Abstract
The TREAT Asia (Therapeutics, Research, Education, and AIDS Training in Asia) Network is building capacity for Human Immunodeficiency Virus Type-1 (HIV-1) drug resistance testing in the region. The objective of the TREAT Asia Quality Assessment Scheme – designated TAQAS – is to standardize HIV-1 genotypic resistance testing (HIV genotyping) among laboratories to permit rigorous comparison of results from different clinics and testing centres. TAQAS has evaluated three panels of HIV-1-positive plasma from clinical material or low-passage, culture supernatant for up to 10 Asian laboratories. Laboratory participants used their standard protocols to perform HIV genotyping. Assessment was in comparison to a target genotype derived from all participants and the reference laboratory’s result. Agreement between most participants at the edited nucleotide sequence level was high (>98%). Most participants performed to the reference laboratory standard in detection of drug resistance mutations (DRMs). However, there was variation in the detection of nucleotide mixtures (0–83%) and a significant correlation with the detection of DRMs (p < 0.01). Interpretation of antiretroviral resistance showed ~70% agreement among participants when different interpretation systems were used but >90% agreement with a common interpretation system, within the Stanford University Drug Resistance Database. Using the principles of external quality assessment and a reference laboratory, TAQAS has demonstrated high quality HIV genotyping results from Asian laboratories.
Keywords: HIV, Drug resistance, TAQAS, Quality assessment, Genotyping
1. Introduction
The introduction and use of effective combination antiretroviral (ARV) therapy for Human Immunodeficiency Virus Type-1 (HIV-1) infection has substantially slowed the progression to AIDS and reduced HIV-related mortality world-wide (Grover et al., 2008; Hirsch et al., 2000; Porter et al., 2003). However, it is clear that the efficacy of ARV therapy can be compromised by the emergence and transmission of HIV-1 drug resistant strains (Geretti, 2006). This is of particular concern in developing countries where there is substantial expansion in the use of ARV therapies (Bennett et al., 2008).
The evolution of drug resistant strains within an individual depends on the generation of genetic variation in HIV-1, which drives selection of ARV-resistant variants during therapy. Certain mutations, alone or in combination may reduce ARV susceptibility compared to wildtype virus (Shafer, 2002). Screening clinical specimens for ARV resistance-related mutations using genotypic assays has become standard of care where resources are unconstrained (Hirsch et al., 2003; Vandamme et al., 2004). However, in resource limited settings public health surveillance and patient management may depend on lower-cost “home brew” methods, which have not been rigorously validated through trials to gain approval from regulatory bodies. Therefore, quality assurance and control measures, and careful evaluation of local genotyping methods are essential for national and individual monitoring for ARV resistance.
HIV-1 genotypic resistance testing (HIV genotyping) is a complex, multi-layered test that involves genetic sequencing of the viral genome, editing of the genetic sequence, detection of the presence, or absence of drug resistance mutations (DRMs) that confer ARV resistance, and the interpretation of resistance to a list of ARV drugs. It is not uncommon for laboratories to use a variety of commercial or in-house methods to generate genetic sequence, and to use different software programs to edit the genetic sequence (Demeter et al., 1998; Descamps et al., 2006; Huang et al., 2003; Kijak et al., 2003; Korn et al., 2003; Neuwald et al., 2002). Furthermore, the edited genetic sequence generated by software programs may require manual, subjective assessment and editing by laboratory personnel when there are ambiguities in the chromatogram because of poor sequence quality, or when two or more nucleotide bases are present at one position indicating a mixed population of wildtype and mutant virus. Frequently this is the case when drug resistance has emerged in the setting of decreased access to drug or diminished adherence. The correct identification of nucleotide mixtures that occur at positions associated with ARV resistance is important because these reflect the evolution of resistant virus under the pressure of ARV therapy, or conversely the re-emergence of sensitive virus when drug pressure is removed.
Laboratories use different systems to interpret ARV resistance from the genotypic data and, in addition, may amend the level of ARV resistance from that generated by the interpretation system in the light of other evidence they consider significant (Huang et al., 2003). The complicated nature of the assay, the variety of testing procedures between laboratories, and risks for cross-contamination and sample mix-up and subjective components in sequencing combine to create the potential for intra- and inter-laboratory variation in the outcome of HIV genotyping (Demeter et al., 1998; Descamps et al., 2006; Erali et al., 2001; Galli et al., 2003; Korn et al., 2003; Neuwald et al., 2002; Sayer et al., 2003; Schuurman et al., 2002; Shafer et al., 2001).
Research studies support the use of HIV genotyping as a tool for epidemiological surveillance of ARV resistance, and for improving clinical management (Grover et al., 2008; Hirsch et al., 2000; Shafer et al., 2001). Both of these are valid concerns in the Asian region where ARV therapy regimens are currently being implemented. Accurate epidemiological surveillance and good clinical management both require optimal laboratory performance of HIV genotyping, and minimal intra- and inter-laboratory differences in test outcome.
External quality assessment schemes (EQAS) are designed to assess the accuracy and reliability of laboratory tests to detect problems in testing procedures, and to identify, and subsequently minimize, discrepancies between results (Fujisaki et al., 2007; Quint et al., 1995; Schirm et al., 2002). Participation involves laboratories processing a panel of samples, using their standard protocols, and sending results for analysis to the quality assessment (QA) provider. Regular participation in an EQAS can improve the quality of laboratory diagnosis by detecting testing problems, deficiencies and disparities in and between participants’ processes (Salkin et al., 1997; Valentine-Thon et al., 2001). Thereby, on-going participation in EQAS facilitates improvement in testing processes where necessary, and leads to standardization of test outcomes across participating laboratories.
The TREAT Asia (Therapeutics, Research, Education, and AIDS Training in Asia) Network, supported by the American Foundation for AIDS Research, is building capacity for the genetic analysis of clinical specimens, across subtypes, in laboratories engaged in surveillance and monitoring of drug resistance in individuals receiving ARV therapy. To this end, the TREAT Asia Network has established an EQAS – designated TAQAS – to assess the accuracy and consistency of HIV genotyping across the network of testing laboratories. The objectives of TAQAS, presented here, were first to establish consistency of sequence data, detection of DRMs associated with ARV resistance and evolving viral mixtures in plasma HIV-1 subtype B and non-B clade HIV-1; secondly to assess parity in the interpretation of ARV resistance between participants who may use different systems to interpret ARV resistance from genotypic data; and thirdly to establish a laboratory network to support the implementation of HIV genotyping in the region.
