Abstract
DNA damage repair and checkpoint responses prevent genome instability and provide a barrier to the development of cancer. Inherited mutations in DNA damage response (DDR) genes such as those that encode the homologous recombination (HR) proteins BRCA1 and BRCA2 cause cancer predisposition syndromes. PARP inhibitors are an exciting new class of targeted therapy for treating patients with HR repair-defective tumors. In this study, we use an RNAi screen to identify genes that when silenced cause synthetic lethality with the PARP inhibitor AZD2281. This screen identified the deubiquitylating enzyme USP11 as a participant in HR repair of DNA double-strand breaks. Silencing USP11 with siRNA leads to spontaneous DDR activation in otherwise undamaged cells and hypersensitivity to PARP inhibition, ionizing radiation, and other genotoxic stress agents. Moreover, we demonstrate that HR repair is defective in USP11-silenced cells. Finally, the recruitment of a subset of double-strand break repair proteins including RAD51 and 53BP1 to repair foci is misregulated in the absence of USP11 catalytic activity. Thus, our synthetic lethal approach identified USP11 as a component of the HR double-strand break repair pathway.
Keywords: Checkpoint Control, DNA Damage, DNA Recombination, DNA Repair, Ubiquitin
Introduction
Faithful DNA replication and repair is critical to maintain genome integrity. An intact DNA damage response (DDR)2 prevents mutations and gross chromosomal instability. Defects in the DDR cause cancer predisposition syndromes, demonstrating the importance of the DDR as a barrier to tumorigenesis (1, 2). Most cancers have defects in some part of the DDR. This difference between normal and cancer cells not only explains the genetic instability associated with cancer, but also provides an opportunity for therapeutic intervention. This concept is the basis for many genotoxic therapies where the DNA damage is repaired in healthy cells but has lethal consequences in cancer cells.
Targeted therapy of tumors is a method of treatment that seeks to maximize response while reducing harmful side effects to healthy cells. Inhibition of the DNA repair enzyme poly(ADP-ribose) polymerase 1 (PARP1) is a recently developed strategy for cancer therapy that exploits DDR defects in a subset of cancers (3). PARP1 is a DNA repair enzyme that is responsible for the sensing and repair of single-strand DNA breaks via short-patch base-excision repair (4). Thousands of single-strand breaks occur per cell per day. When a replication fork encounters a single-strand break, the result is collapse of the fork and formation of a double-strand break (5). In wild-type cells, these replication-associated double-strand breaks are often repaired via homologous recombination (HR). Cells deficient in PARP activity have increased recombination as evidenced by elevated rates of sister chromatid exchange (6) and increased intranuclear foci of the RAD51 recombination protein (7, 8). Cells deficient in HR proteins such as BRCA1 and BRCA2 are unable to repair these double-strand breaks efficiently and die (7, 8). Thus, PARP inhibitors are being successfully used to treat HR-deficient tumors such as those found in the breast of women with inherited mutations in BRCA1 or BRCA2 (9).
PARP inhibitors also preferentially kill cells defective in many other components of the DDR that respond to double-strand breaks including the ataxia-telangiectasia mutated (ATM) kinase (10, 11). ATM is recruited to double-strand breaks by the Mre11-Rad50-Nbs1 (MRN) complex where it phosphorylates the histone variant H2AX on Ser-139 (γH2AX) in nucleosomes surrounding the break. H2AX phosphorylation and the MRN complex itself initiate the recruitment of many additional double-strand break repair and signaling proteins including MDC1, BRCA1, RAD51, and 53BP1 (12–16). Assembly of these proteins within damage-induced foci initiates cell cycle checkpoint signaling and HR. Search for the homologous sister chromatid and strand invasion during HR requires formation of a RAD51 filament on the single-stranded DNA and is facilitated by BRCA2 (17–21).
Ubiquitin modification at the sites of double-strand breaks has emerged as an essential regulator of signaling and repair. Recruitment of BRCA1 and 53BP1 requires ubiquitylation at double-strand break sites catalyzed by the E3 ubiquitin ligases, RNF8 and RNF168 (22–28). First, MDC1 directly binds γH2AX via a BRCT domain and is also a target of phosphorylation by ATM (29, 30). After MDC1 is phosphorylated, it interacts with the FHA domain of RNF8 which ubiquitylates H2A (23–25). Ubiquitylated H2A serves as an interacting partner for RNF168 that further propagates the ubiquitylation of H2A and other unknown targets at the double-strand break site (22, 27). RAP80 binds ubiquitylated H2A and along with Abraxas mediates accumulation of BRCA1 (31–35). Histone ubiquitylation at double-strand break sites is also required for recruitment of 53BP1, although the precise mechanism remains unclear (22). RAD51 recruitment to double-strand breaks requires the E2 enzyme Ubc13, a known interaction partner of RNF8 (26).
