Abstract
The generation of a large number of fully functional hepatocytes from a renewable cell source can provide an unlimited resource for bioartificial liver devices and cell replacement therapies. We have established a directed differentiation system using sodium butyrate treatment to generate an enriched population of hepatocyte-like cells from embryonic stem cells. A metabolic analysis of the hepatocyte populations revealed glycolytic and mitochondrial phenotypes similar to mouse hepatoma cells, implying that these cells represent an immature hepatocyte phenotype. To mediate further differentiation, S-NitrosoAcetylPenicillamine (SNAP), a nitric oxide donor, was utilized to induce mitochondrial development in the precursor populations. A comparative analysis of the different treated populations showed that 500 μM SNAP treatment resulted in the generation of an enriched population of metabolically mature hepatocyte-like cells with increased differentiated function. Specifically, 500 μM SNAP treatment increased glucose consumption, lactate production rates, mitochondrial mass, and potential as compared to untreated populations. In addition, functional analysis revealed that intracellular albumin content, urea secretion rates, and cytochrome P450 7a1 promoter activity were increased in the treated population. The methodology described here to generate an enriched population of metabolically and functionally mature hepatocyte-like cells may have potential implications in drug discovery and regenerative medicine.
Introduction
The liver plays a significant role in coordinating whole-body metabolism. Hepatocytes constitute approximately 70% of the cellular population of the liver and perform major metabolic functions such as plasma protein synthesis and transport, xenobiotic metabolism, glucose homeostasis, urea synthesis, and ketogenesis. From a clinical standpoint, it is known that in cases of irreversible liver failure such as cirrhosis and fulminant hepatic failure, an alternative functional hepatic device to sustain life is vital. While the extracorporeal bioartificial liver device is a promising technology for the treatment of liver failure,1,2 the difficulty in in vitro culture of hepatocytes and scarcity of cells are impending problems. From a diagnostic standpoint, hepatocytes are a useful cell source required for in vitro drug screening and toxicity studies. Thus, generation of fully functional hepatocyte-like cells from a renewable cell source can provide an unlimited resource for clinical and diagnostic applications.
Embryonic stem (ES) cells are highly proliferative, pluripotent cells isolated from the inner cell mass of the embryo.3 These cells, when cultured in vitro under suitable conditions, proliferate indefinitely and have the potential to generate almost any cell type in the body, including hepatocytes,4–7 under controlled differentiation regimens. Currently, the two major problems with obtaining hepatocytes from ES cells are (i) generation of a mixed population and (ii) incomplete functional differentiation into the hepatocyte lineage.
In this regard, we have previously developed a directed differentiation system to obtain an enriched population of hepatocyte-like cells using sodium (Na) butyrate treatment.6 These cells have been shown to possess hepatocyte-specific characteristics. In addition, a metabolic analysis of the differentiated populations revealed a glycolytic phenotype similar to fetal hepatocytes and mouse hepatoma cells. However, from a functional standpoint, these cells have lower urea and albumin secretion rates than mature hepatocytes.8
It is well known that nitric oxide triggers mitochondrial biogenesis in a variety of mammalian cells.9–11 The phenomenon of mitochondrial biogenesis involves the increase in mitochondrial mass, activity, and the increased activity of respiratory enzymes implicated in oxidative metabolism. Further, both nitric oxide (NO) and nitric oxide synthase (NOS) isoforms have been shown to induce differentiation of nerve cells, tumor cells, and cardiomyocytes.12–14 The process of mitochondrial differentiation has been known to be present simultaneously with mitochondrial proliferation during the liver developmental process.15 Biochemically, this phenomenon is associated with an increase in activity of mitochondria and hepatic enzymes involved in energy transduction and liver-specific metabolic pathways as a result of de novo protein synthesis of the mitochondria.
Because hepatic function is tightly linked to intermediary metabolism and S-NitrosoAcetylPenicillamine (SNAP) has been previously shown to induce mitochondria biogenesis in multiple cell lines, we investigated the effects of SNAP on the functional and metabolic capacity of these cells.
Materials and Methods
ES cell culture
The ES-D3 cell line (ATCC, Manassas, VA) was maintained in an undifferentiated state in T-75 gelatin-coated dishes (BD-Biosciences, Bedford, MA) in knockout Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) containing 15% knockout serum (Cat # 10828-028; Gibco), 4 mM L-glutamine (Gibco), 100 U/mL penicillin (Gibco), 100 U/mL streptomycin (Gibco), 10 μg/mL gentamicin (Gibco), 1000 U/mL ESGRO (LIF supplement) (Chemicon, Temecula, CA), and 0.1 mM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO). To maintain cells in the undifferentiated state, media was changed every 2 days until plates were confluent. Cells were dissociated using trypsin-EDTA (Cat # 25200-056; Gibco) and passaged by further replating. Only cells between passages 10 and 20 were used for differentiation. All cell cultures were maintained at 37°C in a humidified 5% CO2 incubator.
Differentiation using Na butyrate and SNAP
On day 0 of differentiation, ES cells were plated at a density of 105 cells/well in 0.1% gelatin-coated six-well plates in Iscove's modified Dulbecco's medium (IMDM) (Cat # 31980-030; Gibco) containing 20% fetal bovine serum (Gibco), 4 mM L-glutamine (Gibco), 100 U/mL penicillin, 100 U/mL streptomycin (Gibco), and 10 μg/mL gentamicin (Gibco). To coat plates with gelatin, 2 mL of 0.1% gelatin solution was added to each well of a polystyrene tissue culture–treated plate. Cells were exposed to 1% dimethylsulfoxide (DMSO) (Sigma-Aldrich) for the next 4 days, followed by exposure to 2.5 mM Na butyrate (Cat # B5887; Sigma-Aldrich) for 6 days. Media was replaced daily. For hepatic functional assessment, cells were removed from the primary culture dish and replated on day 11 of differentiation in polystyrene 12-well plates at a density of 105 cells/well. On day 12, 24 h after replating, cells were exposed to IMDM supplemented with different concentrations of SNAP (Sigma-Aldrich) for 3 days. Serial dilutions of SNAP were prepared by dissolving different amounts of SNAP in a predetermined volume of DMSO concentration adjusted to 5% of total media content to prevent DMSO effects. Days 13 and 15 were chosen as analysis days, and metabolic analysis was performed on these days. In parallel, hepatic functional characteristics, namely, intracellular albumin content, urea and albumin secretion rate, and cytochrome p450 7α1 (cyp7a1) promoter activity, were evaluated.