2. Materials and methods
2.1. Participants
Eleven laboratories participated in TAQAS; 10 from Asia and one from South Africa. An American laboratory with expertise in HIV genotyping participated in a reference laboratory capacity. Participants supplied their contact details and a brief outline of their HIV genotyping methods to the QA provider (Table 1). To ensure confidentiality, the QA provider assigned each participant a unique identifier. Where necessary, participants obtained permits to import biological samples of human origin containing infectious virus.
Table 1.
Methods used as described by participants, and mean length of nucleotide sequence returned by participants testing TAQAS panels.
| Participant ID | Sequencing method or commercial assay | Editing Software | ARV resistance interpretation system | Edited nucleotide sequence length (mean) |
|---|---|---|---|---|
| 1a | BigDye Terminator, ABI | Sequencher 4.8, Gene Codes Corporation | Stanford | 1016 |
| 2 | BigDye Terminator, ABI | Sequence Navigator, ABI | Stanford | 949 |
| 3 | TruGene, Bayer HealthCare | TruGene version 11 | TruGene version 12 | 909 |
| 4 | BigDye Terminator version 1.1, ABI | Staden Package version 1.6.0 | Stanford | 1012 |
| 5 | BigDye Terminator, ABI | BioEdit version 7.052 | Stanford | 1004 |
| 6 | BigDye Terminator version 3.1, ABI | GENETYX version 8 | IAS-USA and Standford | 879 |
| 7 | BigDye Terminator version 3.1, ABI | SeqScape version 2.5, ABI | Stanford | 956 |
| 8 | 3730 XL system, ABI | CEQ TM 8000 Genetic Analysis system | Stanford | 1014 |
| 9 | ViroSeq HIV-1 Genotyping kit, Abbott Molecular | Sequencher 4.5, Gene Codes Corporation | Stanford | 1017 |
| 10 | BigDye Terminator version 3.1, ABI | SeqScape version 2.5, ABI | French ANRS | 1012 |
| 11 | ViroSeq HIV-1 Genotyping kit version 2.0, Abbott Molecular | ViroSeq software version 2.6 | Stanford | 962 |
| 12 | PRISM® 3100-Avant Genetic Analyzer, ABI | BioEdit Version 7.0.9, Chromas Lite version 2.01 | Stanford | 1017 |
Manufacturer details: Abbott Molecular Diagnostics, Abbott Park, IL, USA; Applied Biosystems (ABI), Foster City, CA, USA; Bayer HealthCare, Tarrytown, NY, USA; Beckman Coulter Inc., Fullerton, CA, USA; BioEdit, www.mbio.ncsu.edu/BioEdit/bioedit.html; bioMérieux, France; Celera Diagnostics, Rockville, MD, USA; Chromas Lite, Technelysium, Queensland, Australia, www.technelysium.com.au; Gene Codes Corporation, Ann Arbor, MI, USA; GENETYX CORPORATION, Japan; Hastings Software Inc., NY, USA; Invitrogen, Carlsbad, CA, USA; QIAGEN Science MD, USA; TAKARA BIO Inc., Shiga, Japan.
Reference Laboratory.
2.2. Panels
Panels of five coded samples were prepared by the QA provider (Table 2). Sequential panels were designated TAQAS 1, 2 and 3. Samples were either plasma sourced from HIV-1-infected, ARV-treated or -naive individuals, or supernatant harvested from virus co-cultures presented in normal human plasma.
Table 2.
Details of samples including type, country of origin, HIV-1 subtype, prior ARV exposure and estimated viral load of sample source; description of HIV-1 isolates; number of nucleotide mixtures, insertions, and drug resistance mutations in TAQAS 1, 2 and 3.
| TAQAS panel—sample | Sample type | Country of origin | HIV-1 Subtype | Estimated HIV RNA load (log10 copies/ml) | ARV drugs to which source has been exposed | No. NMs/insertions per TAQAS | No. DRMS per TAQAS |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Complete | Partial | DRMs as NMs (%) | |||||||
| 1-A | P | Australia | B | 3.1 | d4T, RTV, AZT, 3TC, NFV, SQV, KAL | |||||
| 1-B | P | Australia | B | 5.1 | 3TC, ddI, d4T, T20, SQV, TDF, KAL, RTV, AZT, IDV, NFV, ABV, EFV, AZT | |||||
| 1-C | P | Australia | B | 4.0 | EFV, d4T, ABV, RTV, CBV, SQV, TPV, TDF, KAL, 3TC, AZT | 73 | 46 | 35 | 11 | 31 |
| 1-D | P | Australia | B | 4.7 | LPV, ABV, NVP, RTV, IDV | |||||
| 1-E | P | Australia | B | 5.0 | None | |||||
| 2-A | P | Australia | B | 5.3 | 3TC, ddI, d4T, T20, SQV, TDF, KAL, RTV, AZT, IDV, NFV, ABV, EFV, AZT | |||||
| 2-B | P | Australia | B | 4.7 | None | |||||
| 2-C | P | Australia | B | >4.0 | LPV, TDF, 3TC | 48 | 59 | 48 | 11 | 23 |
| 2-D | P | Australia | B | >4.0 | KAL, 3TC, ABV | |||||
| 2-E | SN | Uganda | D | ND | Not known | |||||
| 3-A | P | Australia | B | 4.7 | NVP, ddI, ABV, TDF, NFV | |||||
| 3-B | P | Australia | B | 4.7 | SQV, LPV, ABV, T20 | |||||
| 3-C | P | Australia | B | 4.7 | RTV, SQV, IDV, AZT, 3TC, ddI, d4T, NFV, EFV | 109/1 | 42 | 23 | 19 | 45 |
| 3-D | P | Australia | B | 4.5 | ABV, IDV, CBV, T20, RTV, TPN, 3TC, TDF | |||||
| 3-E | SN | Zimbabwe | C | ND | Not known | |||||
P, plasma; SN supernatant for virus co-culture; ND, not done; d4T, stavudine; RTV, ritonavir; AZT, zidovudine; 3TC, lamivudine; NFV, nelfivanir; SQV, saquinavir; KAL, kaletra; ddI, didanosine; T20, enfurviritide; TDF, tenofovir; IDV, indinavir; Abacavir; ABV; EFV, efavirenz; CBV, combivir; TPV, tipranavir; LPV, lopinavir; NVP, nevirapine; NMs, nucleotide mixtures; DRMs, drug resistance mutations.
2.3. Data analysis
Phylogenetic analyses were performed for each sample in comparison with the reference laboratory sequence by the construction of neighbour-joining trees of the protease and reverse transcriptase regions from the edited nucleotide sequences returned by each participant. Bootstrap values >70% (1000 replicates) were applied to confirm the validity of the trees.
