Abstract
An effective immune response to vaccination is, in part, a complex interaction of alleles of multiple genes regulating cytokine networks. We conducted a genotyping study of Th1/Th2/inflammatory cytokines/cytokine receptors in healthy children (n=738, 11–19 years) to determine associations between individual single-nucleotide polymorphisms (SNPs)/haplotypes and immune outcomes after two doses of rubella vaccine. SNPs (n=501) were selected using the ldSelect-approach and genotyped using Illumina GoldenGate™ and TaqMan assays. Rubella-IgG levels were measured by immunoassay and secreted cytokines by ELISA. Linear regression and post hoc haplotype analyses were used to determine associations between single SNPs/haplotypes and immune outcomes. Increased carriage of minor alleles for the promoter SNPs (rs2844482 and rs2857708) of the TNFA gene were associated with dose-related increases in rubella antibodies. IL-6 secretion was co-directionally associated (p≤0.01) with five intronic SNPs in the TNFRSF1B gene in an allele dose-related manner, while five promoter/intronic SNPs in the IL12B gene were associated with variations in IL-6 secretion. TNFA haplotype AAACGGGGC (t-statistic=3.32) and IL12B promoter haplotype TAG (t-statistic=2.66) were associated with higher levels of (p≤0.01) rubella-IgG and IL-6 secretion, respectively. We identified individual SNPs/haplotypes in TNFA/TNFRSF1B and IL12B genes that appear to modulate immunity to rubella vaccination. Identification of such “genetic fingerprints” may predict the outcome of vaccine response and inform new vaccine strategies.
Keywords: Rubella, Vaccine, Antibody, Cytokine, SNP, Haplotype
Introduction
In the United States, the licensed rubella vaccine contains the attenuated RA 27/3 strain. While safe and effective (the current two-dose vaccination schedule results in seroconversion rates over 95%), a small minority of individuals do not seroconvert or develop suboptimal antibody titers after vaccination (Boulianne et al. 1995; Singh et al. 1994; Mitchell et al. 1999; Chu et al. 1988; Hillary and Griffith 1984). These individuals may not be protected against rubella. In spite of the widespread use of rubella vaccine, individual cases and outbreaks continue to occur, including a recent case in Minnesota (Reef et al. 2002) (http://www.health.state.mn.us/news/pressrel/2009/rubella041709.html).
Humoral responses to rubella immunization have been well-studied, and their role in disease protection is well-established (Kurtzman et al. 1989). However, cell-mediated immunity (CMI) can also confer protection against disease and is crucial in the resolution of many viral infections (Horstmann et al. 1985; Ovsyannikova et al. 2005; Bautista-López et al. 2000). One hallmark of CMI is the production of key cytokines which shape and coordinate immune responses. Cytokines are essential for the development and functioning of both the innate and adaptive immune responses (Haring et al. 2006; Chabalgoity et al. 2007). Cytokine networks are tightly regulated as they mediate their immunomodulatory effects through specific receptors which are regulated by negative and positive feedback mechanisms (Haring et al. 2006; Chabalgoity et al. 2007; Smith and Humphries 2009). Innate immunity is regulated by inflammatory cytokines such as IL-6, TNF-α, and GM-CSF. Adaptive immunity is characterized by two broad patterns of cytokine production by T cells: Th1 responses are marked by secretion of IFN-γ and IL-2 and robust cytotoxic T cell activity; and Th2 responses are defined by the production of IL-4, IL-5, and IL-10 with the subsequent generation of humoral immunity. There is considerable cross-regulation of humoral and CMI responses by cytokines, and variations in cytokine levels can have a profound impact on these responses (Rolph and Ramshaw 2003; Smith et al. 1998). Comparatively little is known about how cytokines control immune responses to rubella virus.
Genetic variations such as single-nucleotide polymorphisms (SNPs) in the cytokine and cytokine receptor genes can result in imbalances in the cytokine milieu by affecting multiple aspects of cytokine biology, such as transcriptional activity, protein production, receptor binding, and functional activity (Keen 2002; Hollegaard and Bidwell 2006). The coding region SNPs can interfere with protein transcription and translation by altering the amino acid sequences and subsequent protein structure, which, in turn, can result in a nonfunctional downstream product. Promoter and regulatory regions SNPs may alter the transcriptional activity of corresponding genes. Cytokine receptor SNPs can similarly alter attributes of the receptor proteins and impact cytokine functionality (van Deventer 2000; Hackstein et al. 2001; van de Vosse et al. 2003). Functional SNPs and their haplotypes that affect gene expression and activity can subsequently influence disease outcome (Hollegaard and Bidwell 2006; van Deventer 2000; Bidwell et al. 1999, 2001; Haukim et al. 2002).
In the current study, we examined associations between SNPs and the resulting haplotypes in key cytokine and cytokine receptor genes with both humoral and CMI responses to rubella in healthy, school-age children.
Materials and methods
Study subjects
The details of subject consent and recruitment have been previously described (Ovsyannikova et al. 2004, 2009a, b). Briefly, the study cohort consisted of a combination of two independent age-stratified random cohorts (n=738, ages 11–19 years) of healthy children and young adults from all socioeconomic strata identified from the Minnesota Independent School District 535, Rochester, MN. Inclusion criteria required each participant to have a written medical record of receiving two doses of measles–mumps–rubella-II (MMR-II) vaccine containing the attenuated RA27/3 Wistar strain (TCID50≥1000) of rubella virus (Merck, Whitehouse Station, NJ). The study was approved by the Mayo Clinic Institutional Review Board. Written, informed consent was obtained from subjects’ parents/guardians as well as written assent from age-appropriate children at the time of recruitment in the study. Blood was drawn for sera and peripheral blood mononuclear cell isolation.
Rubella-specific circulating IgG antibody levels
We used the Beckman Coulter Access® Rubella IgG assay on a UniCel® DxI 800 Access® Immunoassay System (Beckman Coulter; Fullerton, CA) to determine rubella-specific IgG antibody levels in serum as previously described (Greenwood et al. 2009). The WHO reference serum was used as a calibrator for the standard curve, and the limit of detection and the cut-off for seronegativity of the assays was 0.5 and 10 IU/mL, respectively. The coefficient of variation of this assay in our laboratory was 6%.
Rubella-specific secreted cytokines
We performed ELISAs to determine rubella-specific secreted cytokines (IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p40, IFN-γ, TNF-α, and GM-CSF) in cell-free culture supernatants using commercially available kits (BD Biosciences Pharmingen, San Diego, CA, USA) using a response-surface methodology approach to pre-optimize conditions for each cytokine as previously described (Ovsyannikova et al. 2009a, b; Ryan et al. 2009).
TagSNP selection from candidate cytokine and cytokine receptor genes
We generated a list (n=32) of candidate cytokine (IL2, IL4, IL5, IL6, IL10, IL12A, IL12B, IFNA1, IFNA2, IFNA21, IFNB1, IFNG, TNFA, and CSF2) and cytokine receptor (IL2RA, IL2RB, IL2RG, IL4R, IL6R, IL6ST, IL10RA, IL10RB, IL12RB1, IL12RB2, IL18R1, IFNAR1, IFNAR2, IFNGR1, IFNGR2, TNFRSF1A, TNFRSF1B, and CSF2RB) genes involved in Th1, Th2, and inflammatory responses to rubella (Penn and Williams 1984; Tilles et al. 1987; Nakayama et al. 1988; van der Logt and van Loon 1980; Pukhalsky et al. 2003; Akaboshi et al. 2001; Adamo et al. 2008). TagSNPs within and 10 kb upstream and downstream of the 32 candidate genes were selected using the linkage disequilibrium (LD) tagSNP selection approach (Carlson et al. 2004) from the Hapmap Phase II (http://www.hapmap.org), Seattle SNPs (http://pga.mbt.washington.edu/), and NIEHS SNPs (http://egp.gs.washington.edu/). The selection criteria included: SNPs with validation data, successful predictive genotyping scores for Illumina GoldenGate assays, a minor allele frequency (MAF) ≥0.05 and a pairwise LD threshold of r2≥0.90 for Caucasians. Using ldSelect, we binned the tagSNPs by priority of best source of genotypes (the source with the higher number of LD bins for Caucasian samples). In case of a tie, HapMap was used as the best source. The ldSelect selection output files were further refined by post-processing using the SNPPicker program developed to optimize and overcome the Illumina platform constraints on SNPs closer than 60 bp. Finally, we preferentially forced a set of functional “obligate” SNPs to the final list based on a literature review with the preference order of: nonsynonymous coding, synonymous coding, and 5′ or 3′ untranslated regions using SNPPicker. Our SNP selection algorithm resulted in 501 candidate SNPs for genotyping. We used the nomenclature described by den Dunnen and Antonarakis (den Dunnen and Antonarakis 2001) for the genotype variants.
