Abstract
MERIT40 is a recently identified BRCA1 and RAP80 interacting protein that is essential for protein–protein interactions of a BRCA1 complex also containing Abraxas, BRCC36 and BRCC45. It is a mediator of checkpoint functions and DNA damage signaling through a (de)ubiquitination cascade. Based on its interaction with BRCA1 and its role in genome integrity maintenance, MERIT40 is a novel candidate gene for being involved in hereditary susceptibility to breast cancer. Here, we report to our knowledge the first comprehensive mutation screening of this gene in affected cases of breast cancer families. Only a number of sequence variants were found, four of which are novel. None of the observed variants appeared to be disease related, suggesting that germline mutations in MERIT40 are rare or absent in familial breast cancer patients.
Keywords: MERIT40, Breast cancer susceptibility, Germline mutation
Introduction
BRCA1 and BRCA2 are the two major susceptibility genes involved in hereditary predisposition to breast cancer. However, germline mutations in these tumor suppressors account for a maximum 20% of the familial breast cancer cases worldwide [1]. The involvement of further high penetrance breast cancer susceptibility genes is questionable [2], and it has been suggested that if they exist, they would account only for a very small fraction of familial disease risk [3]. Instead, most of the remaining cancer cases are expected to be explained by mutations in moderate and low penetrance cancer susceptibility genes together with environmental factors [2–5]. Genome-wide association studies and candidate gene approaches with direct mutation analysis of tumor suppressor genes have proven to be successful approaches to identify susceptibility genes for the remaining familial breast cancer cases. A candidate gene approach based on physical interaction of the protein product with BRCA1 or BRCA2 has led to the identification of several important breast cancer susceptibility genes, such as PALB2 [6], BRIP1 [7] and BARD1 [8, 9].
MERIT40 (Mediator of RAP80 Interactions and Targeting 40 kDa, alias HSPC142/NBA/c19orf62) is a newly identified BRCA1 interacting protein [10–12] that is essential for RAP80 interaction with BRCA1 [13–15]. By using a systems-wide interactome mapping approach, MERIT40 was previously shown to interact with another BRCA1 interacting protein, BRCC45 [16, 17]. MERIT40 also associates with Abraxas (alias ABRA1 or CCDC98) [15, 18, 19] and BRCC36 [20, 21]. Interaction with MERIT40 is essential to maintain BRCA1-RAP80 complex integrity, protein stability and DNA double-strand break targeting. Consequently, MERIT40 is required for G2 checkpoint execution and viability responses to ionizing radiation [10–12].
The DNA damage response through BRCA1-RAP80 complex signaling is achieved in part through a (de)ubiquitination cascade that emerges as a central event in DNA damage signaling [22–25]. MERIT40 appears to control the cell cycle checkpoint and resistance to DNA-damaging agents by mediating interactions between the core BRCA1-RAP80 complex and BRCC36, thus targeting lysine-63-ubiquitin (K63-Ub) deubiquitination activity to DNA damage sites [10–12]. The MERIT40 gene product consists of 329 residues and possesses a von Willebrand factor A (VWA) domain at its N-terminus that is homologous to the VWA domain of the S5A proteasome subunit. To date, MERIT40 appears to primarily play a potential scaffolding role for the BRCA1 complex, although other activities cannot be ruled out, such as proteasome-linked protein processing activities through the VWA domain. The central region (amino acids 93–320) of MERIT40 is evolutionarily conserved down to invertebrates and plants. Deletion of the last 30 residues prevents MERIT40 nuclear accumulation, formation of ionizing radiation-induced foci, as well as interaction with BRCA1, RAP80, Abraxas and BRCC36 [10].
Given the dependence of DNA damage signaling on MERIT40, and its association with BRCA1, we examined whether mutations in MERIT40 occur in the context of hereditary susceptibility to breast cancer. Here, we report the result of a comprehensive mutation screening of the MERIT40 gene in affected index cases of 125 Finnish breast cancer families. The present investigation revealed 7 sequence variants, 4 of which are not currently reported in public databases. None of the observed germline changes, however, appeared to be disease related. Furthermore, the performed mutation analysis did not indicate any dosage variation in the MERIT40 gene.