Subsequently, the edited nucleotide sequences returned by each participant, including the reference laboratory, were aligned and a target genotype (TG) was deduced for each sample. The TG was a rules based algorithm that generated the most likely consensus quasi-species detectable by DNA sequencing (Table 3) (Sayer et al., 2003). The TG derivation assumed that it was extremely unlikely that two participants incorrectly sequenced the same nucleotide at any one position. The participants’ edited nucleotide sequences were compared with TG and complete and partial differences were distinguished. Complete differences occurred when the participant reported a nucleotide that was different from that defined in the TG, e.g. the participant reported G and the TG was C. Partial differences predominantly occurred when a single nucleotide was reported at a position that was defined as a nucleotide mixture (two or more nucleotides at the same position) in the TG. Partial differences also occurred when a nucleotide mixture that was different from that defined in the TG was reported by the participant, e.g. the TG was a mixture of C and T, and the participant reported a mixture of A and T; and when a nucleotide mixture was reported at a position where there was a single nucleotide in the TG.
Table 3.
Deduction of target genotype.
| Participant ID | Sequence at nucleotide positions 1–7 |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| I | A | A | A | G | C | T | T |
| II | A | T | A | G | C | T | A |
| III | A | A | W | G | C | C | W |
| IV | A | A | A | A | C | T | T |
| V | A | A | A | G | Y | T | T |
| VI | A | A | A | A | C | T | T |
| VII | A | A | A | G | Y | A | T |
| Target genotype | A | A | A | R | Y | T | W |
The target genotype (TG) represents the most likely consensus quasi-species detectable by DNA sequencing from each of the TAQAS panel samples. If one participant sequenced a different nucleotide from that sequenced by the all other participants, either as a partial or as a complete difference, then the TG is the consensus nucleotide sequenced by the other participants (e.g. Table 3 positions 2 and 3: TG=AA). If two or more participants reported an identical nucleotide which was different from the nucleotide reported by the other participants, the TG is a mixture of the two different nucleotide sequences reported (e.g. Table 3 positions 4 and 5: TG = RY). If two participants reported nucleotides different from those reported by the other participants and different from those reported by each other, the TG is the sequence reported by the majority (e.g. Table 3 position 6: TG = T). However, if two participants reported sequences different from those reported by the majority and one is a mixture that is present and contains the sequence reported by the majority and the other participant, the TG is the mixture (e.g. Table 3 position 7; TG = W). The ambiguity codes defined by the International Union of Biochemistry and Molecular Biology were interpreted to indicate mixtures. For example, if a participant reported a “W”, the interpretation was that the position had a mixture of adenine (A) and thymine (T).
The participants’ edited nucleotide sequences were further compared with the TG for the presence of DRMs, for nucleotide mixtures of wildtype and mutant virus, and for the proportion of DRMs present as nucleotide mixtures.
For the purpose of comparing the participants’ interpretation of ARV resistance to 14 drugs, four levels of ARV susceptibility were ascribed: susceptible or no evidence of resistance; potential low-level resistance or low-level resistance; intermediate or possible resistance; resistance or high-level resistance. The interpretation of ARV resistance to each drug was considered to agree when all, or all but one participant, agreed on the level of resistance. Where DRMs for a particular class of ARV drugs were absent, the sample was not included in the analysis. Furthermore, results were excluded for boosted drugs or combinations of drugs, and when the inter-laboratory difference in ARV susceptibility was the consequence of a participant’s failure to detect a DRM(s). The comparison was made between all participants using various interpretation systems, and between a subgroup of participants using the one within the Stanford University Drug Resistance Database (Stanford Database; http://hivd.stanford.edu).
Statistical comparisons were made using Pearson’s correlation coefficient or the chi-squared test.
2.4. Study design
TAQAS panels were shipped frozen with an integrity control to each participant. On receipt, participants were advised to visually inspect the panels and advise the QA provider if the samples had thawed during shipment or were thawed on receipt, then to store the panel at ≤−20 °C prior to testing.
Participants were asked to test the samples using their standard protocol for HIV genotyping, and to interpret the level of ARV resistance using their laboratory’s standard interpretation method (Table 1). For each sample, participants were asked to return a complete, edited nucleotide sequence, the DRMs related to ARV resistance, and their interpretation of the level of resistance to a list of ARV drugs. Participants returned results to the QA provider for analysis electronically or on a CD-ROM sent by the standard postal service. Data analyses and the edited nucleotide sequences aligned against the TG were posted on a secured world-wide-website, and participants were encouraged to review and compare results.
Participants met with the TAQAS provider, experts in HIV genotyping, and the TREAT Asia Laboratory Network, in an annual workshop focused on testing differences and difficulties, methods for improvement of HIV genotyping, and the future directions of TAQAS.
3. Results
Over a 19-month period the participants tested up to three 5-member panels in the TAQAS. TAQAS 1 was tested by 8 Asian laboratories; TAQAS 2 and 3 were tested by 10 Asian laboratories. In addition, all panels were tested by one South African laboratory and one American laboratory (ID: 1) as a reference standard.
All but one of the participants returned complete data sets suitable for analysis. One participant (ID: 12) was unable to amplify sequences from three samples in TAQAS 3 (3-A, 3-B and 3-D); shipment of TAQAS 3 to this participant was delayed, and as a consequence sample integrity may have been compromised.
Participants differed in the methods that they used to perform DNA sequencing, the software programs used to edit nucleotide sequence, and the systems used to interpret ARV resistance (Table 1). Primers sequences were not known to the QA provider. Most participants (9 of 12 testing TAQAS 3) used the Stanford Database to interpret ARV resistance. Three participants used either TruGene (Bayer HealthCare, Tarrytown, NY), the French ANRS (National Agency for AIDS Research) AC11 Resistance Algorithm, or the Stanford Database in combination with recommendations from the International AIDS Society-USA (Johnson et al., 2006).
3.1. Phylogenetic analyses
Construction of neighbour-joining trees found that all sequences, except those mentioned below, clustered closest to other sequences from the same sample compared with other samples, with high bootstrap values (data not shown). This confirmed the accuracy of sample handling and processing as well as sequence integrity, and excluded sample contamination.
Sequences returned by two participants testing two of the samples were significantly distant from the sequences submitted by the other participants testing these samples. The protease sequence returned by one participant (ID: 12) testing TAQAS 2-C, and the protease sequence returned by one participant (ID: 2) testing TAQAS 3-C were outliers. These sequences were removed from further analysis. Participants were informed of these errors via the report prepared by the QA provider, and advised to investigate the source of the error and to consider re-sequencing the respective samples.