Primary mouse hepatocyte culture
Freshly isolated cryopreserved plateable mouse hepatocytes were maintained at a density of ∼90,000 cells/cm2 in a collagen double-gel sandwich configuration in 12-well plates per manufacturer's instructions (Celsis In Vitro Technologies, Baltimore, MD). Media was changed daily using InVitroGRO HI medium for 7 days, and supernatants were collected for glucose, lactate, urea, and albumin measurements.
Cell number
Cultures were treated with phosphate-buffered saline (PBS) followed by incubation with trypsin-EDTA for 3 min. Dissociated cells from each condition were suspended in differentiation media and centrifuged at 1000 rpm for 5 min. The media with trypsin was decanted, and cells were resuspended in fresh media. Cells were counted using a hemacytometer using 1% trypan blue exclusion.
Urea analysis
Media samples were collected directly from cell cultures on days 13 and 15 postdifferentiation for the different experimental conditions and stored at − 20°C for subsequent analysis for urea content. Urea synthesis was assayed using a commercially available kit (Cat # 2050-450; StanBio, Boerne, TX). Absorbance readings for standards and samplers were obtained using a Biorad (Hercules, CA) Model 680 plate reader with a 585 nm emission filter. A standard curve was generated by creating serial dilutions of a urea standard from 0 to 300 μg/mL, and a linear fit of the standards was used to determine the urea concentration in each sample.
Albumin ELISA
To detect secreted albumin within the media supernatants obtained on each of the analysis days, we used a commercially available mouse albumin ELISA kit (#E90-134; Bethyl Laboratories, Montgomery, TX). A standard curve was generated by creating serial dilutions of an albumin standard from 7.8 to 10,000 ng/mL. Absorbance readings were obtained using a Biorad Model 680 plate reader with a 490 nm emission filter. Albumin values were normalized to the cell number recorded on the day of media sample collection.
In situ indirect immunofluorescence for intracellular albumin
On evaluation days as indicated for the urea assay, cells were washed with PBS and fixed in 4% paraformaldehyde (Sigma-Aldrich) in PBS for 15 min at room temperature. The cells were washed twice in cold PBS and then twice in cold saponine/PBS (SAP) membrane permeabilization buffer containing 1% bovine serum albumin (Cat # A7906; Sigma-Aldrich), 0.5% saponine (Cat # 54521; Sigma-Aldrich), and 0.1% sodium azide (Cat # 58032; Sigma-Aldrich). The cells were subsequently incubated for 30 min at 4°C in a SAP solution containing rabbit anti-mouse albumin antibody (150 μg/mL) (MP Biomedicals, Irvine, CA), or normal rabbit serum (150 μg/mL) (MP Biomedicals) as an isotype control, washed twice for 10 min in cold SAP buffer, and then treated for 30 min at 4°C with the secondary antibody, fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit, diluted 1:500 (Jackson Immuno Labs, Westgrove, PA). Cells were then washed once with cold SAP buffer and once with cold PBS. Fluorescent images were acquired with an Olympus IX70 microscope (Olympus, Center Valley, PA) and Olympus digital camera using an excitation filter of 515 nm. Image quantification was conducted using Olympus Microsuite. To generate intensity values for each of the samples, 15–20 cells were randomly chosen from each image taken (three images per well per experiment). The average intensity value for these cells was measured in the normal rabbit serum fluorescent (control) case and was subtracted from each of 15–20 randomly selected cells in the test (antialbumin) case to quantify the number of albumin-positive cells per image. This was done in triplicate experiments yielding a total of 135–180 cells per condition.
Cloning of the cyp7a1 promoter driven pDsRedExpress1 vectors
The pDsRedExpress1 plasmid vector was attained from BD Biosciences Clontech (Mountain View, CA). The cyp7a1 vector was a gift in the form of a PGL3-promoter vector from Dr. Gregorio Gil (Virginia Commonwealth University, Richmond, VA). The promoter regulatory elements were each excised at a blunt and a sticky end and inserted via ligation into respective blunt and sticky sites in the parent pDsRedExpress1 vector. Correct insertion of the regulatory elements into the pDsRedExpress1 vector was confirmed by screening bacterial clones via test transfections in mouse Hepa 1–6 cells and through DNA sequencing. The vector is hereby referred to as pcyp7a1-dsRedExpress1. An additional vector, pDsRed2-C1, driven by the constitutive cytomegalovirus, was used as a control for positive transfection of different cell types.
Transient transfection of liver-specific vectors into differentiated stem cells
On day 13 of differentiation, the liver-specific expression vector pcyp7a1-dsRedExpress1, along with the constitutive pDsRed2-C1 plasmid, was transiently transfected into the distinct differentiated stem cell populations. A control plate of murine Hepa 1–6 cells was used to assess transient transfection efficiency. After 48 h, red fluorescent activity was detected via flow cytometry and imaged for fluorescent activity using a computer-interfaced inverted Olympus IX70 microscope.
Glucose and lactate measurements
Supernatants (1 mL) were collected in triplicate for differentiated cells in secondary culture on days 13 and 15 of differentiation and tested using a Bioprofile Bioanalyzer 400 (Nova Biomedical, Waltham, MA) for metabolite measurements of glucose and lactate. To confirm that replating does not change the metabolic properties in control cell populations, mouse hepatoma cells were cultured for 5 days in six-well plates. Media was replaced daily and on the fifth day, and supernatants were collected for lactate measurements. Cells were trypsinized, resuspended, and 105 cells were replated in each well of a 12-well plate, and media was changed after 24 h. After 48 h, we found that mouse hepatoma cells have a similar lactate production rate in both primary culture (6.6 ± 0.5 μmol/million cells/day) and replated conditions (6.3 ± 0.7 μmol/million cells/day).