Genotyping methods
Genomic DNA was extracted from frozen blood clots using the Puregene extraction kit (Gentra Systems Inc., Minneapolis, MN) and was quantified using the Picogreen method (Molecular Probes).
The 501 candidate SNPs from cytokine and cytokine receptor genes were genotyped using a custom-designed 768-plex Illumina GoldenGate™ assay (Illumina Inc., San Diego, CA) along with SNPs from other gene families of interest. All SNPs had Illumina design scores >0.4, and all DNA samples (n=738) were genotyped following the manufacturer’s instructions as previously described (Dhiman et al. 2008). The BeadStudio 2 software was used to call genotypes. We used Corriel Trio DNA and two other genomic DNA controls for the replicate and inheritance data to review and refine clustering. For initial laboratory quality assurance, we qualified only the samples that had Illumina 10% GenCall scores >0.4 and call rates >90%. BeadStudio genotype data were transferred electronically to SAS for further analysis.
SNPs (n=46) that failed genotyping on the Illumina platform were genotyped using PCR-based TaqMan custom-designed assays (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. The reactions were analyzed on the ABI Prism 7900 using Sequence Detection Software (Applied Biosystems).
Statistical methods
We assessed associations between genetic variation in candidate SNPs and levels of rubella-specific IgG antibody levels and cytokine secretion. Assessments of antibody levels resulted in one recorded value per individual. Assessments of cytokine secretion resulted in six recorded values per individual for each outcome: three prior to stimulation with rubella virus and three post-stimulation. A single summary measurement per individual was obtained for each outcome by subtracting the median of the three unstimulated values from the median of the three stimulated values. Medians were used instead of means because they tend to be better measures of central tendency in the presence of outlying values typically seen in immune response outcomes such as ours. Data based on these summary measures were descriptively summarized across individuals using frequencies and percentages for categorical variables and medians and interquartile ranges for continuous variables.
Observed genotypes were used to estimate allele frequencies for each SNP and departures from Hardy–Weinberg equilibrium (HWE) were assessed using either a Pearson goodness-of-fit test or, for SNPs with a MAF of less than 5%, a Fisher exact test (Weir 1996). Estimates of pairwise LD based on the r-squared statistic were obtained using Haploview software version 3.32 (Barrett et al. 2005). SNP associations with immune response outcomes were individually evaluated using linear regression models. Simple linear regression was used for rubella antibody levels, which had only one measured value per individual. Repeated measure approaches were implemented for cytokine secretion, simultaneously modeling all six observed measurements. The primary test of significance assessed the degree to which the SNP was associated with stimulation-induced differences in the response and was obtained from the covariate reflecting the genotype-by-stimulation status interaction. We accounted for within-subject correlations without imposing any constraints on the nature of the correlations. Primary tests of association assumed an ordinal SNP effect, based on the number of copies of the minor allele. Due to the relatively large number of comparisons, we expect some of these tests to be statistically significant based on chance alone. We addressed this issue using the false discovery rate (FDR), supplementing the per-SNP p values with corresponding q values. Separate sets of q values were calculated for each outcome of interest.
To further explore genomic regions containing statistically significant single-SNP effects for one or more outcomes of interest, we performed post hoc haplotype analyses. Posterior probabilities of all possible haplotypes for an individual, conditional on the observed genotypes, were estimated using an expectation-maximization algorithm, similar to the method outlined by Schaid et al. (Schaid et al. 2002). This information was used to define haplotype design variables that reflected the number of each of the haplotypes that were expected to be carried by each subject. Analyses were performed on these haplotype design variables using the simple least squares regression approach for antibody levels and the repeated measures approach for the cytokine secretion described above. Because of the imprecision involved in estimating the effects of low-frequency haplotypes, we considered only those occurring with an estimated frequency of greater than 1%. Differences in immune response among common haplotypes were first assessed globally and simultaneously tested for statistical significance using a multiple degree-of-freedom test. Following these global tests, we examined individual haplotype effects. These were performed in the spirit of Fisher’s protected least significant difference test; individual associations were not considered statistically significant in the absence of global significance. Each design variable (and, for the cytokine secretion data, stimulation status, and the corresponding interaction term) was included in a separate regression analysis, effectively comparing immune response levels for the haplotype of interest against all others combined. Due to phase ambiguity, haplotype-specific medians and interquartile ranges could not be calculated. Thus, descriptive summaries were represented using the t-statistics corresponding to the haplotype main effect term for antibody levels or the haplotype-by-stimulation status interaction term for cytokine secretion. Using the above approach, three haplotype regions were examined: TNFA and rubella antibodies, TNFRSF1B and IL-6 cytokine secretion levels, and IL12B and IL-6 cytokine secretion levels, as detailed in the results section.
All of the association analyses described above were adjusted for age at enrollment, race, gender, age at first rubella vaccination, age at second rubella vaccination, and cohort status (cohort 1 versus cohort 2) in order to account for their potential impact on the measured immune responses. Data transformations were used to correct for data skewness in all linear regression models. An inverse normal transformation was used for all cytokine secretion outcome variables, and a log transformation was used for the antibody response measure. Because of the large number of statistical tests, we considered associations statistically significant only if the resulting p value was ≤0.01. All statistical tests were two-sided and, unless otherwise indicated, all analyses were carried out using the SAS software system (SAS Institute, Inc., Cary, NC).
Results
Overall genotyping performance of the study subjects
Our overall genotyping success rate using a combination of the Illumina 768-plex platform and TaqMan custom assays was 94.53% (726 SNPs yielded genotyping data out of 768), and the study sample success rate was 96.75% (714 samples yielded genotyping data out of 738). Forty-six SNPs failed genotyping in Illumina and were re-genotyped in TaqMan custom assays. Overall, three SNPs failed completely for Illumina and TaqMan, resulting in 765 from which 39 SNPs had to be excluded either due to lower call rates (n=6; call rates <95%) or MAF<5% (n=33), resulting in 726 SNPs, of which 501 were cytokine/cytokine receptor SNPs and were included in final analysis for this report. Subject exclusions were made on the basis of inadequate or poor DNA samples (n=6), complete genotyping failure on both platforms (n=4), or low call rates (n=14, call rates <95%). The post-exclusion mean subject call rate was 99.8%, the mean Illumina SNP call rate was 99.9%, the mean TaqMan SNP call rate was 98.2% and the concordance of genotypes among duplicated individuals (including CEPH controls) was 100%. We observed no SNP-specific deviation (p<0.01) from the HWE among the SNPs significantly associated (p≤0.01) with immune response.
Demographic and immunological characterization of the study population
The demographic and immunological characteristics of our combined study cohort of 738 healthy children have been previously described (Ovsyannikova et al. 2009a). Briefly, the cohort was primarily Caucasian (91%) with 54% males and the median age at enrollment was 15 years (Table 1). The median age of the study cohort at first and second rubella immunization was 15 months and 11 years, respectively. The median time elapsed between the second rubella immunization, and enrollment was 5.8 years.
Table 1.
Demographic and immunological variables of the study population (© Mayo Clinic 2010)
| Variable | Median (IQR)a |
|---|---|
| Age at enrollment, years | 15.0 (13.0, 17.0) |
| Median age at first rubella immunization, months (IQRa) | 15.0 (15.0,16.0) |
| Median age at second rubella immunization, years (IQRa) | 11.0 (5.0,12.0) |
| Median time from second rubella immunization to enrollment, years (IQR) | 5.8 (3.8,7.4) |
| Gender, [N (%)] | |
| Males | 396 (53.7) |
| Race, [N (%)] | |
| White | 672 (91.1) |
| Antibody, IU/mL | 34.4 (19.2, 63.7) |
| Inflammatory cytokine responseb, pg/mL | |
| IL-6 (n=713) | 3,681.0 (3,160.0, 4,052.0) |
| TNF-α (n=713) | 29.7 (−7.0, 89.2) |
| GM-CSF (n=711) | 28.0 (23.6, 32.6) |
| Th1 cytokine responseb, pg/mL | |
| IL-2 (n=713) | 17.6 (7.7, 30.5) |
| IFN-γ (n=713) | 8.5 (3.0, 23.4) |
| IL-12p40 (n=711) | 0.0 (−7.1, 7.2) |
| Th2 cytokine responseb, pg/mL | |
| IL-4 (n=691) | 0.3 (−0.3, 1.0) |
| IL-5 (n=691) | 0.5 (0.0, 1.1) |
| IL-10 (n=713) | 4.2 (2.3, 6.7) |
All results are in median (IQR interquartile range within 1st quartile–3rd quartile)
Unless indicated otherwise
Response is defined as the subject-specific median rubella-stimulated response (measured in triplicate) minus the median unstimulated response (also measured in triplicate). Negative values indicated that unstimulated responses were, on average, higher than the response to stimulation
IL-12p40 and GM-CSF were assayed at 18 h, IL-4, IL-5, IL-6, and IL-10 at 24 h, IFN-γ at 2 days and IL-2 at 8 days in response to in vitro stimulation with rubella virus at MOI of 5. TNF-α was also assayed at 8 days in response to in vitro stimulation with rubella virus at MOI of 0.05. PHA (5 μg/mL) was used as a positive control.