3.2. Agreement over entire nucleotide sequence
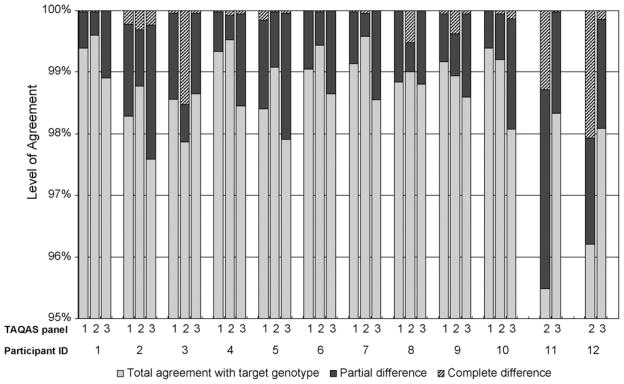
There was a high level of agreement between the participants’ edited nucleotide sequences and the TG for each TAQAS panel (Fig. 1). Comparison with the TG took into account the different sequence lengths returned by participants (Table 1). For the majority of participants the level of agreement with the TG was >98%, and <0.5% of the differences were complete differences (Fig. 1). Three participants reported a lower level of agreement for the first panel they tested: Participants ID: 11 and ID: 12 respectively reported 95.49% and 96.20% agreement for TAQAS 2 but both participants reported >98% agreement in TAQAS 3. Participant ID: 3 reported 1.53% complete differences in TAQAS 2, but 0.04% complete differences in TAQAS 1 and 3. Two remaining participants that reported >0.5% complete differences in their nucleotide sequence compared with the TG when testing their first panel in TAQAS: Participants ID: 11 and ID: 12 respectively reported 1.28% and 2.07% complete differences in TAQAS 2; these participants respectively reported 0.02% and 0.15% complete differences in TAQAS 3.
Fig. 1.
Level of agreement between the participants’ entire edited nucleotide sequences and the target genotype (TG) in up to three TAQAS. Most participants reported >98% agreement with the TG (grey bars) (10 of 10 participants that tested TAQAS 1,9 of 12 participants that tested TAQAS 2, and 10 of 12 participants that tested TAQAS 3). Most of the differences reported from the TG were partial differences (black bars). Less than 0.5% of the differences reported from the TG were complete differences (hatched bars) for the majority of participants (all participants that tested TAQAS 1 and 3, and 8 of 12 participants that tested TAQAS 2).
3.3. Detection of DRMs associated with ARV resistance
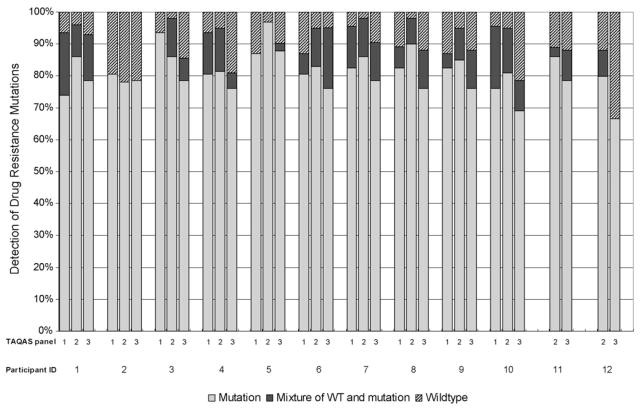
Comparison of the edited nucleotide sequences at positions associated with ARV resistance found that participants varied in their detection of DRMs and in the proportion of DRMs they detected as nucleotide mixtures. There were a total of 46, 59 and 42 DRMs in the TG of all the samples in TAQAS 1, 2 and 3, respectively. The percent of DRMs in each participant’s sequences compared with the TG was determined for each of the three TAQAS (Fig. 2). Sequences from most participants contained >80% of the DRMs. The reference laboratory reported an average of 94% of the DRMs, and sequences from eight of the participants (IDs: 3, 4, 5, 6, 7, 8, 9 and 10) matched the reference laboratory’s performance in at least one of the TAQAS panels. For many participants (7 of 11) and the reference laboratory, detection of DRMs in TAQAS 3 declined compared with previous performance.
Fig. 2.
The participants’ detection of drug resistance mutations (DRMs) at nucleotide sequences positions associated with ARV drug resistance in the target genotype (TG). Most participants reported >80% of the DRMs present in the TG as mutations (grey bars) or nucleotide mixtures of wildtype and mutation (black bars). Participants’ sequences varied in the percent of DRMs reported as nucleotide mixtures (black bars). Some participants reported wildtype at >20% of the nucleotide sequences positions associated with ARV drug resistance in the TG (hatched bars).
Sequences from three participants testing TAQAS 2 and/or 3 contained fewer than 80% of the DRMs. Lower detection of DRMs was paralleled by a lack of detection of nucleotide mixtures at DRMs by two of these participants (ID: 2 and ID: 12). The other participant (ID: 10) detected a portion of DRMs as nucleotide mixtures.
3.4. Detection of nucleotide mixtures
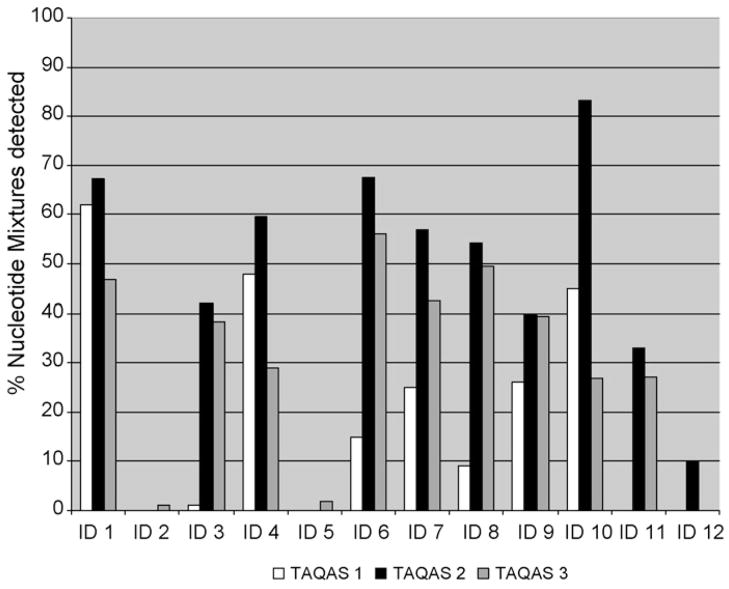
Inclusive of the reference laboratory’s data, participants’ detection of nucleotide mixtures over the entire nucleotide sequence varied in all TAQAS panels (Fig. 3). Most participants (8 of 10) (IDs: 1,3,4,6,7,8,9 and 10) increased their detection of nucleotide mixtures in TAQAS 2 compared with TAQAS 1. However, most (9 of 12) (IDs: 1,3,4,6,7,8,10,11 and 12) participants’ detection of nucleotide mixtures decreased in TAQAS 3 compared with TAQAS 2. Three participants (IDs: 2,5 and 12) consistently detected low levels of, or no, nucleotide mixtures in all the TAQAS panels tested (ID: 2 and ID: 5 tested three TAQAS; ID: 12 tested TAQAS 2 and TAQAS 3).