Mitochondrial mass and membrane potential (Δψ)
On each analysis day of secondary culture, cells from different experimental conditions in each well of a polystyrene 48-well plate were washed with PBS and exposed to 10 μM N-acridine orange (NAO) dye (Molecular Probes, Eugene, MO) for 10 min at room temperature for mitochondrial mass measurements. Experimental analysis was performed with the FL3 filter using the Olympus IX70 microscope, and image analysis was performed using Microsuite software. For mitochondrial membrane potential measurements, on each day of analysis, cells in each well were washed with PBS and then exposed to 30 μM JC-1 dye (Molecular Probes) and incubated for 30 min at 37°C, 10% CO2 at a density of 2.5 × 104 cells/mL in polystyrene 48-well plates. Because JC-1 dye forms a monomer at low mitochondrial potential with a green fluorescence while at high mitochondrial membrane potential, it aggregates and exhibits an orange fluorescence, image acquisition was performed with the FL2 and FL1 filter using the Olympus microscope, and image analysis was performed using the Microsuite software. On each day of analysis, undifferentiated ES cells and mouse hepatoma cells were stained as negative and positive controls, respectively. To generate intensity values for each of the samples, 15–20 cells were randomly chosen from each image taken (three images per well per experiment). The average intensity value for these cells was measured in the particular condition and subtracted from each of 15–20 randomly selected cells in the undifferentiated ES cell fluorescent case to quantify the number of NAO and JC-1–positive cells per image. This was done in triplicate experiments yielding a total of 135–180 cells per condition.
Statistical analysis
Each data point represents the mean of three experiments (each with three biological replicates), and the error bars represent the standard error of the mean. Statistical significance was determined using the Student's t-test for unpaired data. Differences were considered significant when the probability was less than or equal to 0.05.
Results
Energetics of hepatic differentiation
We have previously developed a Na butyrate–treated ES cell differentiation system to generate an enriched population of hepatocyte-like cells.6 In addition, we performed a metabolic analysis of the populations to characterize the energy status of the cells. We have shown that hepatocyte-like cells have reduced function with the expression of a glycolytic phenotype similar to immature hepatocytes. In the current work, we employed a procedure to alter the energetic profile of the cells toward a mature hepatocyte phenotype to upregulate hepatocyte-specific function.
On day 11 of differentiation, 2.5 mM Na butyrate–treated ES cells were replated in polystyrene 12-well plates. On day 12, cells were exposed to 0, 50, 100, 250, or 500 μM SNAP for 3 additional days. In primary culture, exposure to DMSO resulted in about 3 × 106 cells/well on day 5. On day 8, cell number reduced to 106 cells/well and remained at that on day 11. After replating on day 11, cell number remained constant after 48 h of replating for all conditions. On day 15 of differentiation, we observed an increase in cell number for the “No SNAP,” 50 μM and 100 μM SNAP–treated conditions. However, for the 250 and 500 μM conditions, we observed 50% and 70% decrease in cell number implying cell death at higher SNAP concentration treatments (data not shown). On days 13 and 15 of differentiation, functional and metabolic analyses were performed on the differentiated cell populations.
Glucose and lactate metabolism
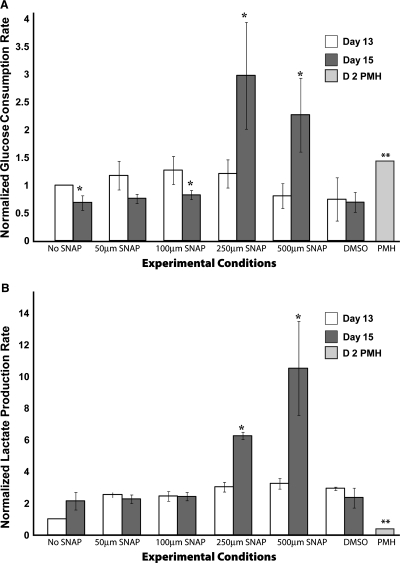
Primary hepatocytes have higher glucose consumption rates than the fetal phenotype. We investigated this phenomenon in SNAP-treated hepatocyte-like cells as shown in Figure 1. No SNAP and DMSO conditions correspond to Na butyrate–treated ES cells exposed to differentiation media only and media supplementation with DMSO only in secondary culture, respectively. Figure 1A shows that hepatocyte-like cells have comparable glucose consumption rates on day 13 independent of SNAP exposure. On the other hand, there was a significant decrease in the consumption rate on day 15 for the No SNAP condition, whereas there was a significant increase in glucose consumption rate for 250 and 500 μM SNAP (∼3-fold)–treated hepatocyte-like cells as compared to day 13 of differentiation.
FIG. 1.
Metabolic analysis of differentiated ES cells. (A) Glucose consumption rate. (B) Lactate production rate. Data are normalized to day 13 No SNAP condition. D2 primary mouse hepatocytes (PMH) condition corresponds to primary mouse hepatocytes on day 2 of culture. *p < 0.05 versus the same experimental condition on day 13 of differentiation. **p < 0.05 for the primary mouse hepatocytes as compared to day 13 No SNAP condition.
For lactate production rate, as shown in Figure 1B, irrespective of the SNAP concentration, day 13 SNAP–treated cells showed a higher lactate production rate than the No SNAP condition. Further, there was no significant change in lactate production rate on day 15 of differentiation for all conditions except the 250 and 500 μM SNAP–treated cells as compared to day 13 of differentiation.
As a positive control, we also measured glucose consumption and lactate production rates for primary murine hepatocyte cultures over a 7-day period. As shown in Figure 1, a comparison of the normalized glucose consumption rate of primary mouse hepatocytes with the No SNAP and SNAP-treated hepatocyte-like cells shows that while the glucose consumption rate is significantly higher, the lactate production rate is significantly lower for the primary mouse hepatocytes than the day 13 control conditions (No SNAP). However, in comparison to the day 15, 500 μM SNAP–treated hepatocyte-like cells, both the glucose consumption rates and lactate production rates are significantly lower. The absolute value for glucose consumption rate for day 2 primary mouse hepatocytes is 4.9 ± 2.5 μmol/million cells/day and lactate production rate is 4.74 ± 0.6 μmol/million cells/day. On the other hand, the values for the 500 μM SNAP–treated hepatocyte-like cells for glucose consumption and lactate production rates are 7.43 ± 2.18 μmol/million cells/day and 45.02 ± 13.21 μmol/million cells/day, respectively. Overall, 500 μM SNAP treatment results in an increase in glucose consumption rate and lactate production rates as compared to the No SNAP condition; in addition, the metabolite clearance rates are significantly higher than primary mouse hepatocytes.