A number of de-ubiquitylating enzymes (DUBs) also function at double-strand breaks including USP3 and BRCC36. BRCC36 is part of the RAP80-Abraxas-BRCA1 complex and antagonizes RNF8-dependent ubiquitylation to maintain steady state levels required for appropriate signaling (31, 36,36–38). USP3 is a chromatin-associated DUB that also antagonizes RNF8-mediated ubiquitylation (39).
Because HR deficiency causes cellular sensitivity to PARP inhibitors, PARP inhibitor sensitivity may indicate a defect in HR repair. Indeed, RNAi screening for PARP inhibitor sensitivity has identified new participants in the DDR (40, 41). In this study, we used siRNA screening to identify and validate the DUB USP11 as a regulator of double-strand break repair involved in modulating sensitivity to DNA damage.
EXPERIMENTAL PROCEDURES
Cell Culture, siRNA, and Plasmids
U2OS cell were maintained in Dulbecco's modified Eagle's medium (DMEM) with 7.5% fetal bovine serum at 37 °C and 5% CO2. siRNA transfections were performed with HiPerfect reagent (Qiagen) with a final siRNA concentration of 5 nm. Target siRNA sequences (listed in supplemental Table S1) and the non-targeting control sequence AUGAACGUGAAUUGCUCAA were purchased from Dharmacon and Qiagen. Full-length cDNA for USP11 (clone ID: 2961383) was obtained from Open Biosystems. A plasmid resistant to USP11 siRNA was created by introducing wobble mutations through PCR amplification of the target sequences. Primers used were as follows: USP11_6: sense: TGTTCAGTTCAGCCATACCGATTCTATCGGTCTGGTGTTGCGCACAGCTCGG; antisense: CCGAGCTGTGCGCAACACCAGACCGATAGAATCGGTATGGCTGAACTGAACA; USP11_10: sense: AGCCAGAGATGAAGAAGCGTTACTACGATGAAGTTGAGGCTGAGGGCTA; antisense: TAGCCCTCAGCCTCAACTTCATCGTAGTAACGCTTCTTCATCTCTGGCTC. The C318S, mutation was made using the following primers: sense: CAATCTGGGCAACACGAGCTTCATGAACTCGGC; antisense: GCCGAGTTCATGAAGCTCGTGTTGCCCAGATTG (mutated site underlined). Stable U2OS cells infected with siRNA-resistant constructs of USP11 were selected and maintained in 1 μg/ml of puromycin.
Drug Treatment and DNA Damage
PARP inhibitor (AZD2281) was dissolved in DMSO at a concentration of 10 mm. Ionizing radiation (IR) treatment was done using a Cs137 source at a rate of 1.8 Gy/min.
Viability Assay and Statistical Analysis
Reverse transfection was performed by spotting siRNA in 96-well plates followed by addition of HiPerfect reagent in Optimem media. After 5 min of incubation at room temperature, 5000 cells were added to each well and incubated 24 h at 37 °C. Cells were split 1:4 with two plates treated with 5 μm AZD2281 and two plates as an untreated control. After 96 h of incubation, medium was replaced with fresh medium containing 80 μl of a 1:10 dilution of WST-1 reagent (Roche) in DMEM. Plates were incubated 1 h at 37 °C, then assayed at 450 nm on a spectrophotometer. Sensitivity was calculated by normalizing absorbance readings to the average of the non-targeting controls within each plate. The mean of two replicates for each experiment was used to calculate a viability ratio by dividing treated by untreated wells for each gene. The log was calculated for these ratios and normalized to the non-targeting control, which represents the relative sensitivity to AZD2281. Multiple replicates of the log ratio were used to calculate a p value using unpaired, two-tailed t test. The sensitivity index was also calculated as previously described (42).