The limits of detection for cytokines, based on the lowest standard, were 4.7 pg/mL (IL-6, GM-CSF, IFN-γ, and TNF-α), 7.8 pg/mL (IL-2, IL-4, IL-5, and IL-10) and 31.3 pg/mL (IL-12p40).
The median (IQR) for rubella-specific antibody levels for our study cohort was 34.4 IU/mL (19.2, 63.7 IU/mL). The overall cytokine profile was skewed towards a predominant inflammatory response and has been previously described (Dhiman et al. 2009). The median (IQR) secretion levels of IL-6 and GM-CSF were 3,681.0 (3,160.0; 4,052.0) and 28.0 pg/mL (23.6, 32.6 pg/mL), respectively, and were detected in virtually all the subjects. TNF-α was detected in 69% of the subjects with median (IQR) secretion levels of 29.7 pg/mL (−7.0, 89.2 pg/mL). Rubella-specific secretion of the Th1 cytokines IL-2 and IFN-γ was modest, while IL-12p40 was undetectable and hence excluded from association analysis. We observed a suppression in rubella-specific Th2 response (IL-4 and IL-5) and very low levels of IL-10 secretion (Table 1). Due to extremely low levels of Th2 cytokines (below the level of detection), we did not perform an association analysis between cytokine and cytokine receptor SNPs and the Th2 cytokines (Table 1).
Cytokine and cytokine receptor SNP associations with rubella-specific antibodies
We found six cytokine and cytokine receptor SNPs belonging to inflammatory and Th2 cytokine subsets that were significantly (p≤0.01) associated with an increase/decrease in rubella-specific antibody levels (Table 2). After adjusting for FDR, only four SNPs remained significant at the q-value=0.20. Major allele C for two SNPs located in the 5′ intergenic region containing the promoter and regulatory regions of the TNFA gene [rs2844482 (TNFA-3752 C>T) and rs2857708 (TNFA-9913C>T)], and one SNP located downstream in the 3′ intergenic region of the TNFA gene [rs2256974 (TNFA+11873C>A)] were associated with variations in rubella antibody levels (Table 2). The two TNFA SNPs rs2844482 (p=0.0002; q=0.09) and rs2857708 (p=0.001; q=0.16) in the 5′ promoter region demonstrated significant linkage (r2=0.82) in inheritance. Increased carriage of major allele G for rs7801617 (IL6-8800G>A; p=0.0005; q=0.11) and major allele T for rs4787947 (IL4R-32760T>G) located in the 5′ intergenic regions of IL6 and IL4R genes, respectively, were also associated with a dose-related decrease in rubella antibodies. A propensity towards lower antibody response (p=0.004) was seen with increasing carriage of minor allele A for rs9610 (IL10RA+ 14903G>A) located in the 3′UTR of IL10RA (Table 2).
Table 2.
SNPs in cytokine and cytokine receptor genes associated with rubella-specific antibody responses post-MMR-II vaccination (© Mayo Clinic 2010)
| Gene (variant name) | SNP ID | Location | Genotype | Na | Median, Ab IU/ml (IQR)b | p valuec | q valuec |
|---|---|---|---|---|---|---|---|
| TNFA | rs2844482 | 5′ Intergenic (Promoter) | GG | 505 | 32 (18.6, 59.3) | 0.0002 | 0.09 |
| -3752 C>T | GA | 197 | 41.2 (21.3, 72.6) | ||||
| AA | 12 | 72.5 (41.5, 108.7) | |||||
| IL6 | rs7801617 | 5′ Intergenic (Promoter) | GG | 586 | 33.2 (18.3, 61.1) | 0.0005 | 0.11 |
| -8800G>A | GA | 110 | 43.2 (25.8, 83.2) | ||||
| AA | 8 | 48.0 (31.3, 119.6) | |||||
| TNFA | rs2857708 | 5′ Intergenic (Promoter) | GG | 535 | 32.2 (18.6, 60.1) | 0.001 | 0.16 |
| -9913C>T | GA | 170 | 42.8 (21.3, 73.2) | ||||
| AA | 9 | 60.1 (26.2, 102.4) | |||||
| IL4R | rs4787947 | 5′ Intergenic (Promoter) | AA | 596 | 32.7 (18.4, 62.4) | 0.002 | 0.20 |
| -32760T>G | AC | 112 | 43.2 (26.4, 68.8) | ||||
| CC | 6 | 79.4 (48.5, 107) | |||||
| IL10RA | rs9610 | 3′UTR | GG | 221 | 39.9 (21.3, 69.7) | 0.004 | 0.45 |
| 14903G>A | GA | 356 | 33.2 (18.6, 66.3) | ||||
| AA | 137 | 29.6 (18.3, 52.4) | |||||
| TNFA | rs2256974 | 3′ Intergenic | CC | 474 | 36.7 (20.6, 68.2) | 0.006 | 0.49 |
| 11873C>A | CA | 208 | 31.3 (16.9, 53.3) | ||||
| AA | 16 | 44.3 (19.4, 109.9) |
Values are presented as homozygous major allele/heterozygous/homozygous minor allele
IQR interquartile range
One degree-of-freedom ordinal p value from the linear regression analyses adjusting for age at enrollment, gender, race, age at first and second MMR-II immunizations, and cohort status
Only associations with p≤0.01 are included in the table
TNFA haplotype associations with rubella-specific antibodies
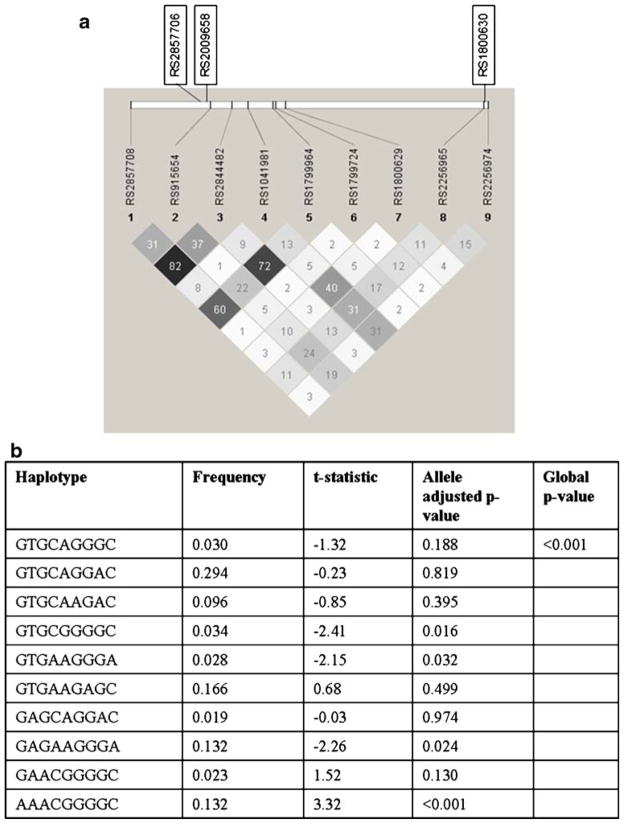
The Haploview output of SNPs genotyped within the TNFA gene and their common haplotypes associated with rubella antibodies are shown in Fig. 1. Carriage of TNFA haplotype AAACGGGGC was significantly associated (p<0.001) with higher (t-statistic=3.32) rubella antibodies, while the presence of GTGCGGGGC, GTGAAGGGA, and GAGAAGGGA were associated with lower levels of (p values between 0.02 and 0.03) rubella antibody.
Fig. 1.