Fig. 3.
The participants’ detection of nucleotide mixtures over the entire sequence in three TAQAS. Most (8 of 10) participants increased their detection of nucleotide mixtures in TAQAS 2 compared with TAQAS 1. Ten participants detected proportionally less nucleotide mixtures in TAQAS 3 compared with their detection of nucleotide mixtures in TAQAS 2. Three participants (ID: 2, 5 and 12) consistently detected low levels of or no nucleotide mixtures in all the TAQAS tested.
A moderate linear relationship was demonstrated between the participants’ detection of DRMs and nucleotide mixtures in samples that contained DRMs in TAQAS 1 with Pearson’s r =(N = 10) .61, p<0.0001 and TAQAS 3 with Pearson’s r =(N =12) .49, p<0.0001. The correlation between detection of DRMs and nucleotide mixtures approached significance in TAQAS 2 with Pearson’s r =(N = 12) .25, p = 0.054.
3.5. Interpretation of ARV resistance
The ARV resistance profiles to 14 ARV drugs reported by the participants and the reference laboratory were compared using the criteria described in the methods (Table 4). The comparison was made between all participants using various interpretation systems, and between a subgroup of participants who used a single (most common) interpretation system, the Stanford Database. When data from all participants were compared the level of the inter-laboratory agreement for ARV resistance was 71%, 64% and 76% for TAQAS 1, 2 and 3, respectively; between only those participants that used the Stanford Database the level of agreement was 97%, 95% and 83% for TAQAS 1, 2 and 3, respectively. The level of agreement between the participants’ interpretation of ARV resistance in TAQAS 1 and TAQAS 2 was significantly higher when all participants used the same system (TAQAS 1: p = 0.003; TAQAS 2: p<0.0001).
Table 4.
Agreement between ARV resistance profiles reported by participants testing three TAQAS. The panel identifier and the number of participants (n) are shown at the top of the table. The interpretation system is given as “various” when participants used different interpretation systems or “Stanford” for the subgroup of participants that used the Stanford Database. Interpretation of resistance was considered to agree when all, or all but one participant agreed, and results were excluded for boosted or combinations of drugs, when samples did not contain drug resistance mutations (DRMs), and where disparities resulted from the participants’ failure to detect DRM(s). The level of agreement is indicated as the proportion of panel samples where participants agreed on the ARV resistance interpretation as a fraction of the number of samples tested.
| ARV drugs | Interpretation systems used (level of agreement (%)) |
|||||
|---|---|---|---|---|---|---|
| TAQAS 1 (n = 10) Various | TAQAS 1 (n = 5) Stanford | TAQAS 2 (n= 12) Various | TAQAS 2 (n = 6) Stanford | TAQAS 3 (n = 12) Various | TAQAS 3 (n = 9) Stanford | |
| Amprenavir | 2/3 | 3/3 | 2/3 | 3/3 | 2/3 | 2/3 |
| Indinavir | 2/3 | 3/3 | 2/3 | 3/3 | 2/3 | 2/3 |
| Nelfinavir | 2/3 | 3/3 | 3/3 | 3/3 | 2/3 | 2/3 |
| Ritonavir | 2/3 | 3/3 | 3/3 | 3/3 | 2/3 | 2/3 |
| Saquinavir | 2/3 | 3/3 | 2/3 | 3/3 | 2/3 | 2/3 |
| Lopinavir | 2/3 | 3/3 | 0/3 | 1/3 | 2/3 | 2/3 |
| Atazanavir | 2/3 | 3/3 | 2/3 | 3/3 | 1/3 | 2/3 |
| Abacavir | 2/2 | 2/2 | 3/3 | 3/3 | 2/3 | 3/3 |
| Lamivudine | 2/2 | 2/2 | 3/3 | 3/3 | 3/3 | 3/3 |
| Stavudine | 2/2 | 2/2 | 0/3 | 3/3 | 3/3 | 3/3 |
| Zidovudine | 1/2 | 1/2 | 1/3 | 3/3 | 3/3 | 3/3 |
| Tenofovir | 0/2 | 2/2 | 2/3 | 3/3 | 3/3 | 3/3 |
| Efavirenz | 2/2 | 2/2 | 1/3 | 3/3 | 2/3 | 3/3 |
| Nevirapine | 2/2 | 2/2 | 3/3 | 3/3 | 3/3 | 3/3 |
| Total (%) | 25/35 (71)a | 34/35 (97)a | 27/42 (64)a | 40/42 (95)a | 31/42 (74) | 35/42 (83) |
Level of inter-laboratory agreement when participants used various systems was lower than when all participants used the Stanford Database in TAQAS 1 and 2 (p < 0.01).
In TAQAS 3 there were discrepancies between the Stanford Database users’ interpretation of the ARV resistance to many of the protease inhibitors. Most of these discrepancies occurred when some participants reported no evidence of protease inhibitor resistance while other participants reported potential low-level resistance. As a consequence, no difference in the level of agreement between the participants’ interpretation of ARV resistance in TAQAS 3 was found when either various systems or the Stanford Database was used (p >0.05).
4. Discussion
The goal of TAQAS is to use quality assessment to build capacity in laboratories in the Asian region, by assuring the quality of their HIV genotyping processes and by ensuring parity of test outcome between participating laboratories including a reference laboratory. A further goal is to use TAQAS as a vehicle to develop a collaborative laboratory network, and thereby to encourage regional dissemination of laboratory know-how and expertise, to facilitate troubleshooting of testing problems, and to support laboratories setting up HIV genotyping.
TAQAS is an ongoing program. The present study reports the results of participants testing the first three TAQAS panels. Most (13 of 15) of the samples were plasma derived from HIV-1-infected, ARV-treated or -naive individuals; two samples were normal plasma spiked with co-culture-derived HIV-1. Others offering quality assessment for HIV genotyping have used constructed samples (Neuwald et al., 2002; Schuurman et al., 1999, 2002). Their samples were characterised so that the DRMs presented and the proportion of nucleotide mixtures were known to the QA provider, and therefore the participants’ detection of DRMs and nucleotide mixtures could be quantified. However, it has been argued that the use of constructed samples does not adequately assess the HIV genotyping process because naturally occurring viral quasi-species are not present in constructed samples (Huang et al., 2003; Sayer et al., 2003). Furthermore, intra-laboratory differences have been demonstrated to occur at the RT-PCR level of the process, and therefore DNA was considered an inappropriate starting template for assessing an RT-PCR assay (Galli et al., 2003). For these reasons, the aim of TAQAS is to use clinical plasma samples and low passage virus isolates as analytes in the program.