Mitochondrial mass and activity
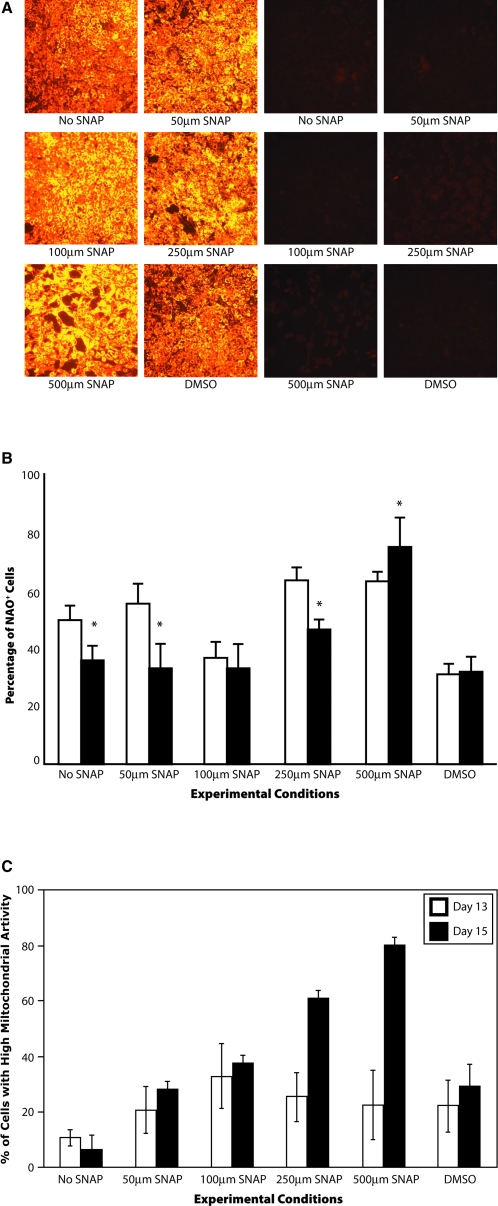
As shown in Figure 2A, on day 15, microscopic evaluation indicated that the mitochondrial activity was highest for 500 μM SNAP–treated cells. Similarly, the mitochondrial mass was the highest for 500 μM SNAP–treated cells. Additionally, we quantified the percentage of cells with higher mitochondrial mass (Fig. 2B) and activity (Fig. 2C). As shown, 500 μM SNAP resulted in about 80% of cells with a higher mitochondrial mass and activity than the No SNAP condition on day 15 of differentiation. For 250 μM SNAP–treated cells, increase in percentage of cells with higher mitochondrial activity (∼60%) was not consistent with a similar increase in percentage of cells with high mitochondrial mass (∼45%).
FIG. 2.
Evaluation of mitochondrial parameters. (A) Microscopic evaluation of mitochondrial mass and activity, (B) percentage of NAO-positive cells, and (C) percentage of cells with high mitochondrial activity. NAO, 5,5′,6,6′-tetrachloro-1,1′,3,3′ tetraethylbenzimidazolylcarbocyanine iodide/chloride (JC-1). *p < 0.05 versus the No SNAP condition on the same day. Color images available online at www.liebertonline.com/ten.
Evaluation of hepatic function
The next step was to assess the effects of different concentrations of SNAP on hepatocyte-specific function. Our hypothesis is that an induction of mitochondrial development in hepatocyte-like cells is associated with an increase in cellular function. As a result, we assessed two extracellular functions, urea and albumin secretion, and two intracellular markers, namely, the presence of intracellular albumin and cyp7a1 promoter activity to identify, if a particular SNAP concentration was effective in increasing function.
Urea and albumin secretion
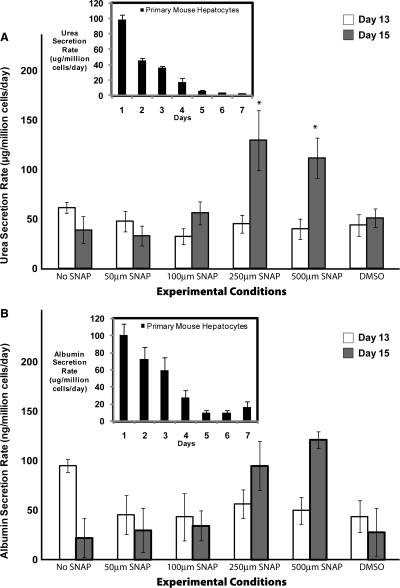
As shown in Figure 3A, urea synthesis rate was similar for different experimental conditions on day 13 of differentiation. However, on day 15, there was an increase in secretion rates for 250 and 500 μM SNAP–treated hepatocyte-like cells. This increase was about twofold higher than No SNAP condition. Figure 3B showed a similar trend for albumin secretion rate on day 13 of differentiation. However, while there was a decrease in albumin secretion on day 15 for the No SNAP condition, there was a stabilization of function in the presence of 250 or 500 μM SNAP as compared to day 13 No SNAP condition.
FIG. 3.
Dynamic profile of hepatocyte-specific function for differentiated ES cells on days 13 and 15 of differentiation for different concentrations of SNAP. (A) Urea secretion rate. (B) Albumin secretion rate. The data correspond to three independent experiments. The insets in the figures correspond to urea and albumin secretion rate for primary mouse hepatocytes over a 7-day period. *p < 0.05 versus the No SNAP condition on the same day.
Overall, the mitochondrial and functional assessments indicate that 500 μM SNAP treatment resulted in the generation of an enriched population of cells with increased mitochondrial mass and activity and higher glucose consumption rates on day 15 of differentiation. More importantly, the alteration in metabolic characteristics was concomitant with increase in urea and maintenance of albumin secretion rates.