The linkage disequilibrium output for SNPs for TNFA gene from Haploview. a The r2 plots with the r2 values inside each box are shown for TNFA gene. Two promoter region SNPs (rs2844482 and rs2857708) in LD (r2=0.82) and a 3′ intergenic SNP (rs2256974) were found to be associated with rubella-specific antibody responses. Three other SNPs (rs2857706, rs2009658, and rs1800630) not genotyped in the current study were identified as tgSNPs with rs2844482 (r2≥0.9) by the SNPPicker algorithm. The r2 color scheme is as follows: white (r2=0), shades of gray (0<r2<1), and black (r2=1). b Common TNFA haplotypes and association analysis with rubella antibodies, based on the SNPs reported in panel a (© Mayo Clinic 2010)
Cytokine and cytokine receptor SNP associations with rubella-specific inflammatory (IL-6, TNF-α, and GM-CSF) secreted cytokines
Specific cytokine and cytokine receptor SNPs associated (p ≤ 0.01) with secreted levels of rubella-specific inflammatory cytokines, IL-6, TNF-α, and GM-CSF, are shown in Table 3. SNPs in the TNFRSF1B and IL12B genes were associated with rubella-specific IL-6 secretion levels. Minor alleles for five intronic SNPs (rs5745993, rs17882988, rs472093, rs5746059, and rs590977) in the TNFRSF1B gene were co-directionally associated with an allele dose-related increase in rubella-specific secreted IL-6 levels, while homozygous minor allele variant for intronic SNP rs474247 in the same gene was associated with higher IL-6 secretion in response to rubella stimulation. Similarly, minor alleles for an intronic (rs2569253) and promoter region (rs1422876 and rs6868898) SNPs in the IL12B gene were associated with an allele dose-related increase in secreted IL-6 levels, while minor alleles in an intronic region (rs730691) in the same gene were associated with an allele dose-related decrease in rubella-specific IL-6 secretion. Another intronic SNP in the IL12B (rs2546893) gene was significantly (p=0.009) associated with variable levels of IL-6 secretion. However, after adjusting for FDR, none of the SNPs remained significant.
Table 3.
SNPs in cytokine and cytokine receptor genes associated with rubella-specific responses of inflammatory (IL-6, TNF-α, and GM-CSF) secreted cytokines (© Mayo Clinic 2010)
| Secreted cytokine | SNP ID-gene (variant name) | Location | Genotype | Na | Median (IQR)b, pg/ml | p valuec | q valuec |
|---|---|---|---|---|---|---|---|
| IL-6 | rs5745993-TNFRSF1B | Intron | GG | 478 | 3,668.5 (3,098.7, 4,080.6) | 0.001 | 0.33 |
| 19859G>C | GC | 192 | 3,690.2 (3,174.4, 4,014.5) | ||||
| CC | 22 | 3,802.8 (3,650.4, 4,280.7) | |||||
| rs474247-TNFRSF1B | Intron | GG | 435 | 3,679.4 (3,110.4, 4,090) | 0.001 | 0.33 | |
| 19026C>T | GA | 223 | 3,670.1 (3,186.9, 4,024.9) | ||||
| AA | 34 | 3,734.2 (3,090.5, 4,062.6) | |||||
| rs730691-IL12B | Intron | GG | 238 | 3,734 (3,266.6, 4,123.8) | 0.003 | 0.33 | |
| -2437C>T | GA | 345 | 3,671.8 (3,064.6, 4,024.2) | ||||
| AA | 109 | 3,609.8 (3,182.7, 4,030) | |||||
| rs17882988-TNFRSF1B | Intron | GG | 478 | 3,670.8 (3,099, 4,087.5) | 0.003 | 0.33 | |
| 15721G>A | GA | 193 | 3,681 (3,161.8, 3,994.4) | ||||
| AA | 21 | 3,788 (3,650.4, 4,280.7) | |||||
| rs2569253-IL12B | Intron | AA | 179 | 3,629.3 (3,098.7, 4,107.1) | 0.004 | 0.33 | |
| 2797T>C | AG | 345 | 3,684.3 (3,124.8, 4,030.4) | ||||
| GG | 168 | 3,741.3 (3,234.4, 4,112.9) | |||||
| rs472093-TNFRSF1B | Intron | AA | 471 | 3,670.1 (3,098.7, 4,087.5) | 0.004 | 0.33 | |
| 3+49A>T | AT | 196 | 3,687.2 (3,177.7, 3,980.2) | ||||
| TT | 24 | 3,784.3 (3,378.3, 4,281) | |||||
| rs1422876-IL12B | 5′ Intergenic (Promoter) | CC | 179 | 3,591.1 (3,081.3, 3,964.1) | 0.005 | 0.33 | |
| -7509C>T | CT | 344 | 3,694.1 (3,134.6, 4,062.7) | ||||
| TT | 161 | 3,777.7 (3,290.2, 4,126.2) | |||||
| rs5746059-TNFRSF1B | Intron | AA | 414 | 3,669.4 (3,110.4, 4,062.6) | 0.005 | 0.33 | |
| 35643A>T | AG | 232 | 3,699.7 (3,163.6, 4,088.5) | ||||
| GG | 46 | 3,756.5 (3,231.9, 3,971.6) | |||||
| rs590977-TNFRSF1B | Intron | AA | 458 | 3,659.8 (3,099, 4,064.3) | 0.008 | 0.46 | |
| 28211A>C | AC | 206 | 3,730.7 (3231.9, 4,031.3) | ||||
| CC | 28 | 3,784.3 (3,165.7, 4,293.7) | |||||
| rs2546893-IL12B | Intron | GG | 182 | 3,630.7 (3,049.7, 4,146.6) | 0.009 | 0.46 | |
| -2170G>A | GA | 345 | 3,701.1 (3,115.1, 4,043.7) | ||||
| AA | 165 | 3,675.5 (3,320.2, 4,062.6) | |||||
| rs6868898-IL12B | 5′ Intergenic (Promoter) | AA | 313 | 3,641.8 (3,090.5, 4,041.3) | 0.01 | 0.48 | |
| 10630T>C | AG | 296 | 3,692.8 (3,125.7, 4,032.1) | ||||
| GG | 83 | 3,820.1 (3387.2, 4165) | |||||
| GM-CSF | rs2834211-IFNGR2 | Intron | AA | 579 | 28.4 (23.5, 32.8) | 0.0003 | 0.13 |
| 13958T>C | AG | 107 | 27 (23.3, 30.7) | ||||
| GG | 4 | 25 (24.1, 27.2) | |||||
| rs2069778-IL2 | Intron | GG | 487 | 28.4 (23.7, 32.8) | 0.002 | 0.36 | |
| 13958T>C | GA | 186 | 27 (23.4, 32.2) | ||||
| AA | 17 | 27 (22.9, 30.1) | |||||
| rs9610-IL10RA | 3′ UTR | GG | 214 | 28.8 (23.8, 32.5) | 0.005 | 0.71 | |
| 14903G>A | GA | 342 | 27.6 (23.4, 32.5) | ||||
| AA | 134 | 27.8 (23.3, 32.9) | |||||
| rs739718-IL5 | 3′ Intergenic | AA | 586 | 28 (23.7, 32.5) | 0.006 | 0.71 | |
| 6097C>T | AG | 94 | 27.4 (23.2, 33.0) | ||||
| GG | 6 | 25.1 (24, 34.3) | |||||
| TNF-α | rs12483293-IFNAR1 | Intron | AA | 365 | 21.6 (−9.8, 73.5) | 0.005 | 0.80 |
| 19428T>G | AC | 272 | 37.1 (−10.4, 99.2) | ||||
| CC | 55 | 42.3 (20.5, 129.1) |
Values are presented as homozygous major allele/heterozygous/homozygous minor allele
IQR interquartile range
One degree-of-freedom ordinal p value from the repeated measures regression analysis adjusting for age at enrollment, gender, race, age at first and second MMR-II immunizations, and cohort status
Only associations with p≤0.01 are included in the table
Four significant (p≤0.01) associations were found between cytokines (rs2069778-IL2 and rs739718-IL5) and cytokine receptors (rs2834211-IFNGR2 and rs9610-IL10RA) SNPs and secreted levels of GM-CSF in response to rubella stimulation (Table 3). All associations had a predisposition for a decrease in GM-CSF levels with increasing counts of minor alleles in a dose-related manner.
A single intronic SNP (rs12483293) of the IFNAR1 gene was associated with a minor allele dose-related increase (p=0.005) in rubella-specific TNF-α levels (Table 3).