Antibodies and Immunoblotting
Cells were lysed for 20 min on ice in 50 mm Tris, pH 7.5, 150 mm NaCl, 0.5% Igepal, 10 mm NaF supplemented with 1 mm phenylmethylsulfonyl fluoride, 20 mm β-glycerophosphate, 1 mm sodium vanadate, 1 mm dithiothreitol, 5 μg/ml aprotinin, and 5 μg/ml leupeptin. Lysates were cleared by centrifugation prior to Bradford protein concentration determination (Bio-Rad). Total cellular protein was separated by SDS-PAGE and transferred to nitrocellulose membranes. Protein detection was done using infrared fluorescent-conjugated secondary antibodies on an Odyssey imaging system (LI-COR). Antibodies to γH2AX were purchased from Cell Signaling. RAD51 and BRCA1 antibodies were purchased from EMD BioSciences. 53BP1 and USP11 antibodies were purchased from Bethyl Laboratories. GAPDH antibody was purchased from Millipore. HA antibody was purchased from Covance. ORC2 antibody was obtained from BD Pharmingen.
Immunofluorescence
Cells were plated on coverslips and allowed to attach before treatment with IR. After incubation, cell were fixed in 3% paraformaldehyde and permeabilized with 0.5% Triton X-100 solution before incubation with primary antibodies. Fluorescein isothiocyanate and rhodamine red-X-conjugated secondary antibodies were obtained from Jackson Immunoresearch. Cells were visualized and foci counted on a Zeiss Axioplan 2.
Clonogenic Survival Assay
Sensitivity to IR was determined by transfecting U2OS cells with non-targeting and USP11 siRNA for 24 h followed by plating in 60-mm dishes at increasing cell densities. Treatment with 3 and 5 Gy IR was carried out 72 h after siRNA knockdown. Colonies were allowed to grow for 7–10 days and stained with 2% methylene blue in a 50:50 solution of methanol/water. Colonies of >50 cells were counted, and the surviving fraction was calculated and normalized to untreated control.
Chromosomal Homologous Recombinational Repair (HR) Analysis
HR repair assay was carried out as previously described (43). HEK293DRGFP cells carrying a chromosomally integrated single copy of homologous recombinational repair (HR) substrate were used to test USP11 role in HR. DSB-induced HR results in restoration and expression of GFP and was quantified by FACS. Briefly, 48 h after one repeat of transfection of control or USP11-targeting siRNA, chromosomal DSBs were induced through the expression of I-SceI. 48 h later, cells were subjected to two-color fluorescence analysis, which revealed the percentage of green fluorescent cells relative to the total viable cell number. For each analysis, 100,000 cells were processed.
RESULTS
RNAi Screen for PARP Inhibitor (PARPi) Hypersensitivity Identifies USP11
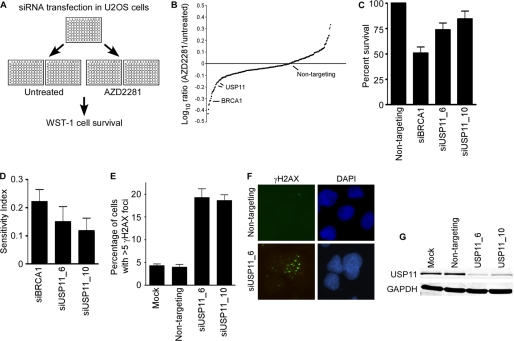
We previously used activation of the DNA damage response as a reporter to identify genome maintenance activities in mammalian cells (44). 73 genes were identified that caused increased DDR signaling when silenced by RNAi even in the absence of any added genotoxic agent. We expected that a subset of these genes were likely to function in HR repair and predicted that any HR deficiency would cause synthetic lethality with PARP inhibition. Therefore, we examined whether silencing each of these 73 genes would cause hypersensitivity to a PARPi (AZ2281). U2OS cells were transfected with siRNAs targeting each gene in a one siRNA/well format then split into untreated and PARPi-treated groups. After allowing 96 h of growth, wells were measured for cell viability (Fig. 1A). The log ratio of PARPi-treated divided by untreated provided a measurement of sensitivity for each gene screened. (Fig. 1B and supplemental Table S1). Sensitivity index was also calculated, to determine the combined contribution of siRNA along with drug treatment to cell viability (42) (supplemental Table S1). Any gene that was determined as significant by two or more independent siRNAs by either method was considered a potential positive.
FIGURE 1.
USP11 silencing causes AZD2281 hypersensitivity and spontaneous DDR activation. A, diagram of AZD2281 synthetic lethal screen. U2OS cells were split 24 h after transfection with siRNA and treated with AZD2281 for 96 h prior to measurement of cell survival. B, graph of all siRNAs screened. C, survival of USP11-silenced cells compared with non-targeting or BRCA1 siRNA controls after PARP inhibition. D, sensitivity index was calculated as previously described (42). Genes scored as >0.1 were considered as positive for hypersensitivity to AZD2281. E and F, non-targeting and USP11 siRNA were transfected into U2OS cells and γH2AX foci were counted 72 h after transfection (Mock, mock transfection with no siRNA). Graph represents percentage of cells with >5 foci, and error bars are S.D. (n = 3). G, immunoblot analysis of protein knockdown 72 h after transfection of control and USP11 siRNA.