Comparison with a TG, generated for each sample using a rules based algorithm from the edited protease and reverse transcriptase nucleotide sequences submitted by all participants, including a reference laboratory, was considered to be an impartial method of assessing inter-laboratory agreement (Sayer et al., 2003). All participants achieved a high level of agreement at the nucleotide sequence level (>98%) when they tested their second and, in many cases, first TAQAS panel. In an analogous QA program, 97.9% agreement at the nucleotide sequence level was reported between nine laboratories testing four plasma samples (Sayer et al., 2003). Therefore, TAQAS participants were considered to have achieved a level of inter-laboratory agreement at the level of edited nucleotide sequences comparable with a group of laboratories testing in well-resourced settings.
Consistent with the findings of other studies, only a minority of the nucleotide sequence differences (<0.5%) were complete differences (Demeter et al., 1998; Sayer et al., 2003; Shafer et al., 2001). Differences reported were either a single nucleotide at a position that was defined as a nucleotide mixture in the TG, or a nucleotide mixture different from the mixture in the TG. TAQAS participants used different sequencing methods, and therefore presumably different sequencing primers, to produce their sequences. Furthermore, they were sequencing samples that contained viral quasi-species. Therefore a low level of partial nucleotide sequence differences between participants’ sequences should be expected, but it should be ensured that the level remains low and does not affect the participants’ detection of DRMs. Importantly, participants that reported a lower level of agreement with the TG in their first TAQAS increased their level of agreement in subsequent panels.
Most participants detected at least 80% of the DRMs in each of the three TAQAS panels, and many participants matched the performance of the reference laboratory in detecting >90% of the DRMs. Two participants that detected fewer DRMs also detected few nucleotide mixtures. Substantial inter-laboratory variation in detection of DRMs has been reported in other QA programs. Participants testing constructed samples in a world-wide proficiency testing program were reported to have detected between 0% and 88% of DRMs (Schuurman et al., 1999, 2002). However, participants testing clinical samples, similar to those presented by TAQAS, detected ≥82% of the DRMs (Sayer et al., 2003). Thereby, the detection of DRMs by TAQAS participants in samples containing multiple DRMs and nucleotide mixtures was comparable to other laboratories’ performances.
Increased sample complexity potentially explained the decreased detection of DRMs by many of the participants in TAQAS 3 compared with previous TAQAS panels. Others have demonstrated that complexity of sequence interpretation is increased by the presence of mixed virus populations, and that sample complexity influences the detection of DRMs (Descamps et al., 2006; Erali et al., 2001; Neuwald et al., 2002). Furthermore, laboratories may fail to detect DRMs presented as viral mixtures when they made up only a portion of the viral population (Schuurman et al., 1999).
A relationship was demonstrated between the detection of DRMs and nucleotide mixtures in the participants’ edited nucleotide sequences. The same correlation has been found by another HIV genotyping quality assessment program, supporting the contention that under-detection of nucleotide mixtures could potentially lead to underreporting of DRMs (Erali et al., 2001; Land et al., 2004; Sayer et al., 2003). Therefore, in TAQAS, of nucleotide mixtures was considered an indicator of the quality of the HIV genotyping outcome. The participants’ detection of all nucleotide mixtures in the three TAQAS panels varied. Most participants demonstrated an increased detection of nucleotide mixtures between the first and second TAQAS panels, but in general this increase was not maintained in TAQAS 3. The complexity of TAQAS 3, as previously discussed, may have influenced this outcome. In addition, several participants consistently detected low levels of or no nucleotide mixtures in all the TAQAS panels. No common aspects in the testing methods limited to the participants who under detected nucleotide mixtures were identified however, participants supplied only limited information about their respective HIV genotyping methods. Under-detection of DRMs by two participants (ID: 2 and ID: 12) may be explained by their low detection of nucleotide mixtures. However, suboptimal detection of nucleotide mixtures did not always correlate with low detection of DRMs, which may indicate that some participants reported only the mutant nucleotide base when a mixture was present (ID: 5 and ID: 10). Because of their clinical significance, the importance of the detection of nucleotide mixtures has been a focus of TAQAS. Identifying nucleotide mixtures, which relies in part on visual inspection of electropherograms, has been the subject of training at past TAQAS workshops, and further training in this aspect of the test process is planned.
Not surprisingly, concordance in the interpretation of ARV resistance was higher when all participants used the same, as compared to various, interpretation system. Importantly, the extent of parity between participants when they used various systems, and the extent when they used the same system, were the same as those achieved in a national EQAS conducted in Australia (Land et al., 2004). Studies have found differences between systems to interpret ARV resistance profiles to combinations of DRMs (de Luca et al., 2003; Fox et al., 2007; Huang et al., 2003; Ravela et al., 2003; Ross et al., 2005; Snoeck et al., 2006). The findings of TAQAS support the previous evidence that different interpretations of ARV resistance will occur when different systems are used, arguing for a single standard.
The limitations of the TAQAS program reported here include the use of predominantly Subtype B virus in a geographical region where non-B subtypes are the norm, a small number of laboratory participants, limited information about the participants’ HIV genotyping methods, and the lack of defined criteria to assess laboratory performance. It is a future objective of TAQAS to address the deficiencies that have been identified in the initial stages of the program.
To date participation in TAQAS by 10 Asian laboratories has shown that these laboratories produce quality output from HIV genotyping that is on par with the performance of a reference laboratory, and with the performances of laboratories in well-resourced settings. Low levels of inter-laboratory differences were demonstrated between edited nucleotide sequences, detection of DRMs, and interpretation of ARV resistance to multiple drugs.
Over the course of testing up to three quality assessment panels, participants in TAQAS have improved or maintained a high standard of quality in HIV genotyping outcome. The impact of the variations that exist as a result of the differences in test methods could be minimised by adopting standardized approaches in, for example, interpretation of edited nucleotide sequence and editing raw sequence. These options will be explored as future TAQAS initiatives.