Intracellular albumin and cyp7a1 promoter activity
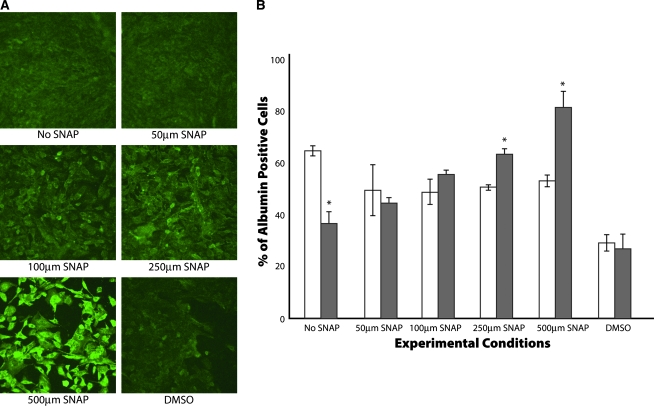
While urea secretion and albumin secretion are representative of an average population, these characteristics do not provide information about the intracellular characteristics within the cell population. As a result, we performed an immunofluorescent assay to test for intracellular albumin. As shown in Figure 4A, there was a drastic difference in intracellular albumin content between the different conditions on day 15 of differentiation. It was evident from the microscopic analysis that 500 μM SNAP–treated hepatocyte-like cells showed a higher intracellular albumin intensity than any other experimental condition. Figure 4B shows the percentage of cells positive for albumin under different experimental conditions. As shown, 500 μM SNAP treatment resulted in an enriched population of ∼80% albumin-positive cells on day 15 of differentiation. On the other hand, 250 μM SNAP treatment resulted in a heterogeneous population (∼65% albumin-positive cells), while there are fewer positive cells in the other conditions. In the absence of SNAP, percentage of albumin expressing cells decreased to 35% by day 15 postdifferentiation.
FIG. 4.
Intracellular evaluation of albumin function. (A) Intracellular albumin content on day 15. (B) Percentage of albumin-positive cells on days 13 and 15 of differentiation. The data are representative of three independent experiments. *p < 0.05 versus the No SNAP condition on the same day. Color images available online at www.liebertonline.com/ten.
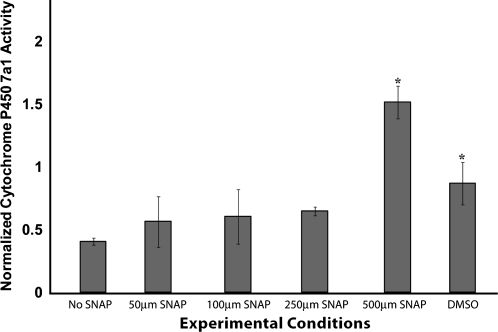
To confirm the presence of a hepatocyte-specific population, cyp7a1 promoter activity was determined in the different conditions on day 15 of differentiation. cyp7a1 (cholesterol 7α-hydroxylase) is a rate-limiting enzyme implicated in cholesterol metabolism in the liver.16 As shown in Figure 5, the highest promoter activity was observed in 500 μM SNAP–treated cells.
FIG. 5.
cyp7a1 Promoter activity on day 15 of differentiation. The data are normalized to the cytomegalovirus promoter activity for the experimental condition and are representative of three independent experiments. *p < 0.05 versus the No SNAP condition.
Thus, even though the metabolic and functional secretion rates were similar for 250 and 500 μM SNAP exposure on day 15 of differentiation, 500 μM SNAP treatment was the only experimental condition that resulted in the generation of an enriched population of albumin-positive cells with highest mitochondrial mass and higher relative percentage of mitochondrial mass and activity with improved cyp7a1 activity closer to the mature hepatic phenotype than other experimental conditions.
Discussion
We previously established a directed differentiation system wherein Na butyrate–treated ES cells generated an enriched population of hepatocyte-like cells. A metabolic analysis of the population showed that hepatocyte-like cells have a glycolytic energy status similar to fetal hepatocytes with incomplete metabolic and quantitative hepatic differentiated function. In this context, we investigated the effects of SNAP, a nitric oxide donor molecule implicated in mitochondrial development on Na butyrate–treated hepatocyte-like cells derived from ES cells.
From a mechanistic standpoint, numerous studies have identified PGC-1α, a downstream target of SNAP, as the key regulator of energy metabolism. PGC-1α is a transcriptional coactivator and is implicated in the switch from glycolytic (anaerobic) to oxidative phosphorylation (aerobic) metabolism for ATP synthesis. PGC-1α docks on specific transcription factors increasing the affinity of the transcription complex to coactivators possessing histone acetyltransferase activity. The end result is acetylation of histone proteins that increases the accessibility of DNA to the transcription complex.17 Although there are three members of the PGC-1 family (PGC-1α, PGC-1β, and PGC-1α related coactivator18), PGC-1α has specifically gained interest because it is a powerful regulator of energy metabolism under conditions of both health and disease.19
While the effects of NO and NOS isoforms in association with PGC-1α are well known in primary and transformed cell lines, the utilization of these molecules in cellular differentiation is less studied. The effect of PGC-1α has been extensively investigated in liver cell systems.20–25 In vivo liver developmental studies have been shown to involve complex set of events related to mitochondrial development. From a hepatic standpoint, it has been shown that fetal hepatocytes have a more compromised (glycolytic) energy state and lower functionality than highly functional mature hepatocytes with complex, functional mitochondria. The embryonic liver is primarily responsible for hematopoiesis during fetal development and undergoes many metabolic changes just before and after birth.26
This phenomenon, known as mitochondrial biogenesis, has been well studied in liver cells, and it has been shown that PGC-1α is implicated in hepatic mitochondrial biogenesis wherein an increase in mitochondrial mass, number, and activity is observed.10 This increase, which occurs immediately after birth in vivo, is associated with an increase in oxygen consumption, induction of mitochondrial respiratory enzymes, and increased/induced terminally differentiated function. In summary, the identification of the mechanism of NO induction of PGC-1α in mitochondrial biogenesis holds tremendous promise for applications in hepatocyte differentiation from ES cells.