We identified USP11 as a candidate based on significant PARPi-sensitivity induced by two independent siRNAs (Fig. 1, C and D). The same two siRNAs cause a 4-fold increase in the percentage of cells staining for γH2AX foci in the absence of any added genotoxic stress agent compared with non-targeting siRNA-transfected cells (Fig. 1, E and F). Successful silencing of USP11 expression was confirmed with Western blot analysis (Fig. 1G). Cell cycle profiles were not significantly altered in control or USP11-depleted cells indicating indirect effects of cell cycle status were unlikely to account for our results (supplemental Fig. S1).
USP11 Silencing Impairs HR Repair at Double-strand Breaks
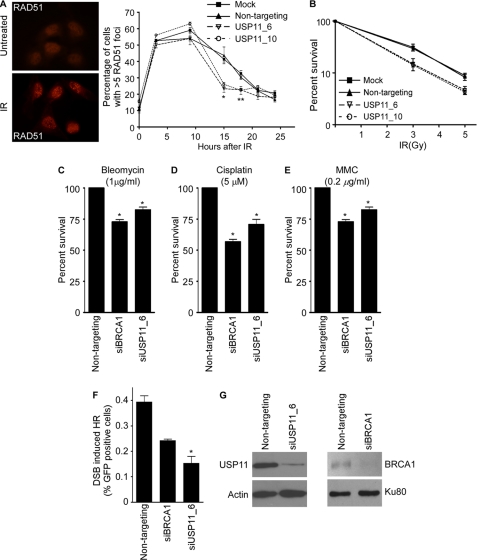
We next examined whether the PARPi hypersensitivity and spontaneous DDR activation in USP11-silenced cells was due to defective double-strand break sensing and repair. We first performed immunofluorescence to measure formation of RAD51 foci after ionizing radiation, which is an indirect measure of HR repair. The initial rate of RAD51 foci formation was similar in USP11-silenced and control cells (Fig. 2A). However, the RAD51 foci were not maintained at later time points after IR exposure in the absence of USP11 compared with control. Statistically significant differences were observed at both 15 and 18 h after IR (Fig. 2A). Eventually, both the control and USP11-silenced cell populations reached the same low level of RAD51 foci.
FIGURE 2.
USP11 silencing impairs HR repair at double-strand breaks. A, RAD51 foci were scored by immunofluorescent imaging at the indicated times after treatment with IR (*, p = 0.001; **, p = 0.038). B, mock, non-targeting and USP11-silenced cells were plated in 60-mm dishes and treated with the indicated doses of IR. Colonies of >50 cells were scored 7–10 days after irradiation. Error bars represent S.D. (n = 3). C–E, non-targeting, BRCA1- and USP11-silenced U2OS cells were treated with C, bleomycin (1 μg/ml), D, cisplatin (5 μm), or E, mitomycin C (0.2 μg/ml) for 96 h followed by measurement of viability (*, p < 0.05). Error bars are S.D. (n = 4). F, non-targeting, BRCA1- and USP11-silenced HEK293 cells containing an integrated HR repair substrate were transfected with I-SceI to induce a double-strand break in the reporter gene. The percentage of GFP+ cells was scored by flow cytometry. (Error bars are S.D., n = 3, *, p = 0.046). G, USP11 and BRCA1 protein levels in the cells used in F were measured by immunoblotting.
The reduced RAD51 foci in the USP11 cells combined with the PARPi sensitivity suggested that there may be reduced efficiency of HR repair of double-strand breaks in the absence of USP11. Consistent with this interpretation, we found that USP11-silenced cells are hypersensitive to ionizing radiation in clonogenic survival assay compared with controls (Fig. 2B). Furthermore, USP11-silenced cells are hypersensitive to other genotoxic agents that induce DNA damage repaired via HR-dependent pathways including bleomycin, cisplatin, and mitomycin C (MMC) (Fig. 2, C–E). In all cases, silencing USP11 yielded hypersensitivity similar to that observed when BRCA1 is silenced.