Patients and methods
Cases and controls
Mutation screening of the MERIT40 gene was performed on 125 breast and breast-ovarian cancer families originating from northern Finland. For statistical purposes, one index patient from each family was chosen according to the youngest age of breast cancer onset. Inclusion criteria for the 72 high-risk families were the following: (1) three or more cases of breast cancer, potentially in combination with single ovarian cancers in first- or second-degree relatives, or (2) two cases of breast cancer in first- or second-degree relatives, of which at least one with early disease onset (<35 years), bilateral breast cancer or multiple primary tumors including breast or ovarian cancer in the same individual. The remaining 53 families were indicative of moderate disease susceptibility, presenting either two cases of breast cancer in first- or second-degree relatives or one case of breast cancer under the age of 35 or one breast cancer and other types of cancer in the same family. Altogether fifteen of the studied index cases had previously been tested positive for known breast cancer-associated germline mutations in BRCA1 or BRCA2 (eleven), TP53 (one) and PALB2 (three). All of the biological specimens and clinical information of the familial breast cancer cases investigated were collected at the Oulu University Hospital, with the informed consent of the patients. The used control samples (N = 192) derived from anonymous female cancer-free Finnish Red Cross blood donors (age ≥45 years) originating from the same geographical region as the studied cancer families. Approval to perform the study was obtained from the Ethical Board of the Northern Ostrobothnia Health Care District and the Finnish Ministry of Social Affairs and Health.
DNA isolation and mutation analysis
DNA from blood lymphocytes was extracted using the standard phenol–chloroform method or the Puregene D-50 K purification kit (Gentra). The entire coding region and exon–intron boundaries of the MERIT40 gene were screened for germline mutations by conformation sensitive gel electrophoresis (CSGE). Samples with deviating band patterns were sequenced with the Li-Cor IR2 4200-S DNA Analysis system (Li-Cor Inc.) using the SequiTherm EXEL™II DNA Sequencing Kit-LC (Epicentre Technologies). Besides sequence variants, also possible aberrations in DNA dosage were monitored for. Oligonucleotides for CSGE and sequencing were designed by the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) based on sequence information obtained from publicly available databases. Oligonucleotide sequence information is shown in the Supplementary Table S1, and PCR conditions are available upon request.
Statistical and bioinformatical analysis
Carrier frequencies between patients and healthy controls were compared by using Pearson Chi-Square or Fisher’s exact test (two-sided) (SPSS version 17.0 for Windows). All alterations were checked with NNSplice software for potential splicing effects (http://www.fruitfly.org/seq_tools/splice.html). The missense alteration Lys274Arg was tested for possible pathogenicity by using PolyPhen software (http://genetics.bwh.harvard.edu/pph).
Results
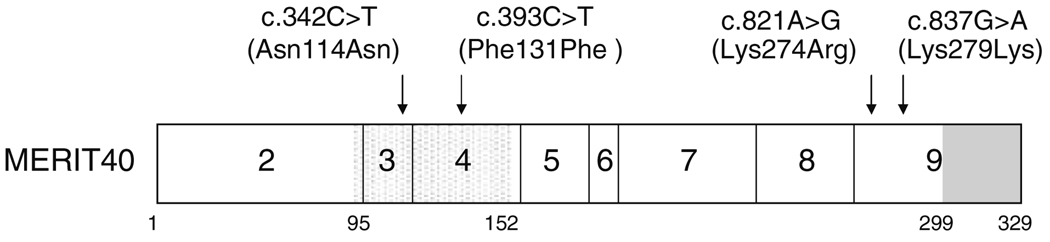
Screening of index patients of breast cancer families for germline alterations in MERIT40 revealed 2 intronic and 4 exonic variants, and a variant in the 3′ UTR of the gene, but did not uncover any DNA dosage aberrations. Four of the observed sequence changes are not reported in the NCBI SNP database (http://www.ncbi.nlm.nih.gov/SNP/). Details and occurrence of the observed nucleotide variants are shown in Table 1, and the positions of the exonic changes in the protein are depicted in Fig. 1. Only one alteration resulted in an amino acid change (Lys274Arg). However, according to the result of PolyPhen software analysis, the change is not predicted to affect protein function. c.*87G > A was observed in one family but not in the controls, whereas all of the other variants were found at similar frequencies in the control population. The nucleotide alterations were also assessed for possible effects on consensus splice sites. Of the observed changes, c.*87G > A might be implicated in introducing a new splice acceptor site (score 0.68, vs. 0.53 for the wild-type sequence), the effect of which would be unclear. None of the variants located to the last 30 amino acids of the protein that is implicated in nuclear localization and protein–protein interactions with various members of the BRCA1 complex.