Many studies have shown the influence of NO inducers on mitochondrial development. These studies have been implicated in diverse cell lines, such as HeLa cells, brown adipocytes, 3T3-L1, and U937 cells.10 In addition, SNAP, a nitric oxide donor, had been successfully shown to induce cardiomyogenesis in ES cells.27 About 250 μM SNAP induced cardiomyogenesis in embryoid body aggregates based on the hanging drop culture protocol. In the experimental setup, ES cells were exposed to the molecule at earlier time points that differed from our experimental analysis wherein SNAP was supplemented after Na butyrate treatment. It is well documented that SNAP, though shown to increase oxygen consumption28 and mitochondrial development in hepatocytes, leads to cell death.29 While the exact mechanism for this phenomenon in our system is unknown, we observed a decrease in cell number at higher SNAP concentrations (data not shown).
Metabolic assessment of SNAP-treated cells showed that SNAP increased glucose clearance rates at higher concentrations of 250 and 500 μM on day 15 of differentiation (Fig. 1). This observation is known to be prevalent in mature hepatocytes to consume higher glucose rates than fetal hepatocytes for performing metabolic functions. It is worth mentioning that higher SNAP concentration treatment increased glucose and lactate metabolism concomitant with increased mitochondrial mass and activity (Fig. 2A) that is indicative of mitochondrial biogenesis. While the glucose and lactate production rates were significantly higher for the 500 μM SNAP treatment conditions, the levels were lower for primary mouse hepatocytes, possibly implying a higher glycolytic rate during hepatic differentiation. This increase was associated with an increased percentage in mitochondrial mass (Fig. 2B) and activity (Fig. 2C) for 500 μM SNAP–treated cells on day 15 of differentiation. These increased mitochondrial parameters were shown in various mammalian cell lines after 3–4 days of SNAP treatment.11 From a concentration standpoint, while 100 μM SNAP was used for different cell lines,10 250 μM SNAP was shown to induce ES-cardiomyocyte differentiation.26 In our population, we showed highest mitochondrial mass and activity in 500 μM SNAP–treated cells. Beyond this concentration, we observed a significant decrease in cell viability and loss of mitochondrial function (data not shown).
Figure 3 shows the functional evaluation of hepatic characteristics after SNAP exposure. It is noteworthy that both 250 and 500 μM SNAP exposure increased urea secretion by ∼2-fold on day 15 of differentiation with comparative secretion rates with day 1 primary mouse hepatocyte cultures. This was associated with a stabilization of albumin secretion for the two conditions (Fig. 3B), which is significantly lower than albumin secretion rates of primary mouse hepatocytes. An intracellular assessment of albumin showed reduced intracellular albumin over time for the No SNAP condition, which was consistent with albumin secretion rates. As shown in Figure 4, while the 250 μM SNAP–treated cells revealed a mixed population indicative of different lineages, the 500 μM SNAP treatment resulted in an enriched population of albumin-positive cells (∼85%, Fig. 4B). A comparison of the functional and metabolic analysis showed that the enriched population of hepatocyte-like cells also had an increased percentage of mitochondrial mass and activity (∼80%). In addition to albumin, cyp7a1 promoter activity analysis, a hepatocyte-specific enzyme implicated in cholesterol metabolism30 showed highest activity in 500 μM SNAP–treated cells (Fig. 5). Numerous studies have shown the effects of DMSO as a differentiation agent in hepatocyte precursors, immortalized cell lines, and ES cells toward hepatocytes.31,32 A human hepatoma cell line, HepaRG, has been shown to differentiate into a hepatocyte morphology using 2% DMSO supplementation with the mRNA expression of cytochrome P450 enzymes, namely, cyp3a4 and cyp7a1. The presence of DMSO alone in the media on days 12–14 of differentiation might explain a significantly higher induction of cyp7a1 activity in the differentiated stem cell populations than controls.33
From a mechanistic standpoint, previous work34 indicates the application of nitric oxide based inhibition of urea secretion of hepatocytes derived from Na butyrate–treated immortalized cell lines. While the nitric oxide inhibition seems logical, the experimental system comprises immortalized human hepatocyte cell line that differs from murine ES cell–treated cells. This might indicate different mechanisms inherent in ES cell differentiation; in addition, there have been recent reports that IL-1β–mediated inhibition of urea secretion is only partially dependent on NO in primary rat hepatocytes.35 Moreover, NO, an inducible stress molecule, is implicated in both increase and decrease in rate of urea synthesis from immobilized perfused hepatocytes and anoxia/reperfused livers. Thus, dependent on the concentration of the NO produced, lower NO concentrations have been shown to increase,36 while higher concentrations reduce the rate of urea synthesis in hepatocyte cultures.34
Thus, using an optimal nitric oxide supplementation, we have generated an enriched population of albumin-positive cells that have higher albumin and urea secretion levels and possess improved metabolic characteristics closer to the mature hepatic phenotype than other experimental conditions.
While the above data show hepatic functional characteristics prevalent for the 500 μM SNAP–treated Na butyrate differentiated ES cells, a suitable comparison with cell differentiation systems provides a standard of comparison for the different systems. While most ES cell hepatic differentiation systems comprise hepatic differentiation characteristics using PCR for intracellular mRNA expression,37–41 very few systems quantify the percentage of hepatic protein-positive cells and protein secretion levels. In fact, one report claims that albumin secretion rate is not detected until day 30 of differentiation with a value of ∼1 ng/million cells/day,42 which is approximately 100-fold lower than 500 μM SNAP–treated differentiated ES cells. On the other hand, embryoid body mediated murine ES cell differentiation and subsequent replating on collagen coated dishes in hepatocyte culture medium resulted in 29% albumin positive cells with 240 ng/million cells/day albumin secretion rates.43 Further, alginate encapsulation of ES cells resulted in hepatic differentiation with albumin secretion rates of 160 ng/million cells/day on day 23 of differentiation.44 In summary, the literature fails to include quantification of cells positive for combination of mature hepatic markers such as intracellular albumin and cytokeratin 18, and simultaneous functional secretion rates for urea and albumin. Moreover, there is very little information on metabolic characterization of the ES cell–derived hepatocytes.45
For future work, we plan to develop a comprehensive metabolic reaction network model for identifying potential targets of metabolic manipulation of cells differentiating toward a mature phenotype. Further, this methodology can also be applied to ES cell differentiation systems involving other metabolically active cells such as of the neuronal, pancreatic, and cardiac lineages.
Acknowledgment
Contract Grant Sponsor: National Institutes of Health, Hepatic Tissue Engineering Grant. Contract Grant Number: R01DK43371.