We then directly assessed HR repair in the USP11-silenced cells using HEK293 cells stably expressing a DR-GFP reporter plasmid, which is interrupted by a single SceI restriction site (43). Transient expression of the SceI endonuclease introduces a site specific double-strand break within the reporter. Proficient HR repair in these cells results in restoration of the GFP gene and a positive signal. Cells were analyzed by two-color flow cytometry to quantify GFP-positive cells relative to the total number of cells. Results indicated that USP11-silenced cells have a significant defect in HR repair activity compared with non-targeting and BRCA1 positive control (Fig. 2F). Western blot analysis confirmed USP11 and BRCA1 silencing in these cells (Fig. 2G). Taken together, these results indicate USP11-silencing contributes to hypersensitivity to multiple DNA-damaging agents and PARPi, most likely through defects in HR repair.
USP11 Regulates 53BP1 Localization
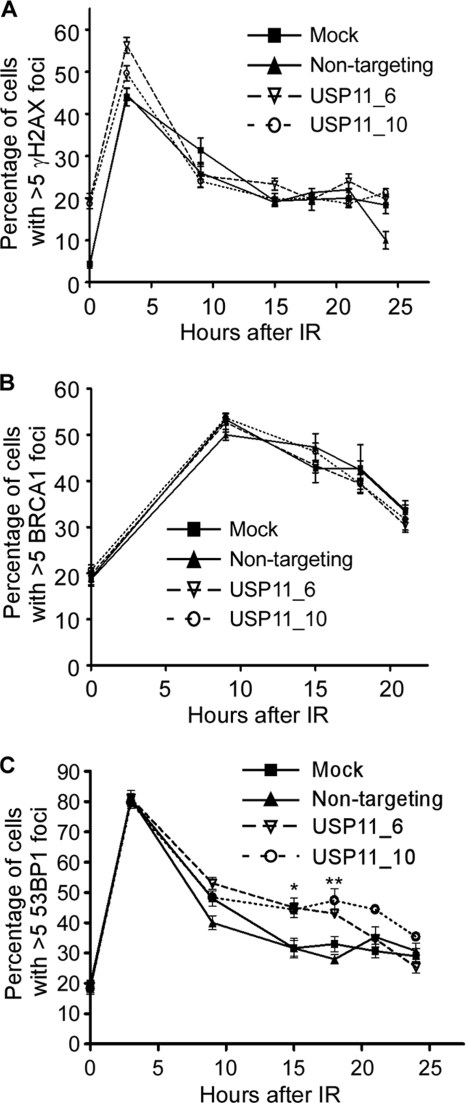
The HR double-strand break repair defect in USP11-silenced cells correlates with a defect in maintaining RAD51 foci. Given that USP11 is a DUB and ubiquitylation of proteins at double-strand break sites is critical for the assembly of repair and signaling complexes, we hypothesized that the loss of USP11 may perturb the dynamics of these complexes. Therefore, we examined the focal localization of DNA repair proteins at double-strand breaks. Although γH2AX foci are more abundant in USP11-silenced cell populations in the absence of IR, we did not observe a difference in the kinetics of γH2AX after IR (Fig. 3A). We also found no difference in the recruitment of BRCA1 to double-strand break foci after IR (Fig. 3B). However, when we examined 53BP1 foci formation, we observed a significant increase after IR at exactly the same time points that we observed a decrease in RAD51 foci (Fig. 3C). These results suggest that USP11 acts upstream in the regulation of RAD51 and 53BP1 but downstream of the initial γH2AX signal and independently of the BRCA1 localization mechanism.
FIGURE 3.
USP11 regulates 53BP1 foci after IR. A–C, mock, non-targeting and USP11-silenced U2OS cells were treated with IR and allowed to recover for the indicated time prior to fixation and staining with antibodies to A, γH2AX; B, BRCA1; or C, 53BP1 (*, p = 0.0006; **, p = 0.074). The percentage of cells with >5 foci were scored at each time point. Error bars are S.D. (n = 3).
The DUB Activity of USP11 Is Necessary for Its Function in Double-strand Break Repair
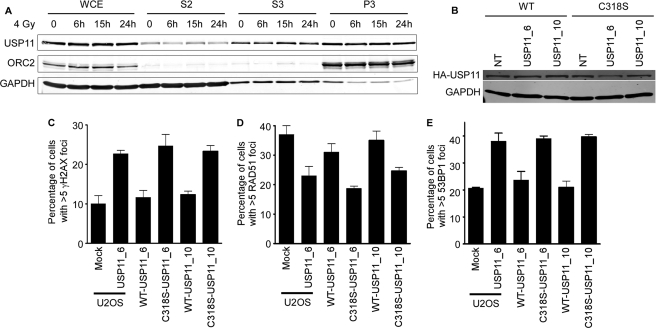
To gain further insights into the mechanism of action of USP11 in double-strand break repair we first sought to determine if it localized to double-strand break sites. Immunofluorescence localization of endogenous USP11 did not reveal an accumulation of the protein in damage-induced foci. However, we did observe a significant fraction of USP11 on chromatin in cells both before and after treatment with ionizing radiation (Fig. 4A).