TNFRS1B and IL12B haplotype associations with rubella-specific IL-6 secretion
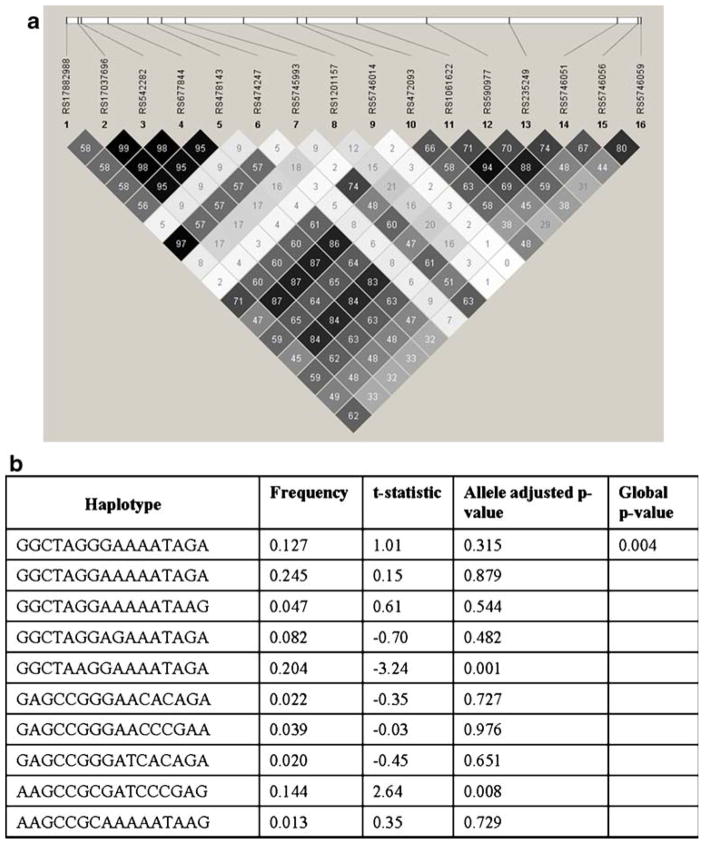
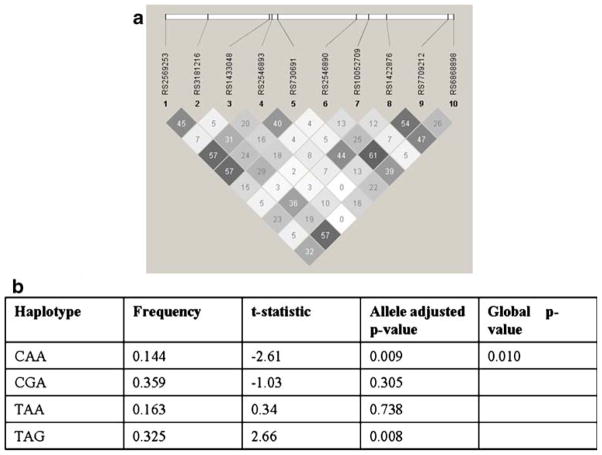
The Haploview output for the genotyped SNPs in the TNFRSF1B and their common haplotypes associated with rubella-specific IL-6 secretion are shown in Fig. 2. TNFRS1B haplotype GGCTAAGGAAAATAGA was significantly associated (p=0.001) with lower levels of (t-statistic=−3.24) rubella-specific IL-6 secretion, while the presence of AAGCCGCGATCCCGAG was associated (p=0.008) with higher (t-statistic=2.64) secretory IL-6 levels. Additionally, a promoter region IL12B haplotype TAG was associated (p= 0.008) with higher (t-statistic=2.66), while CAA haplotype was associated (p=0.009) with lower (t-statistic=−2.61) IL-6 secretion in response to rubella virus stimulation (Fig. 3).
Fig. 2.
The linkage disequilibrium output for TNFRSF1B SNPs from Haploview. a The r2 plots with the r2 values inside each box are shown for TNFRSF1B gene. Six intronic SNPs (rs5745993, rs17882988, rs472093, rs5746059, rs474247, and rs590977) in the TNFRSF1B gene were associated with rubella-specific secreted IL-6 levels. Strong LD (r2=0.97) was observed between rs5745993 and rs17882988. The r2 color scheme is as follows: white (r2=0), shades of gray (0<r2<1), and black (r2=1). b Common TNFRSF1B haplotypes and association analysis with rubella-specific IL-6 secretion, based on the SNPs reported in panel a (© Mayo Clinic 2010)
Fig. 3.
The linkage disequilibrium output for IL12B SNPs from Haploview. a The r2 plots with the r2 values inside each box are shown for IL12B gene. Two promoter (rs1422876 and rs6868898) and three intronic (rs2569253, rs730691, and rs2546893) SNPs from the IL12B gene were associated with IL-6 secretion. No strong linkage was observed among the genotyped SNPs in this gene. The r2 color scheme is as follows: white (r2=0), shades of gray (0<r2<1), and black (r2=1). b Common IL12B promoter region SNP (rs1422876, rs7709212, and rs6868898) haplotypes and association analysis with rubella-specific IL-6 secretion (© Mayo Clinic 2010)
Cytokine and cytokine receptor SNP associations with rubella-specific Th1 secreted cytokines
Specific cytokine and cytokine receptor SNPs associated with rubella-specific Th1 cytokines (IL-2 and IFN-γ) are shown in Table 4. Increased carriage of major allele T for rs2243115 (IL12A-564T>G) located in the 5′ intergenic region containing the promoter, and other regulatory elements of IL12A was associated with a dose-related decrease (p= 0.002) in IL-2 secretion levels. The SNPs rs6870828 (IL12B+ 5278T>C) and rs4787947 (IL4R-32760T>G) were associated with variations in IL-2 secretion response. Major alleles for rs1800693 located in the intronic region of TNFRSF1A and rs10811469 located in the promoter region of the IFNB1 gene were also associated with an allele dose-related increase in IL-2 secretion levels. These SNPs did not remain significant after adjusting for FDR.
Table 4.
SNPs in cytokine and cytokine receptor genes associated with rubella-specific responses of Th1 (IL-2 and IFN-γ) secreted cytokines (© Mayo Clinic 2010)
| Secreted cytokine | SNP ID-gene (variant name) | Location | Genotype | Na | Median (IQR)b, pg/ml | p valuec | q valuec |
|---|---|---|---|---|---|---|---|
| IL-2 | rs2243115- IL12A | 5′ Intergenic (Promoter) | AA | 551 | 16.2 (6.3, 28.8) | 0.002 | 0.78 |
| -564T>G | AC | 129 | 21.4 (11.4, 36.9) | ||||
| CC | 12 | 28.2 (8.6, 35.6) | |||||
| rs6870828- IL12B | 3′ Intergenic | AA | 203 | 16.6 (6.1, 26.8) | 0.003 | 0.78 | |
| 15278T>C | AG | 345 | 18 (7.7, 34.1) | ||||
| GG | 144 | 17.8 (8.0, 31.5) | |||||
| rs1800693-TNFRSF1A | Intron | AA | 258 | 19.1 (9.1, 31.2) | 0.008 | 0.81 | |
| 6+10T>C | AG | 324 | 17.3 (6.9, 31.4) | ||||
| GG | 110 | 13.7 (5.7, 28.6) | |||||
| rs10811469- IFNB1 | 5′ Intergenic (Promoter) | AA | 526 | 18.1 (8.0, 31.4) | 0.009 | 0.81 | |
| -8472A>G | AG | 152 | 15.6 (7.7, 26.8) | ||||
| GG | 14 | 12.8 (1.8, 24.5) | |||||
| rs4787947- IL4R | 5′ Intergenic (Promoter) | AA | 577 | 17.9 (8.2, 31.2) | 0.01 | 0.81 | |
| -32760T>G | AC | 109 | 14.7 (5.7, 26.6) | ||||
| CC | 6 | 34.2 (4.4, 48.2) | |||||
| IFN-γ | rs3024560-IL4R | Intron | AA | 299 | 9.0 (3.1, 24) | 0.005 | 0.43 |
| 5142T>G | AC | 300 | 9.0 (3.2, 27) | ||||
| CC | 92 | 8.4 (2.1, 19.5) | |||||
| rs4787948-IL4R | Intron | AA | 343 | 9 (3.1, 24.2) | 0.006 | 0.43 | |
| -10466A>G | AG | 269 | 9.2 (3, 26.3) | ||||
| GG | 79 | 6.8 (2, 1 6.6) | |||||
| rs4252279-IL10RA | Intron | GG | 554 | 7.8 (2.9, 23.5) | 0.007 | 0.43 | |
| 10004C>T | GA | 128 | 12.2 (3.2, 23.4) | ||||
| AA | 9 | 16.1 (6, 37.8) | |||||
| rs9976971-IFNGR2 | 5′ Intergenic (Promoter) | GG | 224 | 7.6 (2.5, 26.7) | 0.008 | 0.43 | |
| -7753G>A | GA | 335 | 8.7 (2.9, 23.6) | ||||
| AA | 132 | 12 (4.6, 23.3) | |||||
| rs4252287-IL10RA | Intron | GG | 553 | 7.8 (2.9, 23.5) | 0.009 | 0.43 | |
| 11455G>A | GA | 130 | 12.2 (3.1, 23.4) | ||||
| AA | 9 | 16.1 (6.0, 37.8) | |||||
| rs17860160-IFNAR2 | Intron | CC | 493 | 9.2 (3.4, 28.0) | 0.009 | 0.43 | |
| -2908G>T | CA | 174 | 7.7 (2.1, 21.3) | ||||
| AA | 25 | 6.3 (3.9, 13.6) |
Values are presented as homozygous major allele/heterozygous/homozygous minor allele
IQR interquartile range
One degree-of-freedom ordinal p value from the repeated measures regression analysis adjusting for age, gender, race, age at first and second MMR-II immunizations, and cohort status
Only associations with p≤0.01 are included in the table
Overall, six SNPs were significantly associated (p≤0.01) with rubella-specific secreted IFN-γ levels (Table 4). Minor alleles for two intronic SNPs, rs3024560 and rs4787948, located in the IL4R gene were associated with overall lower IFN-γ secretion. The minor allele for intronic SNP rs4252279 (IL10RA+10004C>T) was associated with an allele dose-related increase (p=0.007) in IFN-γ secretion in response to rubella stimulation. Major alleles for rs9976971 located in the promoter region of IFNGR2 and rs4252287 located in the intronic region of IL10RA were associated with an allele dose-related decrease in IFN-γ secretion. Finally, minor allele T for intronic SNP rs17860160 from the IFNAR2 gene was associated with an allele dose-related decrease (p=0.009) in rubella-specific IFN-γ secretion (Table 4). However, after adjusting for FDR, none of the SNPs remained significant.