Table 1.
Observed sequence variations in the MERIT40 gene
| Location | Nucleotide change | Effect on protein | rs number | Frequency of heterozygotes, % (n/N) | P (OR; 95% CI) | |
|---|---|---|---|---|---|---|
| Familial cases | Controls | |||||
| Exon 3 | c.342C > T | Asn114Asn | – | 4.0 (5/125) | 3.7 (7/192) | 1.0 (1.1; 0.3–3.6) |
| Intron 3 | c.344 + 41A > T | – | rs10420922 | 49.6 (62/125) | 44.3 (85/192) | 0.4 (1.2; 0.8–1.9) |
| Exon 4 | c.393C > T | Phe131Phe | – | 0.8 (1/125) | 1.6 (3/192) | 1.0 (0.5; 0.1–4.9) |
| Intron 8a | c.787 − 6C > T | – | rs10406920 | 39.2 (49/125) | 37.5 (72/192) | 0.8 (1.1; 0.7–1.7) |
| Exon 9 | c.821A > G | Lys274Arg | – | 1.6 (2/125) | 1 (2/192) | 0.6 (1.5; 0.2–11.1) |
| Exon 9a | c.837G > A | Lys279Lys | rs8170 | 39.2 (49/125) | 37.5 (72/192) | 0.8 (1.1; 0.7–1.7) |
| Exon 9b | c.*87G > A | – | – | 0.8 (1/125) | − (0/192) | 0.4 (1.0; 0.98–1.01) |
The following sequence information was used: NC_000019 (genomic DNA), NM_001033549.1 (mRNA), and NP_001028721 (protein) OR odds ratio, CI confidence interval
rs10406920 and rs8170 were in linkage
3′UTR change
Fig 1.
Schematic diagram of the MERIT40 protein and effect of the observed nucleotide changes in the coding region of the gene. Boxes represent exons. The only currently known functional domain, the von Willebrand factor A domain (aa 95–152) is dotted. Deletion of the last 30 amino acids of MERIT40 prevents its nuclear accumulation and association with BRCA1, RAP80, Abraxas and BRCC36 (region shown in gray)
Discussion
MERIT40 is involved in the integrity of aBRCA1 containing complex also including RAP80, Abraxas, BRCC36 and BRCC45 [10–12]. MERIT40 is required for the recruitment of these interaction partners to DNA double-strand breaks upon ionizing radiation and for the participation through RAP80 and BRCC36 in a (de)ubiquitination signaling cascade. Interestingly, a potentially pathogenic amino acid deletion was recently reported in RAP80 in two breast cancer patients [26]. This mutation abrogated ubiquitin binding and DNA damage response signaling for RAP80 and other members of the BRCA1 complex. These data suggest that MERIT40, similar to RAP80, may play a role in breast tumorigenesis via the interaction with BRCA1.
In our present study, the whole coding region and exon–intron boundaries of MERIT40 were screened for germline mutations. Although several new alterations were found, they are unlikely to be pathogenic. Two nucleotide changes, however, may be functionally important. c.*87G > A was found in one index patient, but not in the control population, suggesting that it is either a rare polymorphism or it might exert a detrimental effect if it affected splicing or transcriptional stability of MERIT40. The only amino acid change observed, Lys274Arg, is also a novel variant, and though statistically its frequency was not significantly elevated compared to that of the controls, we cannot currently fully exclude the slight possibility that it may represent a low penetrance susceptibility allele. Large-scale association studies would be needed to address this issue. A matter that could be of further interest is the observation that all of the amino acid changes seen in the current study are silent ones, except for Lys274 that is substituted in the most conservative fashion. This notion could reflect the functional essentiality of these amino acid residues. On the other hand, it might be speculated that Lys274 or Lys279 are potential post-translational modification sites and that a switch from Lys to Arg at position 274 in a heterozygous manner could subtly modulate MERIT40’s role in DNA damage signaling. Consequently, a better understanding of MERIT40 functions, and particularly the role of its first 300 residues, would be helpful in evaluating the significance of genetic alterations in MERIT40 regarding normal cellular functions and in human disease.