FIGURE 4.
USP11 is a chromatin-associated protein, and its DUB activity is required for its activity in double-strand break repair. A, whole cell extracts (WCE), soluble fractions (S2 and S3) and chromatin fractions (P3) from cells treated with IR and allowed to recover for the indicated times were immunoblotted for USP11, ORC2, and GAPDH. B–E, HA-tagged wild-type (WT), and catalytic inactive (C318S) USP11 cDNAs that are insensitive to the USP11 siRNAs were stably expressed in U2OS cells. B, expression levels of HA-tagged proteins after non-targeting and USP11 siRNA transfections were determined by immunoblotting. C, parental-, WT-, and C318S USP11-expressing cells were transfected with the indicated siRNAs and scored for γH2AX foci in undamaged cells. D and E, parental-, WT-, and C318S-expressing cells were transfected with the indicated siRNAs, irradiated with 1 or 4 Gy of IR, allowed to recover for 15 h, then scored for RAD51 and 53BP1 foci by immunofluorescent imaging. Error bars in C–E are S.D. (n = 3).
We next asked whether the DUB activity of USP11 is required for its function in double-strand break repair. We generated stable cell lines expressing either HA-tagged wild type (WT) or catalytically inactive USP11 (C318S) that are resistant to both USP11 siRNAs because of wobble base pair changes in their target recognition sequences. These proteins were expressed at equal levels and unaffected by transfection with the siRNAs (Fig. 4B). We first determined whether the spontaneous activation of the DDR following silencing of USP11 was complemented by either the WT or C318S proteins. WT USP11 efficiently complemented the γH2AX induced by both USP11 siRNAs (Fig. 4C). In contrast, C318S-USP11 expression was unable to complement this USP11-deficient phenotype (Fig. 4C). Similarly, WT but not C318S-USP11 could complement the reduced RAD51 foci and increased 53BP1 foci in USP11 siRNA-transfected cells following ionizing radiation treatment (Fig. 4, D and E). Thus, USP11 catalytic activity is required for its function in the double-strand break response.
DISCUSSION
Our results demonstrate a role for USP11 in the repair of double-strand breaks via homologous recombination. Depletion of USP11 by introduction of siRNA leads to spontaneous DNA damage in the absence of genotoxic stress. USP11-silenced cells are hypersensitive to IR, bleomycin, cisplatin, mitomycin C, and PARPi treatment. They also exhibit a defect in HR directed double-strand break repair. Proper kinetic regulation of protein accumulation at double-strand breaks is impaired in USP11-depleted cells. RAD51 foci are not maintained and 53BP1 foci are increased at the same time points after IR exposure. Moreover, USP11 is a chromatin-associated protein and its catalytic activity is required for its genome maintenance activities. Taken together, these results indicate that USP11 is a DUB that functions in the DNA damage response to double-strand breaks.
Ubiquitylation of proteins at double-strand breaks is regulated by multiple ubiquitin ligases and DUBs creating a heterogeneous landscape (45). Ubiquitin conjugation regulates the recruitment of multiple proteins including RAD51 and 53BP1 through as yet undefined mechanisms (23, 26, 46). The accelerated disappearance of RAD51 foci in USP11-silenced cells may indicate an inability to maintain a sufficient RAD51 filament to facilitate HR repair. This could be due to changes in the resection of the double-strand breaks or perhaps inappropriate processing of the resected end.
It is unclear whether the persistent 53BP1 foci observed at later times after IR in USP11-silenced cells is a cause or consequence of the repair defect. 53BP1 recruitment is dependent on ubiquitylation (47). However, 53BP1 does not bind directly to ubiquitin-modified proteins. Instead, 53BP1 tudor domains bind methylated histones. The precise role of ubiquitin is unknown. The prolonged 53BP1 foci could be caused by the persistence of unrepaired double-strand breaks or increased levels of ubiquitin-conjugation at the DSB. Interestingly, 53BP1 has been shown to be required for XRCC4-dependent end-joining repair but not required for HR (48). Furthermore, 53BP1 deficiency actually increases HR repair. Thus, the increased 53BP1 retention at DSBs observed in USP11-silenced cells could be causally connected to the decrease in HR repair. In any case, the alterations in 53BP1 and RAD51 foci support a diminished capacity for HR repair in USP11-depleted cells leading to increased spontaneous γH2AX foci, PARPi sensitivity, and reduced survival after IR.