Discussion
Immune responses to vaccination are influenced by genetic heterogeneity in immune response genes. Several recent studies, including our own, emphasize the role of polymorphisms in the non-HLA genes, primarily cytokine and cytokine receptor genes, in mediating differential immunological outcomes in response to viral vaccines and infections (Hollegaard and Bidwell 2006; Dhiman et al. 2007, 2008; Hennig et al. 2008; Hohler et al. 2005; Stanley et al. 2007; Ovsyannikova et al. 2008). In the current study, we focused our efforts on Th1/Th2 and inflammatory cytokines and their corresponding receptor genes, carefully selected based on known or likely influences of polymorphisms in these candidate genes on rubella virus antigen processing and presentation and effector T and B cell responses. Our central hypothesis is that genetic variations in these key immunoregulatory genes translate into increases or decreases in the effector molecules leading to heterogeneity in immune response to rubella vaccination.
Our study identified genetic variations in inflammatory cytokine and cytokine receptor genes significantly associated with variations in antibody and CMI responses to rubella vaccination. Two (rs2844482 and rs2857708) of the four SNPs with significant p and q-value associations with antibody response were located in the 5′ regulatory region of the TNFA gene. Of particular interest is rs2844482 (TNFA-3752 C>T), which is a tagSNP binned together with three other promoter region SNPs (rs2857706, rs2009658, rs1800630) that are in pairwise LD (r2≥0.90) with rs2844482, suggesting they are highly correlated variants with similar allelic frequencies and hence similar associations with the outcome of antibody response to rubella. The minor allele variant for rs1800630 has been reported to enhance promoter activity by ~2-fold via minor allele-specific binding of ubiquitous transcription factor OCT-1, resulting in the subsequent upregulation of TNFA expression (Hohjoh and Tokunaga 2001). A contrasting study determined that the minor allele variant for rs1800630 had 10-fold reduced binding affinity for the immunoregulatory element NF–κB and could translate into lower TNFA gene activity (Udalova et al. 2000). These contradictory functional studies indicate multifaceted effects of SNPs in the TNFA promoter region, with possible cooperative allelic effects of multiple SNPs in functional haplotypes. An additional SNP in the regulatory region of the IL6 gene also had significant p and q-value associations with a rubella-specific antibody response; however, the functional significance of this SNP is unknown. The secreted levels of IL-6 in turn were primarily associated with intronic and promoter region SNPs in the TNFRSF1B and IL12B genes, while those of TNF-α were coupled with genetic variation in the IFNAR1 gene suggesting a predominant role of inflammatory cytokine-genetics in regulating overall response to the rubella vaccine. Additionally, we identified specific haplotypes resulting from SNPs in TNFA, TNFRSF1B, and IL12B genes that were associated with increases or decreases in rubella antibody levels and IL-6 secretion. The overall secretion of IL-2 and IFN-γ was modest with little differences in cytokine secretion at genotypic levels to draw any strong conclusions.
In the same cohort, we previously reported a predominance of inflammatory cytokine response post-vaccination with marked TNF-α and IL-6 production (Dhiman et al. 2009), both of which are central mediators to T cell functions directly, and cross-talk between T cell and other immune cells to mediate B cell functions indirectly (Croft 2009; Jones 2005; Romagnani 2006; Larosa and Orange 2008). TNF-α is the front-line antiviral cytokine that acts as a trigger for “innate immune pathways” and IL-6 is the critical “immunological switch” that directs the innate host immune system to progress towards an adaptive response (Croft 2009; Jones 2005; Romagnani 2006; Larosa and Orange 2008; Bartee et al. 2008). We observed that rubella stimulation leads to overall suppression of Th1/Th2 responses and an immune-deviation towards inflammatory cytokines (IL-6 and TNF-α; Dhiman et al. 2009), a perfect cytokine milieu for Th17 phenotype development, which is an area of interest in numerous viral infections (Steinman 2007; Rowan et al. 2008; Patera et al. 2002; Ndhlovu et al. 2008). In the current study, we have identified key genetic-variants that may play a role in modulating secreted levels of IL-6 and TNF-α and hence the outcome of immune response to rubella vaccination. The underlying mechanism is still unknown; however, several hypotheses may be postulated to explain how differential regulation of secreted inflammatory cytokine levels due to these genetic variations can influence effector immune response. The TNF superfamily has been identified as a potent trigger for NK cell activation, eosinophil recruitment, and Th2 cytokine production (Fang et al. 2008). The inflammatory cytokine milieu is conducive to expanding the cytokine repertoire in lymph nodes and hence boosting effector cell functions (Katzman and Fowell 2008). Induction of the Th17 phenotype prevents viral elimination via inhibiting apoptosis and cytotoxic T cell responses and promotes viral persistence in the host (Hou et al. 2009), thus making it available to induce a protective response. In addition, some specific haplotypes identified in our study within the candidate genes we tested or extended haplotypes with additional immune function genes (e.g., TNFA is in LD with HLA-DQA1 and –DQB1 genes) may confer synergistic effects (Shin et al. 2008; Louka et al. 2003). An effective immune response may require multiple signals and potential “checkpoints” that regulate the levels of transcription and translation within the genetic composition of cytokine and cytokine receptor genes.
The primary strength of our study is the homogenous nature of our study cohort consisting of primarily Caucasians, which is representative of the US white population. Our sampling area, Olmsted County, MN, has exceptionally high vaccine coverage and no circulating wild-type rubella virus giving us confidence that we measured immunity specific to rubella vaccine and not disease. Another key strength of our study is the robust SNP selection approach, SNPPicker. It can post-process SNP panels from multiple sources and populations into a statistical framework that can furnish a comprehensive tagSNP list and circumvent single-SNP assay failures at the same time.
However, without further confirmation, our findings should not be extrapolated to non-Caucasian races due to non-transferability of haplotype tag sets across populations. Another restraint was the modest sample size. This resulted in a lower power to detect relatively small associations. Finally, the number of tests performed in this study is moderately large. Because of this, we chose a more stringent p value of 0.01 as a cut-off to minimize the potential number of false-positive results. Using this cut-off point, we had a slightly higher number of statistically significant associations than expected by chance alone (33 vs. 30), but type I errors are still a legitimate concern. Thus, any results reported here need to be validated in a replication study (Rodriguez-Murillo and Greenberg 2008; Chanock et al. 2007; Ioannidis et al. 2003), and we are in the process of reconfirming these associations in a larger cohort.