Disclosure Statement
No competing financial interests exist.
References
- 1.Yarmush M.L. Toner M. Dunn J.C. Rotem A. Hubel A. Tompkins R.G. Hepatic tissue engineering. Development of critical technologies. Ann NY Acad Sci. 1992;665:238. doi: 10.1111/j.1749-6632.1992.tb42588.x. [DOI] [PubMed] [Google Scholar]
- 2.Chan C. Berthiaume F. Nath B.D. Tilles A.W. Toner M. Yarmush M.L. Hepatic tissue engineering for adjunct and temporary liver support: critical technologies. Liver Transpl. 2004;10:1331. doi: 10.1002/lt.20229. [DOI] [PubMed] [Google Scholar]
- 3.Zandstra P.W. Nagy A. Stem cell bioengineering. Annu Rev Biomed Eng. 2001;3:275. doi: 10.1146/annurev.bioeng.3.1.275. [DOI] [PubMed] [Google Scholar]
- 4.Novik E.I. Maguire T.J. Orlova K. Schloss R.S. Yarmush M.L. Embryoid body-mediated differentiation of mouse embryonic stem cells along a hepatocyte lineage: insights from gene expression profiles. Tissue Eng. 2006;12:1515. doi: 10.1089/ten.2006.12.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavon N. Benvenisty N. Study of hepatocyte differentiation using embryonic stem cells. J Cell Biochem. 2005;96:1193. doi: 10.1002/jcb.20590. [DOI] [PubMed] [Google Scholar]
- 6.Sharma N.S. Shikhanovich R. Schloss R. Yarmush M.L. Sodium butyrate-treated embryonic stem cells yield hepatocyte-like cells expressing a glycolytic phenotype. Biotechnol Bioeng. 2006;94:1053. doi: 10.1002/bit.20936. [DOI] [PubMed] [Google Scholar]
- 7.Maguire T. Novik E. Schloss R. Yarmush M. Alginate-PLL microencapsulation: effect on the differentiation of embryonic stem cells into hepatocytes. Biotechnol Bioeng. 2006;93:581. doi: 10.1002/bit.20748. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi A. Kurihara M. Kobayashi K. Cho C.S. Akaike T. Hara M. Albumin and urea production by hepatocytes cultured on extracellular matrix proteins-conjugated poly(vinyl alcohol) membranes. J Biomater Sci Polym Ed. 2005;16:847. doi: 10.1163/1568562054255727. [DOI] [PubMed] [Google Scholar]
- 9.Nisoli E. Falcone S. Tonello C. Cozzi V. Palomba L. Fiorani M. Pisconti A. Brunelli S. Cardile A. Francolini M. Cantoni O. Carruba M.O. Moncada S. Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci USA. 2004;101:16507. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nisoli E. Clementi E. Paolucci C. Cozzi V. Tonello C. Sciorati C. Bracale R. Valerio A. Francolini M. Moncada S. Carruba M.O. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 11.Bloch W. Fleischmann B.K. Lorke D.E. Andressen C. Hops B. Hescheler J. Addicks K. Nitric oxide synthase expression and role during cardiomyogenesis. Cardiovasc Res. 1999;43:675. doi: 10.1016/s0008-6363(99)00160-1. [DOI] [PubMed] [Google Scholar]
- 12.Magrinat G. Mason S.N. Shami P.J. Weinberg J.B. Nitric oxide modulation of human leukemia cell differentiation and gene expression. Blood. 1992;80:1880. [PubMed] [Google Scholar]
- 13.Peunova N. Enikolopov G. Nitric oxide triggers a switch to growth arrest during differentiation of neuronal cells. Nature. 1995;375:68. doi: 10.1038/375068a0. [DOI] [PubMed] [Google Scholar]
- 14.Davis R.A. Miyake J.H. Hui T.Y. Spann N.J. Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002;43:533. [PubMed] [Google Scholar]
- 15.Cuezva J.M. Ostronoff L.K. Ricart J. Lopez de Heredia M. Di Liegro C.M. Izquierdo J.M. Mitochondrial biogenesis in the liver during development and oncogenesis. J Bioenerg Biomembr. 1997;29:365. doi: 10.1023/a:1022450831360. [DOI] [PubMed] [Google Scholar]
- 16.Puigserver P. Adelmant G. Wu Z. Fan M. Xu J. O'Malley B. Spiegelman B.M. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 17.Puigserver P. Spiegelman B.M. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 18.Finck B.N. Kelly D.P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houten S.M. Auwerx J. PGC-1alpha: turbocharging mitochondria. Cell. 2004;119:5. doi: 10.1016/j.cell.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Liang H. Ward W.F. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 21.Lin J. Tarr P.T. Yang R. Rhee J. Puigserver P. Newgard C.B. Spiegelman B.M. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 22.Naiki T. Nagaki M. Shidoji Y. Kojima H. Imose M. Kato T. Ohishi N. Yagi K. Moriwaki H. Analysis of gene expression profile induced by hepatocyte nuclear factor 4alpha in hepatoma cells using an oligonucleotide microarray. J Biol Chem. 2002;277:14011. doi: 10.1074/jbc.M105403200. [DOI] [PubMed] [Google Scholar]
- 23.Puigserver P. Wu Z. Park C.W. Graves R. Wright M. Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 24.Yoon J.C. Puigserver P. Chen G. Donovan J. Wu Z. Rhee J. Adelmant G. Stafford J. Kahn C.R. Granner D.K. Newgard C.B. Spiegelman B.M. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 25.Duncan S.A. Transcriptional regulation of liver development. Dev Dyn. 2000;219:131. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1051>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Kanno S. Kim P.K. Sallam K. Lei J. Billiar T.R. Shears L.L. Nitric oxide facilitates cardiomyogenesis in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12277. doi: 10.1073/pnas.0401557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farghali H. Martinek J. Masek K. The amelioration of hepatocyte oxidative stress injury by nitric oxide released from S-nitroso-N-acetyl penicillamine: a study in immobilized perfused hepatocytes. Methods Find Exp Clin Pharmacol. 1999;21:395. doi: 10.1358/mf.1999.21.6.541919. [DOI] [PubMed] [Google Scholar]