Our data indicate a requirement for the DUB activity of USP11 in the DNA damage response, suggesting defective ubiquitin de-conjugation or ubiquitin turnover causes the double-strand break repair defect in USP11-silenced cells. However, we cannot rule out that mutation of the catalytically active site of USP11 could cause disruption of protein interactions, accounting for the defects we see in our C318S mutant. The identification of the USP11 target(s) mediating its repair function is an important future goal. USP11 was previously reported to interact with BRCA2 (49). BRCA2 is ubiquitylated in cells; however, Schoenfeld et al. found that catalytic-inactive USP11 had no effect on BRCA2 ubiquitylation or protein levels. It is possible the BRCA2 interaction provides a mechanism to recruit USP11 to double-strand breaks where it can act on other substrates. However, we were unable to find an accumulation of USP11 in foci or a significant increase in its chromatin association after IR.
Although the interaction of USP11 and BRCA2 suggests a direct activity of USP11 at double-strand breaks, other reported USP11 interactions are consistent with indirect effects. Mass spectrometry results indicate that USP11 interacts with a large number of proteins including the transcriptional elongation factors TCEAL1 and TCEAL4, other DUBs including USP7, and the NRF2 regulatory protein KEAP1 (50). Thus, USP11 may have multiple functions within the cell, and we cannot exclude the possibility that the double-strand break repair defects are caused by indirect functions of USP11 in transcription regulation or other cellular pathways.
In summary, we have used a synthetic lethal approach to identify USP11 as a participant in the HR double-strand break repair pathway. PARP inhibitors are a new class of targeted therapy with exciting clinical promise in a subset of patients (9). USP11 status or the status of other HR-repair proteins in tumors may provide biomarkers for use of PARP inhibitors. Furthermore, our approach provides an experimental model to identify other HR proteins and potential biomarkers. The same methodologies could also be applied to determine targets for combination therapy with PARP inhibitors.
Supplementary Material
Acknowledgment
We thank Fei Ye for help with statistical analysis.
This work was supported, in whole or in part, by NCI, National Institutes of Health Grants (R21CA132010 and R01CA136933) and a Susan G. Komen for the Cure grant (to D. C.). Support for research facilities was provided by the Vanderbilt Center for Molecular Toxicology (Grant P30 ES000267) and the Vanderbilt-Ingram Cancer Center (Grant P30 CA068485).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- DDR
- DNA damage response
- PARP
- poly(ADP-ribose) polymerase
- HR
- homologous recombination
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- WT
- wild type
- DSB
- double-strand break
- DUB
- de-ubiquitylating enzymes
- IR
- ionizing radiation.
REFERENCES
- 1.Kastan M. B., Bartek J. (2004) Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 2.Huen M. S., Chen J. (2010) Trends Biochem. Sci. 35, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashworth A. (2008) J. Clin. Oncol. 26, 3785–3790 [DOI] [PubMed] [Google Scholar]
- 4.Herceg Z., Wang Z. Q. (2001) Mutat. Res. 477, 97–110 [DOI] [PubMed] [Google Scholar]
- 5.Saffhill R., Ockey C. H. (1985) Chromosoma 92, 218–224 [DOI] [PubMed] [Google Scholar]
- 6.Wang Z.-Q., Stingl L., Morrison C., Jantsch M., Los M., Schulze-Osthoff K., Wagner E. F. (1997) Genes Dev. 11, 2347–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T. (2005) Nature 434, 913–917 [DOI] [PubMed] [Google Scholar]
- 8.Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C. M., Ashworth A. (2005) Nature 434, 917–921 [DOI] [PubMed] [Google Scholar]
- 9.Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M. J., Ashworth A., Carmichael J., Kaye S. B., Schellens J. H., de Bono J. S. (2009) N. Engl. J. Med. 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 10.McCabe N., Turner N. C., Lord C. J., Kluzek K., Bialkowska A., Swift S., Giavara S., O'Connor M. J., Tutt A. N., Zdzienicka M. Z., Smith G. C., Ashworth A. (2006) Cancer Res. 66, 8109–8115 [DOI] [PubMed] [Google Scholar]