In conclusion, we can say that specific genetic polymorphisms in the TNFA, TNFRSF1B, and IL12B genes appear to be involved in the variations in immune responses to rubella vaccination either directly or indirectly by modulating the activity of other cytokines, such as IL-6. We plan on confirming these variants in a replication cohort, fine-mapping/re-sequencing the regions of interest and determining their functional relevance. We would also like to assess the extended haplotypes of identified genes (e.g., TNFA is in LD with HLA-DQA1 and –DQB1 genes) in order to identify synergistic effects that SNPs in different genes might have on rubella vaccine response. Identification of such “genetic fingerprints” can be used to predict vaccine response and help design appropriate vaccination strategies and are a key to the development of predictive vaccinology and vaccinomics (Poland et al. 2007).
Acknowledgments
We thank the Mayo Clinic Vaccine Research Group nurses for subject recruitment and the children who participated in our studies. We also thank David N. Rider and Hugues Sicotte from the Department of Biomedical Statistics and Informatics for development and optimization of the SNP selection algorithm for this study. This work was supported by NIH grants AI 48793, AI 33144, and 1 UL1 RR024150-01 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health, and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
Financial Disclosure Dr. Poland is the chair of a Safety Evaluation Committee for novel non-rubella vaccines undergoing clinical studies by Merck Research Laboratories. Dr. Jacobson serves on a Safety Review Committee for a post-licensure study of a human papillomavirus vaccine for Kaiser–Permanente. Other authors declare no conflict of interest.
This work was presented as oral presentation (abstract # G1-1110) at the 49th Annual ICAAC Meeting. San Francisco CA, Sep. 12–15, 2009.
References
- Adamo MP, Zapata M, Frey TK. Analysis of gene expression in fetal and adult cells infected with rubella virus. Virology. 2008;370:1–11. doi: 10.1016/j.virol.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaboshi I, Nagayoshi I, Omura M, Iwaki K. Elevated serum levels of interleukin-10 in children with acute rubella infection. Scand J Infect Dis. 2001;33:462–465. doi: 10.1080/00365540152029945. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bartee E, Mohamed MR, McFadden G. Tumor necrosis factor and interferon: cytokines in harmony. Curr Opin Microbiol. 2008;11:378–383. doi: 10.1016/j.mib.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista-López N, Ward BJ, Mills E, McCormick D, Martel N, Ratnam S. Development and durability of measles antigen-specific lymphoproliferative response after MMR vaccination. Vaccine. 2000;18:1393–1401. doi: 10.1016/s0264-410x(99)00396-5. [DOI] [PubMed] [Google Scholar]
- Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1:3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, et al. Cytokine gene polymorphism in human disease: on-line databases: supplement 1. Genes Immun. 2001;2:61–70. doi: 10.1038/sj.gene.6363733. [DOI] [PubMed] [Google Scholar]
- Boulianne N, De Serres G, Ratnam S, Ward BJ, Joly JR, Duval B. Measles, mumps, and rubella antibodies in children 5–6 years after immunization: effect of vaccine type and age at vaccination. Vaccine. 1995;13:1611–1616. doi: 10.1016/0264-410x(95)00098-l. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabalgoity JA, Baz A, Rial A, Grille S. The relevance of cytokines for development of protective immunity and rational design of vaccines. Cytokine Growth Factor Rev. 2007;18:195–207. doi: 10.1016/j.cytogfr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chu SY, Bernier RH, Stewart JA, Herrmann KL, Greenspan JR, Henderson AK, et al. Rubella antibody persistence after immunization. Sixteen-year follow-up in the Hawaiian Islands. JAMA. 1988;259:3133–3166. doi: 10.1001/jama.259.21.3133. [DOI] [PubMed] [Google Scholar]
- Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol. 2009;9:271–285. doi: 10.1038/nri2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- Dhiman N, Ovsyannikova IG, Cunningham JM, Vierkant RA, Kennedy RB, Pankratz VS, et al. Associations between measles vaccine immunity and single nucleotide polymorphisms in cytokine and cytokine receptor genes. J Infect Dis. 2007;195:21–29. doi: 10.1086/510596. [DOI] [PubMed] [Google Scholar]
- Dhiman N, Ovsyannikova I, Vierkant R, Pankratz V, Jacobson R, Poland G. Associations between cytokine/cytokine receptor SNPs and humoral immunity to measles, mumps and rubella in a Somali population. Tissue Antigens. 2008;72:211–220. doi: 10.1111/j.1399-0039.2008.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N, Haralambieva I, Vierkant RA, Pankratz VS, Ryan JE, Jacobson RM, et al. Predominant inflammatory cytokine secretion patterns in response to two doses of live rubella vaccine in healthy vaccinees. Cytokine. 2009 doi: 10.1016/j.cyto.2009.12.002. (Unpublished observation) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Adkins B, Deyev V, Podack ER. Essential role of TNF receptor superfamily 25 (TNFRSF25) in the development of allergic lung inflammation. J Exp Med. 2008;205:1037–1048. doi: 10.1084/jem.20072528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood NP, Ovsyannikova IG, Vierkant RA, O’Byrne MM, Poland GA. A qualitative and quantitative comparison of two rubella virus specific IgG antibody assays. Clin Vacc Immunol. 2009 doi: 10.1089/vim.2010.0026. (Unpublished observation) [DOI] [PubMed] [Google Scholar]
- Hackstein H, Hecker M, Kruse S, Bohnert A, Ober C, Deichmann KA, et al. A novel polymorphism in the 5′ promoter region of the human interleukin-4 receptor alpha-chain gene is associated with decreased soluble interleukin-4 receptor protein levels. Immunogenetics. 2001;53:264–269. doi: 10.1007/s002510100324. [DOI] [PubMed] [Google Scholar]
- Haring JS, Badovinac VP, Harty JT. Inflaming the CD8+ T cell response. Immunity. 2006;25:19–29. doi: 10.1016/j.immuni.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Haukim N, Bidwell JL, Smith AJ, Keen LJ, Gallagher G, Kimberly R, et al. Cytokine gene polymorphism in human disease: on-line databases: supplement 2. Genes Immun. 2002;3:313–330. doi: 10.1038/sj.gene.6363881. [DOI] [PubMed] [Google Scholar]
- Hennig BJ, Fielding K, Broxholme J, Diatta M, Mendy M, Moore C, et al. Host genetic factors and vaccine-induced immunity to hepatitis B virus infection. PLoS ONE. 2008;3:e1898. doi: 10.1371/journal.pone.0001898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary IB, Griffith AH. Persistence of rubella antibody 15 years after subcutaneous administration of Wistar 27/3 strain live attenuated rubella virus vaccine. Vaccine. 1984;2:274–276. doi: 10.1016/0264-410x(84)90043-4. [DOI] [PubMed] [Google Scholar]
- Hohjoh H, Tokunaga K. Allele-specific binding of the ubiquitous transcription factor OCT-1 to the functional single nucleotide polymorphism (SNP) sites in the tumor necrosis factor-alpha gene (TNFA) promoter. Genes Immun. 2001;2:105–109. doi: 10.1038/sj.gene.6363721. [DOI] [PubMed] [Google Scholar]
- Hohler T, Reuss E, Freitag CM, Schneider PM. A functional polymorphism in the IL-10 promoter influences the response after vaccination with HBsAg and hepatitis A. Hepatology. 2005;42:72–76. doi: 10.1002/hep.20740. [DOI] [PubMed] [Google Scholar]
- Hollegaard MV, Bidwell JL. Cytokine gene polymorphism in human disease: on-line databases, supplement 3. Genes Immun. 2006;7:269–276. doi: 10.1038/sj.gene.6364301. [DOI] [PubMed] [Google Scholar]
- Horstmann DM, Schluederberg A, Emmons JE, Evans BK, Randolph MF, Andiman WA. Persistence of vaccine-induced immune responses to rubella: Comparison with natural infection. Rev Infect Dis. 1985;7:S80–S85. doi: 10.1093/clinids/7.supplement_1.s80. [DOI] [PubMed] [Google Scholar]
- Hou W, Kang HS, Kim BS. Th17 cells enhance viral persistence and inhibit T cell cytotoxicity in a model of chronic virus infection. J Exp Med. 2009;206:313–328. doi: 10.1084/jem.20082030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG. Genetic associations in large versus small studies: an empirical assessment. Lancet. 2003;361:567–571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- Jones SA. Directing transition from innate to acquired immunity: defining a role for IL-6. J Immunol. 2005;175:3463–3468. doi: 10.4049/jimmunol.175.6.3463. [DOI] [PubMed] [Google Scholar]