- 28.Kucera T. Canova N.K. Farghali H. Martinek J. The morphological and immunocytochemical evaluation of primary rat hepatocytes undergoing spontaneous cell death: modulation by the nitric oxide donor S-nitroso-N-acetylpenicillamine. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:75. doi: 10.5507/bp.2006.008. [DOI] [PubMed] [Google Scholar]
- 29.Asahina K. Fujimori H. Shimizu-Saito K. Kumashiro Y. Okamura K. Tanaka Y. Teramoto K. Arii S. Teraoka H. Expression of the liver-specific gene Cyp7a1 reveals hepatic differentiation in embryoid bodies derived from mouse embryonic stem cells. Genes Cells. 2004;9:1297. doi: 10.1111/j.1365-2443.2004.00809.x. [DOI] [PubMed] [Google Scholar]
- 30.Massimi M. Lear S.R. Huling S.L. Jones A.L. Erickson S.K. Cholesterol 7alpha-hydroxylase (CYP7A): patterns of messenger RNA expression during rat liver development. Hepatology. 1998;28:1064. doi: 10.1002/hep.510280422. [DOI] [PubMed] [Google Scholar]
- 31.Hay D.C. Zhao D. Fletcher J. Hewitt Z.A. McLean D. Urruticoechea-Uriguen A. Black J.R. Elcombe C. Ross J.A. Wolf R., et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 2008;26:894. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- 32.Jiang J. Kojima N. Guo L. Naruse K. Makuuchi M. Miyajima A. Yan W. Sakai Y. Efficacy of engineered liver tissue based on poly-L-lactic acid scaffolds and fetal mouse liver cells cultured with oncostatin M, nicotinamide, and dimethyl sulfoxide. Tissue Eng. 2004;10:1577. doi: 10.1089/ten.2004.10.1577. [DOI] [PubMed] [Google Scholar]
- 33.Kanebratt K.P. Andersson T.B. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos. 2008;36:1444. doi: 10.1124/dmd.107.020016. [DOI] [PubMed] [Google Scholar]
- 34.Yoon J.H. Lee H.S. Kim T.H. Woo G.H. Kim C.Y. Augmentation of urea-synthetic capacity by inhibition of nitric oxide synthesis in butyrate-induced differentiated human hepatocytes. FEBS Lett. 2000;474:175. doi: 10.1016/s0014-5793(00)01599-4. [DOI] [PubMed] [Google Scholar]
- 35.Kang Y.H. Berthiaume F. Yarmush M.L. Long-term stable cultures of rat hepatocytes: an in vitro model to study acute and chronic hepatic inflammation. Tissue Eng. 2002;8:681. doi: 10.1089/107632702760240599. [DOI] [PubMed] [Google Scholar]
- 36.Farghali H. Hynie S. Vohnikova Z. Masek K. Possible dual role of nitric oxide in oxidative stress injury: a study in perfused hepatocytes. Int J Immunopharmacol. 1997;19:599. doi: 10.1016/s0192-0561(97)00048-9. [DOI] [PubMed] [Google Scholar]
- 37.Drobinskaya I. Linn T. Saric T. Bretzel R.G. Bohlen H. Hescheler J. Kolossov E. Scalable selection of hepatocyte- and hepatocyte precursor-like cells from culture of differentiating transgenically modified murine embryonic stem cells. Stem Cells. 2008;26:2245. doi: 10.1634/stemcells.2008-0387. [DOI] [PubMed] [Google Scholar]
- 38.Shiraki N. Umeda K. Sakashita N. Takeya M. Kume K. Kume S. Differentiation of mouse and human embryonic stem cells into hepatic lineages. Genes Cells. 2008;13:731. doi: 10.1111/j.1365-2443.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 39.Moore R.N. Dasgupta A. Rajaei N. Yarmush M.L. Toner M. Larue L. Moghe P.V. Enhanced differentiation of embryonic stem cells using co-cultivation with hepatocytes. Biotechnol Bioeng. 2008;101:1332. doi: 10.1002/bit.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maezawa K. Miyazato K. Matsunaga T. Momose Y. Imamura T. Johkura K. Sasaki K. Ohmori S. Expression of cytochrome P450 and transcription factors during in vitro differentiation of mouse embryonic stem cells into hepatocytes. Drug Metab Pharmacokinet. 2008;23:188. doi: 10.2133/dmpk.23.188. [DOI] [PubMed] [Google Scholar]
- 41.Ishii T. Fukumitsu K. Yasuchika K. Adachi K. Kawase E. Suemori H. Nakatsuji N. Ikai I. Uemoto S. Effects of extracellular matrixes and growth factors on the hepatic differentiation of human embryonic stem cells. Am J Physiol Gastrointest Liver Physiol. 2008;295:G313. doi: 10.1152/ajpgi.00072.2008. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto K. Mizumoto H. Nakazawa K. Ijima H. Funatsu K. Kajiwara T. Hepatic differentiation of mouse embryonic stem cells in a three-dimensional culture system using polyurethane foam. J Biosci Bioeng. 2008;105:350. doi: 10.1263/jbb.105.350. [DOI] [PubMed] [Google Scholar]
- 43.Kania G. Blyszczuk P. Jochheim A. Ott M. Wobus A.M. Generation of glycogen- and albumin-producing hepatocyte-like cells from embryonic stem cells. Biol Chem. 2004;385:943. doi: 10.1515/BC.2004.123. [DOI] [PubMed] [Google Scholar]
- 44.Maguire T. Davidovich A.E. Wallenstein E.J. Novik E. Sharma N. Pedersen H. Androulakis I.P. Schloss R. Yarmush M. Control of hepatic differentiation via cellular aggregation in an alginate microenvironment. Biotechnol Bioeng. 2007;98:631. doi: 10.1002/bit.21435. [DOI] [PubMed] [Google Scholar]
- 45.Ishii T. Yasuchika K. Fujii H. Hoppo T. Baba S. Naito M. Machimoto T. Kamo N. Suemori H. Nakatsuji N. Ikai I. In vitro differentiation and maturation of mouse embryonic stem cells into hepatocytes. Exp Cell Res. 2005;309:68. doi: 10.1016/j.yexcr.2005.05.028. [DOI] [PubMed] [Google Scholar]