In conclusion, the present data suggest that mutations predisposing to breast cancer are either very rare or absent in the coding region of MERIT40. It would be interesting to know whether large genomic rearrangements, or mutations in transcriptional regulatory regions located further away from the coding region, could represent alternative and more typical ways of dysregulating MERIT40 function. On the other hand, it might also be that mutations in this gene are poorly tolerated, pointing to the essentiality of MERIT40 in the DNA damage response and other functions maintaining genomic integrity. To our knowledge, this is the first study exploring the possible involvement of the MERIT40 gene in hereditary predisposition to breast cancer.
Supplementary Material
Acknowledgments
We wish to thank Dr. Aki Mustonen, Dr. Jaakko Ignatius and nurses Kari Mononen and Outi Kajula for their help in sample and data collection and also in patient contacts, Helmi Konola and Meeri Elina Otsukka for technical assistance and Hannele Erkko for critical reading of the manuscript. We thank all the patients and their family members for volunteering to participate in these studies, as well as the Finnish Red Cross Blood Service for help with collection of population control blood samples. This study was financially supported by the Sigrid Jusélius Foundation, the Academy of Finland, CIMO, the Orion-Farmos Research Foundation, the Northern Ostrobothnia Fund of the Finnish Cultural Foundation, the University of Oulu, and the Oulu University Hospital.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-009-0453-7) contains supplementary material, which is available to authorized users.
Contributor Information
Szilvia Solyom, Laboratory of Cancer Genetics, Department of Clinical Genetics and Biocenter Oulu, University of Oulu, Oulu University Hospital, P.O. Box 5000, 90014 Oulu, Finland.
Jeffery Patterson-Fortin, Department of Cancer Biology, University of Pennsylvania School of Medicine, 421 Curie Blvd, Philadelphia, PA 19104-6160, USA; University of Pennsylvania School of Veterinary Medicine, 3800 Spruce St, Philadelphia, PA 19104-6160, USA.
Katri Pylkäs, Laboratory of Cancer Genetics, Department of Clinical Genetics and Biocenter Oulu, University of Oulu, Oulu University Hospital, P.O. Box 5000, 90014 Oulu, Finland.
Roger A. Greenberg, Department of Cancer Biology, University of Pennsylvania School of Medicine, 421 Curie Blvd, Philadelphia, PA 19104-6160, USA Department of Pathology, Abramson Family Cancer Research Institute, University of Pennsylvania School of Medicine, 421 Curie Blvd, Philadelphia, PA 19104-6160, USA.
Robert Winqvist, Email: robert.winqvist@oulu.fi, Laboratory of Cancer Genetics, Department of Clinical Genetics and Biocenter Oulu, University of Oulu, Oulu University Hospital, P.O. Box 5000, 90014 Oulu, Finland.
References
- 1.Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br J Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 4.Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 5.Wooster R, Weber BL. Breast and ovarian cancer. N Engl J Med. 2003;348:2339–2347. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- 6.Erkko H, Xia B, Nikkilä J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 7.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 8.Ghimenti C, Sensi E, Presciuttini S, et al. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosom Cancer. 2002;33:235–242. doi: 10.1002/gcc.1223. [DOI] [PubMed] [Google Scholar]
- 9.Karppinen SM, Heikkinen K, Rapakko K, et al. Mutation screening of the BARD1 gene: evidence for involvement of the Cys557Ser allele in hereditary susceptibility to breast cancer. J Med Genet. 2004;41:e114. doi: 10.1136/jmg.2004.020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao G, Patterson-Fortin J, Messick TE, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Hurov K, Hofmann K, et al. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 14.Sobhian B, Shao G, Lilli DR, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 18.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol. 2007;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Arciero CA, Wang C, et al. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 2006;66:5039–5046. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y, Hakimi MA, Chen X, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao G, Lilli DR, Patterson-Fortin J, et al. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci USA. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart GS, Panier S, Townsend K, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117:305–317. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikkilä J, Coleman KA, Morrissey D, et al. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009;28:1843–1852. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.