- Katzman SD, Fowell DJ. Pathogen-imposed skewing of mouse chemokine and cytokine expression at the infected tissue site. J Clin Invest. 2008;118:801–811. doi: 10.1172/JCI33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen LJ. The extent and analysis of cytokine and cytokine receptor gene polymorphism. Transpl Immunol. 2002;10:143–146. doi: 10.1016/s0966-3274(02)00061-8. [DOI] [PubMed] [Google Scholar]
- Kurtzman GJ, Cohen BJ, Field AM, Oseas R, Blaese RM, Young NS. Immune response to B19 parvovirus and an antibody defect in persistent viral infection. J Clin Invest. 1989;84:1114–1123. doi: 10.1172/JCI114274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larosa DF, Orange JS. 1. Lymphocytes. J Allergy Clin Immunol. 2008;121:S364–S369. doi: 10.1016/j.jaci.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Louka AS, Lie BA, Talseth B, Ascher H, Ek J, Gudjonsdottir AH, et al. Coeliac disease patients carry conserved HLA-DR3-DQ2 haplotypes revealed by association of TNF alleles. Immunogenetics. 2003;55:339–343. doi: 10.1007/s00251-003-0586-5. [DOI] [PubMed] [Google Scholar]
- Mitchell LA, Tingle AJ, Décarie D, Shukin R. Identification of rubella virus T-cell epitopes recognized in an amnestic response to RA27/3 vaccine: associations with boost in neutralizing antibody titer. Vaccine. 1999;17:2356–2365. doi: 10.1016/s0264-410x(99)00040-7. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Urano T, Osano M, Nakagawa M, Maehara N, Sasaki K, et al. Production of interferon by human peripheral lymphocytes stimulated with rubella virus. Kitasato Arch Exp Med. 1988;61:187–193. [PubMed] [Google Scholar]
- Ndhlovu LC, Chapman JM, Jha AR, Snyder-Cappione JE, Pagan M, Leal FE, et al. Suppression of HIV-1 plasma viral load below detection preserves IL-17 producing T cells in HIV-1 infection. AIDS. 2008;22:990–992. doi: 10.1097/QAD.0b013e3282ff884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. The contribution of HLA class I antigens in immune status following two doses of rubella vaccination. Hum Immunol. 2004;65:1506–1515. doi: 10.1016/j.humimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. Human leukocyte antigen class II alleles and rubella-specific humoral and cell-mediated immunity following measles-mumps-rubella-II vaccination. J Infect Dis. 2005;191:515–519. doi: 10.1086/427558. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova IG, Jacobson RM, Dhiman N, Vierkant RA, Pankratz VS, Poland GA. Human leukocyte antigen and cytokine receptor gene polymorphisms associated with heterogeneous immune responses to mumps viral vaccine. Pediatrics. 2008;121:e1091–e1099. doi: 10.1542/peds.2007-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Ryan JE, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. Influence of host genetic variation on rubella-specific T cell cytokine responses following rubella vaccination. Vaccine. 2009a Feb 5; doi: 10.1016/j.vaccine.2009.01.079. (e-pub ahead of print: PMID19200845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Vierkant RA, Pankratz VS, O’Byrne MM, Jacobson RM, Poland GA. HLA haplotype and supertype associations with cellular immune responses and cytokine production in healthy children after rubella vaccine. Vaccine. 2009b Feb 5; doi: 10.1016/j.vaccine.2009.01.080. (e-pub ahead of print; PMID19200828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patera AC, Pesnicak L, Bertin J, Cohen JI. Interleukin 17 modulates the immune response to vaccinia virus infection. Virology. 2002;299:56–63. doi: 10.1006/viro.2002.1400. [DOI] [PubMed] [Google Scholar]
- Penn LJ, Williams BR. Interferon-induced 2-5A synthetase activity in human peripheral blood mononuclear cells after immunization with influenza virus and rubella virus vaccines. J Virol. 1984;49:748–753. doi: 10.1128/jvi.49.3.748-753.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther. 2007;82:653–664. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- Pukhalsky AL, Shmarina GV, Bliacher MS, Fedorova IM, Toptygina AP, Fisenko JJ, et al. Cytokine profile after rubella vaccine inoculation: evidence of the immunosuppressive effect of vaccination. Mediat Inflamm. 2003;12:203–207. doi: 10.1080/09629350310001599639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reef SE, Frey TK, Theall K, Abernathy E, Burnett CL, Icenogle J, et al. The changing epidemiology of rubella in the 1990s: on the verge of elimination and new challenges for control and prevention. JAMA. 2002;287:464–472. doi: 10.1001/jama.287.4.464. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Greenberg DA. Genetic association analysis: a primer on how it works, its strengths and its weaknesses. Int J Androl. 2008;31:546–556. doi: 10.1111/j.1365-2605.2008.00896.x. [DOI] [PubMed] [Google Scholar]
- Rolph MS, Ramshaw IA. Interleukin-4-mediated downregulation of cytotoxic T lymphocyte activity is associated with reduced proliferation of antigen-specific CD8+ T cells. Microbes Infect. 2003;5:923–932. doi: 10.1016/s1286-4579(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Romagnani S. Regulation of the T cell response. Clin Exp Allergy. 2006;36:1357–1366. doi: 10.1111/j.1365-2222.2006.02606.x. [DOI] [PubMed] [Google Scholar]
- Rowan AG, Fletcher JM, Ryan EJ, Moran B, Hegarty JE, O’Farrelly C, et al. Hepatitis C virus-specific Th17 cells are suppressed by virus-induced TGF-beta. J Immunol. 2008;181:4485–4494. doi: 10.4049/jimmunol.181.7.4485. [DOI] [PubMed] [Google Scholar]
- Ryan JE, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Poland GA. Response surface methodology to determine optimal cytokine responses in human peripheral blood mononuclear cells after smallpox vaccination. J Immunol Methods. 2009;341:97–105. doi: 10.1016/j.jim.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HD, Yang SW, Kim DH, Park Y. Independent association of tumor necrosis factor polymorphism with type 1 diabetes susceptibility. Ann N Y Acad Sci. 2008;1150:76–85. doi: 10.1196/annals.1447.059. [DOI] [PubMed] [Google Scholar]
- Singh R, John TJ, Cherian T, Raghupathy P. Immune response to measles, mumps & rubella vaccine at 9, 12 & 15 months of age. Indian J Med Res. 1994;100:155–159. [PubMed] [Google Scholar]
- Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20:43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Smith GL, Symons JA, Alcamí A. Poxviruses: interfering with interferon. Semin Virol. 1998;8:409–418. [Google Scholar]
- Stanley SL, Jr, Frey SE, Taillon-Miller P, Guo J, Miller RD, Koboldt DC, et al. The immunogenetics of smallpox vaccination. J Infect Dis. 2007;196:212–219. doi: 10.1086/518794. [DOI] [PubMed] [Google Scholar]
- Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- Tilles JG, Balkwill F, Davilla J. 2′,5′-Oligoadenylate synthetase and interferon in peripheral blood after rubella, measles, or mumps live virus vaccine. Proc Soc Exp Biol Med. 1987;186:70–74. doi: 10.3181/00379727-186-42586. [DOI] [PubMed] [Google Scholar]
- Udalova IA, Richardson A, Denys A, Smith C, Ackerman H, Foxwell B, et al. Functional consequences of a polymorphism affecting NF-kappaB p50-p50 binding to the TNF promoter region. Mol Cell Biol. 2000;20:9113–9119. doi: 10.1128/mcb.20.24.9113-9119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vosse E, Lichtenauer-Kaligis EGR, van Dissel JT, Ottenhoff THM. Genetic variations in the interleukin-12/interleukin-23 receptor (β1) chain, and implications for IL-12 and IL-23 receptor structure and function. Immunogenetics. 2003;54:817–829. doi: 10.1007/s00251-002-0534-9. [DOI] [PubMed] [Google Scholar]
- van der Logt JT, van Loon AM, van d V. Replication of rubella virus in human mononuclear blood cells. Infect Immun. 1980;27:309–314. doi: 10.1128/iai.27.2.309-314.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer SJH. Cytokine and cytokine receptor polymorphisms in infectious disease. Intensive Care Med. 2000;26(Suppl 1):S98–S102. doi: 10.1007/s001340051125. [DOI] [PubMed] [Google Scholar]
- Weir B. Genetic data Analysis II: methods for discrete population genetic data. Sinauer Associates, Inc; Sunderland, MA: 1996. pp. 98–99. [Google Scholar]