BF, Giorgi EF, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kircherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Han BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Unbiased estimates of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Li WH, Tanimura M, Sharp PM. Rates and dates of divergence between AIDS virus nucleotide sequences. Mol Biol Evol. 1988;5:313–330. doi: 10.1093/oxfordjournals.molbev.a040503. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- O’Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, Dunphy EJ, Melsaether C, Mothé B, Yamamoto H, Horton H, Wilson N, Hughes AL, Watkins DI. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nature Medicine. 2002;8:493–499. doi: 10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, McDermott AB, Krebs AC, Dodds EJ, Miller JE, Gonzalez EJ, Jacoby TJ, Yant L, Piontkivska H, Pantophlet R, Burton DR, Rehrauer WR, Wilson N, Hughes AL, Watkins DI. A dominant role for CD8+-T-lymphocyte selection in Simian Immunodeficiency Virus sequence variation. J Virol. 2004;78:14012–14022. doi: 10.1128/JVI.78.24.14012-14022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyerl FW, Bazick HS, Newberg MH, Barouch DH, Sodroski J, Letvin NL. Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope. J Virol. 2004;78:13901–13910. doi: 10.1128/JVI.78.24.13901-13910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistello M, Del Santo B, Buttò S, Barganga M, Domenici R, Bendinelli M. Genetic and phylogenetic analyses of HIV-1 corroborate the transmission link hypothesis. J Clin Virol. 2004;30:11–18. doi: 10.1016/j.jcv.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 2. W.H. Freeman; San Francisco: 1981. [Google Scholar]
- Wichman HA, Badgett MR, Scott LA, Bouliane CM, Bull JJ. Different trajectories of parallel evolution during viral adaptations. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yeh WW, Cale EM, Jaru-Amponpan P, Lord CI, Peyerl FW, Letvin NL. Compensatory substitutions restore normal core assembly in simian immunodeficiency virus isolated with gag epitope cytotoxic T-lymphocyte escape mutations. J Virol. 2006;80:8168–8177. doi: 10.1128/JVI.00068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerly S, Jost S, Telent A, Flepp M, Kaiser L, Chave JP, Vernazza P, Battegay M, Furrer H, Chanzy B, Burgisser P, Rickenbach M, Gebhardt M, Bernard MC, Perneger T, Hirschel B, Perrin L Swiss HIV Cohort Study. Infrequent transmission of HIV-1 drug-resistant variants. Antivir Ther. 2004;9:375–384. [PubMed] [Google Scholar]