Acknowledgments
The TREAT Asia Quality Assurance Scheme is an initiative of TREAT Asia, a program of amfAR, The Foundation for AIDS Research, and is supported in part by amfAR with additional support from the Dutch Ministry of Foreign Affairs through a partnership with Stichting Aids Fonds. The National Centre in HIV Epidemiology and Clinical Research is funded by the Australian Government Department of Health and Ageing, and is affiliated with the Faculty of Medicine of The University of New South Wales. The content of this publication is, however, solely the responsibility of the authors and does not necessarily represent the official views of any of the institutions mentioned above.
The TAQAS Laboratory Network: L. Kang, Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China; W.C. Yam and J. Chen, HIV Molecular Biology Laboratory, Department of Microbiology, Queen Mary Hospital, Hong Kong; S. Oka and X. Bi, AIDS Clinical Center, International Medical Center of Japan, Tokyo, Japan; W. Sugiura, National Institute of Infectious Diseases, Tokyo, Japan; N. Kumarasamy, V. Madhavan and S. Saravanan, Infectious Diseases Laboratory, Chennai, India; K.P. Ng and A. Kamarulzaman, HIV Research Laboratory, Medical Microbiology Department, University of Malaya, Kuala Lumpur, Malaysia; W. Stevens and C. Wallis, National Health Laboratory Services (NHLS)—University of Witwatersrand, Johannesburg, S. Africa; Y.M.A. Chen and Y.J. Chen, AIDS Centre of the National Yang-Ming University, Taipei, Taiwan; R. Sutthent, Department of Microbiology, Faculty of Medicine, Siriraj Hospital, Bangkok, Thailand; K. Ruxrungtham and S. Sirivichayakul, Division of Allergy and Clinical Immunology, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand; W. Chantratita, W. Piroj and C. Watitpun, Virology and Molecular Microbiology Unit, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand; M. Winters, D. Katzenstein and M. Balamane, Center for AIDS Research, Division of Infectious Diseases and Geographic Medicine, Stanford University, CA, USA; T.T.X. Lien and T.C. Thanh, HIV/AIDS laboratory, Pasteur Institute in Hochiminh City, Ho Chi Minh City, Vietnam.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit:
References
- Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization’s global strategy for prevention and assessment of HIV drug resistance. Antiviral Ther. 2008;13:1–13. [PubMed] [Google Scholar]
- de Luca A, Antinori A, di Giambenedetto S, Cingolani A, Perno CF, Cauda R. Interpretation systems for genotypic drug resistance of HIV-1. Scand J Infect Dis Suppl. 2003;106:29–34. doi: 10.1080/03008870310009623. [DOI] [PubMed] [Google Scholar]
- Demeter LM, D’Aquila R, Weislow O, Lorenzo E, Erice A, Fitzgibbon J, Shafer R, Richman D, Howard TM, Zhao Y, Fisher E, Huang D, Mayers D, Sylvester S, Arens M, Sannerud K, Rasheed S, Johnson V, Kuritzkes D, Reichelderfer P, Japour A ACTG Sequencing Working Group. AIDS Clinical Trials Group. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. J Virol Methods. 1998;75:93–104. doi: 10.1016/s0166-0934(98)00100-1. [DOI] [PubMed] [Google Scholar]
- Descamps D, Delaugerre C, Masquelier B, Ruffault A, Marcelin AG, Izopet J, Chaix ML, Calvez V, Brun-Vezinet F, Costagliola D The ANTS Resistance Group. Repeated HIV-1 resistance genotyping external quality assessments improve virology laboratory performance. J Med Virol. 2006;78:159–160. doi: 10.1002/jmv.20522. [DOI] [PubMed] [Google Scholar]
- Erali M, Page S, Reimer LG, Hillyard DR. Human Immunodeficiency Virus Type 1 drug resistance testing; a comparison of three sequence-based methods. J Clin Microbiol. 2001;39:2157–2165. doi: 10.1128/JCM.39.6.2157-2165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox ZV, Geretti AM, Kjaer J, Dragsted UB, Phillips AN, Gerstoft J, Staszewski S, Clotet B, von Wyl V, Lundgren JD. The ability of four genotypic interpretation systems to predict virological response to ritonavir-boosted protease inhibitors. AIDS. 2007;21:2033–2042. doi: 10.1097/QAD.0b013e32825a69e4. [DOI] [PubMed] [Google Scholar]
- French ANRS (National Agency for AIDS Research) [accessed 25/07/2008.];AC11 Resistance Algorithm. 2008 http://www.hivfrenchresistance.org/table.html.
- Fujisaki S, Fujisaki S, Ibe S, Asagi T, Itoh T, Yoshida S, Koike T, Oie M, Konda M, Sadamasu K, Nagashima M, Gatanaga H, Matsuda M, Ueda M, Masakane A, Hata M, Mizogami Y, Mori H, Minami R, Okada K, Watanabe K, Shirasaka T, Oka S, Sugiura W, Kaneda T. Performance and quality assurance of genotypic drug-resistance testing for human immunodeficiency Virus Type 1 in Japan. Jpn J Infect Dis. 2007;60:113–117. [PubMed] [Google Scholar]
- Galli RA, Sattha B, Wynhoven B, O’Shaughnessy MV, Harrigan PR. Sources and magnitude of intralaboratory variability in a sequence-based genotypic assay for Human Immunodeficiency Virus Type 1 drug resistance. J Clin Microbiol. 2003;41:2900–2907. doi: 10.1128/JCM.41.7.2900-2907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geretti AM. Clinical implications of HIV drug resistance to nucleoside and nucleotide reverse transcriptase inhibitors. AIDS Rev. 2006;8:210–220. [PubMed] [Google Scholar]
- Grover D, Copas A, Green H, Edwards SG, Dunn DT, Sabin C, Phillips A, Allen E, Pillay D. What is the risk of mortality following diagnosis of multidrug-resistant HIV-1? J Antimicrobial Chemother. 2008;61:705–713. doi: 10.1093/jac/dkm522. [DOI] [PubMed] [Google Scholar]
- Hirsch MS, Brun-Vézinet F, D’Aquila RT, Hammer SM, Johnson VA, Kuritzkes DR, Loveday C, Mellors JW, Clotet B, Conway B, Demeter LM, Vella S, Jacobsen DM, Richman DD. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- Hirsch MS, Brun-Vézinet F, Clotet B, Conway B, Kuritzkes DR, D’Aquila RT, Demeter LM, Hammer SM, Johnson VA, Loveday C, Mellors JW, Jacobsen DM, Richman DD. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis. 2003;37:113–128. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- Huang DD, Eshleman SH, Brambilla DJ, Palumbo PE, Bremer JW. Evaluation of the editing process in Human Immunodeficiency VirusType 1 genotyping. J Clin Microbiol. 2003;41:3265–3272. doi: 10.1128/JCM.41.7.3265-3272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VA, Brun-Vézinet F, Clotet B, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: fall 2006. Top HIV Med. 2006;14:125–130. [PubMed] [Google Scholar]