- 11.Bryant H. E., Helleday T. (2006) Nucleic Acids Res. 34, 1685–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavin M. F. (2008) Nat. Rev. Mol. Cell Biol. 9, 759–769 [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa R., Kastan M. B. (2005) Cold Spring Harbor Symp. Quant. Biol. 70, 99–109 [DOI] [PubMed] [Google Scholar]
- 14.Stucki M., Jackson S. P. (2006) DNA Repair 5, 534–543 [DOI] [PubMed] [Google Scholar]
- 15.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 16.Yuan J., Chen J. (2010) J. Biol. Chem. 285, 1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung P. (1994) Science 265, 1241–1243 [DOI] [PubMed] [Google Scholar]
- 18.Baumann P., Benson F. E., West S. C. (1996) Cell 87, 757–766 [DOI] [PubMed] [Google Scholar]
- 19.Shinohara A., Ogawa H., Ogawa T. (1992) Cell 69, 457–470 [DOI] [PubMed] [Google Scholar]
- 20.Davies A. A., Masson J. Y., McIlwraith M. J., Stasiak A. Z., Stasiak A., Venkitaraman A. R., West S. C. (2001) Mol. Cell 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 21.Yuan S. S., Lee S. Y., Chen G., Song M., Tomlinson G. E., Lee E. Y. (1999) Cancer Res. 59, 3547–3551 [PubMed] [Google Scholar]
- 22.Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 23.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 24.Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao G. Y., Sonoda E., Barber L. J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., Boulton S. J., Takeda S. (2007) Mol. Cell 25, 663–675 [DOI] [PubMed] [Google Scholar]
- 27.Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 28.Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 30.Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. (2003) Nature 421, 961–966 [DOI] [PubMed] [Google Scholar]
- 31.Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 33.Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H., Huang J., Chen J. (2007) Nat. Struct. Mol. Biol. 14, 710–715 [DOI] [PubMed] [Google Scholar]
- 35.Yan J., Kim Y. S., Yang X. P., Li L. P., Liao G., Xia F., Jetten A. M. (2007) Cancer Res. 67, 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Arciero C. A., Wang C., Broccoli D., Godwin A. K. (2006) Cancer Res. 66, 5039–5046 [DOI] [PubMed] [Google Scholar]
- 37.Dong Y., Hakimi M. A., Chen X., Kumaraswamy E., Cooch N. S., Godwin A. K., Shiekhattar R. (2003) Mol. Cell 12, 1087–1099 [DOI] [PubMed] [Google Scholar]
- 38.Shao G., Lilli D. R., Patterson-Fortin J., Coleman K. A., Morrissey D. E., Greenberg R. A. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicassio F., Corrado N., Vissers J. H., Areces L. B., Bergink S., Marteijn J. A., Geverts B., Houtsmuller A. B., Vermeulen W., Di Fiore P. P., Citterio E. (2007) Curr. Biol. 17, 1972–1977 [DOI] [PubMed] [Google Scholar]
- 40.Lord C. J., McDonald S., Swift S., Turner N. C., Ashworth A. (2008) DNA Repair 7, 2010–2019 [DOI] [PubMed] [Google Scholar]
- 41.Turner N. C., Lord C. J., Iorns E., Brough R., Swift S., Elliott R., Rayter S., Tutt A. N., Ashworth A. (2008) EMBO J. 27, 1368–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanton C., Marani M., Pardo O., Warne P. H., Kelly G., Sahai E., Elustondo F., Chang J., Temple J., Ahmed A. A., Brenton J. D., Downward J., Nicke B. (2007) Cancer Cell 11, 498–512 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J., Willers H., Feng Z., Ghosh J. C., Kim S., Weaver D. T., Chung J. H., Powell S. N., Xia F. (2004) Mol. Cell. Biol. 24, 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovejoy C. A., Xu X., Bansbach C. E., Glick G. G., Zhao R., Ye F., Sirbu B. M., Titus L. C., Shyr Y., Cortez D. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 19304–19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Messick T. E., Greenberg R. A. (2009) J. Cell Biol. 187, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart G. S. (2009) Cell Cycle 8, 1532–1538 [DOI] [PubMed] [Google Scholar]
- 48.Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F. W., Chen J., Jackson S. P., Scully R. (2007) Mol. Cell 28, 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoenfeld A. R., Apgar S., Dolios G., Wang R., Aaronson S. A. (2004) Mol. Cell. Biol. 24, 7444–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.