- Kijak GH, Rubio AE, Pampuro SE, Zala C, Cahn P, Galli R, Montaner JS, Salomón H. Discrepant results in the interpretation of HIV-1 drug-resistance genotypic data among widely used algorithms. HIV Med. 2003;4:72–78. doi: 10.1046/j.1468-1293.2003.00131.x. [DOI] [PubMed] [Google Scholar]
- Korn K, Reil H, Walter H, Schmidt B. Quality control trial for human immunodeficiency virus type 1 drug resistance testing using clinical samples reveals problems with detecting minority species and interpretation of test results. J Clin Microbiol. 2003;41:3559–3565. doi: 10.1128/JCM.41.8.3559-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land S, Sayer D, Gizzarelli L, Birch C, Emery S, Hales G, French M, Dax EM. An international external quality assessment scheme to standardise the interpretation of HIV-1 genotypic antiretroviral resistance testing. XV International AIDS Conference abstract MoPeB3086; 11–16 July 2004.2004. [Google Scholar]
- Neuwald PD, Funelas MB, DelCarmen JP, Raybold AW, Jorgensen PA. Results of the 2001 AcroMetrix HIV-1 resistance proficiency program. J Clin Virol. 2002;25:S55–S63. doi: 10.1016/s1386-6532(02)00191-9. [DOI] [PubMed] [Google Scholar]
- Porter K, Babiker A, Bhaskaran K, Darbyshire J, Pezzotti P, Porter K, Walker AS CASCADE Collaboration. Determinants of survival following HIV-1 seroconversion after the introduction of HAART. Lancet. 2003;18:1267–1274. doi: 10.1016/s0140-6736(03)14570-9. [DOI] [PubMed] [Google Scholar]
- Quint WGV, Heijtink RA, Schirm J, Gerlich WH, Niesters HGM. Reliability of methods for Hepatitis B DNA detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravela J, Betts BJ, Brun-Vézinet F, Vandamme AM, Descamps D, van Laethem K, Smith K, Schapiro JM, Winslow DL, Reid C, Shafer RW. HIV-1 protease and reverse transcriptase mutation patterns responsible for discordances between genotypic drug resistance interpretation algorithms. J Acquir Immune Defic Syndr. 2003;1:8–14. doi: 10.1097/00126334-200305010-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L, Boulmé R, Fisher R, Hernandez J, Florance A, Schmit JC, Williams V. A direct comparison of drug susceptibility to HIV type 1 from antiretroviral experienced subjects as assessed by the antivirogram and PhenoSense assays and by seven resistance algorithms. AIDS Res Hum Retroviruses. 2005;21:933–939. doi: 10.1089/aid.2005.21.933. [DOI] [PubMed] [Google Scholar]
- Salkin IF, Limberger RJ, Stasik D. Commentary on the objectives and efficacy of proficiency testing in microbiology. J Clin Microbiol. 1997;35:1921–1923. doi: 10.1128/jcm.35.8.1921-1923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer DC, Land S, Gizzarelli L, French M, Hales G, Emery S, Christiansen FT, Dax EM. Quality assessment program for genotypic antiretroviral testing improves detection of drug resistance mutations. J Clin Microbiol. 2003;41:227–236. doi: 10.1128/JCM.41.1.227-236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm J, van Loon AM, Valentine-Thon E, Klapper P, Reid J, Cleator GM. External quality assessment programme for qualitative and quantitative detection of Hepatitis C virus RNA in diagnostic virology. J Clin Microbiol. 2002;40:2973–2980. doi: 10.1128/JCM.40.8.2973-2980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the Human Immunodeficiency Virus Type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman R, Brambilla D, de Groot T, Huang D, Land S, Bremer J, Benders I, Boucher C. Underestimation of HIV Type 1 drug resistance mutations: results from the ENVA-2 genotyping proficiency program. AIDS Res Hum Retroviruses. 2002;4:243–248. doi: 10.1089/088922202753472801. [DOI] [PubMed] [Google Scholar]
- Shafer RW, Hertogs K, Zolopa AR, Warford A, Bollr S, Betts BJ, Merigan TC, Harrigan R, Larder BA. High degree of interlaboratory reproducibility of Human Immunodeficiency Virus Type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J Clin Microbiol. 2001;39:1522–1529. doi: 10.1128/JCM.39.4.1522-1529.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer RW. Genotypic testing for Human Immunodeficiency Type 1 drug resistance. Clin Micro Rev. 2002;15:247–277. doi: 10.1128/CMR.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck J, Kantor R, Shafer RW, van Laethem K, Deforche K, Carvalho AP, Wynhoven B, Soares MA, Cane P, Clarke J, Pillay C, Sirivichayakul S, Ariyoshi K, Holguin A, Rudich H, Rodrigues R, Bouzas MB, Brun-Vézinet F, Reid C, Cahn P, Brigido LF, Grossman Z, Soriano V, Sugiura W, Phanuphak P, Morris L, Weber J, Pillay D, Tanuri A, Harrigan RP, Camacho R, Schapiro JM, Katzenstein D, Vandamme AM. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob Agents Chemother. 2006;50:694–701. doi: 10.1128/AAC.50.2.694-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [accessed on 25/07/2008.];Stanford University Drug Resistance Database. 2008 http://hivdb.stanford.edu/pages/algs/HIVdb.html.
- Valentine-Thon E, van Loon AM, Schirm J, Reid J, Klapper P, Cleator GM. European proficiency testing programme of molecular detection and Quantitation of Hepatitis B virus DNA. J Clin Microbiol. 2001;39:4407–4412. doi: 10.1128/JCM.39.12.4407-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme AM, Sonnerborg A, Ait-Khaled M, Albert J, Asjo B, Bacheler L, Banhegyi D, Boucher C, Brun-Vézinet F, Camacho R, Clevenbergh P, Clumeck N, Dedes N, de Luca A, Doerr HW, Faudon JL, Gatti G, Gerstoft J, Hall WW, Hatzakis A, Hellmann N, Horban A, Lundgren JD, Kempf D, Miller M, Miller V, Myers TW, Nielsen C, Opravil M, Palmisano L, Perno CF, Phillips A, Pillay D, Pumarola T, Ruiz L, Salminen M, Schapiro J, Schmidt B, Schmit JC, Schuurman R, Shulse E, Soriano V, Staszewski S, Vella S, Youle M, Ziermann R, Perrin L. Updated European recommendations for the clinical use of HIV drug resistance testing. Antiviral Ther. 2004;9:829–848. [PubMed] [Google Scholar]