Abstract
CD39 is a transmembrane enzyme that inhibits platelet reactivity and inflammation by phosphohydrolyzing ATP and ADP to AMP. Cyclic AMP (cAMP), an essential second messenger, is particularly important in regulating genes controlling vascular homeostasis. These experiments test the hypothesis that cAMP might positively regulate the expression of CD39 and thereby modulate important vascular homeostatic properties. Cd39 mRNA was induced by 13.8- fold in RAW cells treated with a membrane-permeant cAMP analogue (8-bromo-cyclic AMP; 8-Br-cAMP), stimulation of adenylate cyclase, or prostanoids known to drive cAMP response. Fluorescence-activated cell sorting, immunofluorescence, and TLC assays demonstrated that both CD39 protein expression and enzymatic activity were increased in cells treated with 8-Br-cAMP but not in cells transfected with short hairpin RNA against CD39. This analogue drove a significant increase in transcriptional activity at the Cd39 promoter although not when the promoter's cAMP-response element sites were mutated. Pretreatment with cAMP-dependent protein kinase (PKA), phosphoinositide 3-kinase (PI3K), or ERK inhibitors nearly obliterated the cAMP-driven increase in Cd39 mRNA, protein expression, and promoter activity. 8-Br-cAMP greatly increased the phosphorylation of CREB1 (Ser133) and ATF2 (Thr71) in a PKA-, PI3K-, and ERK-dependent fashion. Chromatin immunoprecipitation assays demonstrated that binding of phosphorylated CREB1 and ATF2 to cAMP-response element-like sites was significantly increased with 8-Br-cAMP treatment and that binding was reduced with PKA, PI3K, and ERK inhibition, whereas transfection of Creb1 and Atf2 overexpression constructs enhanced cAMP-driven Cd39 mRNA expression. Transfection of RAW cells with mutated Creb1 (S133A) reduced cAMP-driven Cd39 mRNA expression. Furthermore, the cAMP-mediated induction of Cd39 mRNA, protein, and phosphohydrolytic activity was replicated in primary peritoneal macrophages. These data identify cAMP as a crucial regulator of macrophage CD39 expression and demonstrate that cAMP acts through the PKA/CREB, PKA/PI3K/ATF2, and PKA/ERK/ATF2 pathways to control a key vascular homeostatic mediator.
Keywords: Akt PKB, CREB, Cyclic AMP (cAMP), ERK, Macrophage, Phosphatidylinositol 3-kinase, Protein kinase A (PKA), ATF2, CD39
Introduction
CD39 (ectonucleoside triphosphate diphosphohydrolase 1) is an integral membrane protein expressed on the surface of vascular and immune cells. It has an extracellular domain exposed to flowing blood that affects the terminal phosphohydrolysis of ATP and ADP. This ecto-apyrase activity modulates many of the purinergic signaling processes in which ATP or ADP play a role, and is important for the tonic maintenance of vascular homeostatic properties related to inflammation, coagulation, vasodilation, and barrier function. For instance, CD39 can inhibit platelet activation and maintain vascular fluidity in the complete absence of prostacyclin and nitric oxide as well as regulate immune function and provide protection from both cardiac and cerebral ischemia and reperfusion injuries (1–9).
Although its presence is critical for maintaining vascular homeostasis particularly under stress conditions, little is known about the regulation of CD39 expression at the molecular level. In silico analysis of the Cd39 promoter region revealed, among potential regulatory sites, several cAMP-response element (CRE)2-like motifs, one of which was in a region proximal enough to the transcriptional start point to be of interest. cAMP, an essential second messenger that acts largely through its downstream effector, protein kinase A (PKA), regulates a diverse array of physiologic processes, ranging from metabolic control to cellular proliferation, by altering basic patterns of gene expression (10). cAMP is also known to be a critical regulator of vascular homeostasis that parallels the function of CD39 in regulating inflammation, coagulation, vasodilation, and barrier function.
PKA resides in the cytoplasm as an inactive heterotetramer of paired regulatory and catalytic subunits. The presence of cAMP liberates the catalytic subunits, allowing their passive diffusion into the nucleus. Once catalytic subunit translocation has occurred, it subsequently phosphorylates a serine residue of CREB (cAMP-response element-binding protein) at position 133 (Ser133), an important step for the activation and dimerization of CREB (11). CREB dimers bind to CRE sequence elements (TGACGTCA) (12, 13) in the promoter of various target genes (14), where phosphorylated Ser133 of CREB can act as a scaffold for the transcriptional co-activator CREB-binding protein and its paralogue p300. This CREB-CREB-binding protein-p300 complex ultimately associates with RNA polymerase II and thereby stimulates transcription (13).
CREB and activating transcription factor (ATF) are members of a large basic leucine zipper (bZIP) superfamily of transcription factors. Members of the ATF/CREB subfamily, including ATF-1, ATF-2, CREB, and CREM, among others, coalesce as homo- or heteromultimers to bind to CREs within the promoter and enhancer sequences of target genes. ATF-1, ATF-2, CREB, and CREM may act as either positive or negative regulators of transcription. Modulation and indeed fine tuning the transcription of genes containing CRE regulatory elements is thereby possible. CREB and CREM have been shown to play important roles in basal and hormone-regulated transcription and differentiation, whereas the role of ATF-1 is less well defined. ATF-1 homodimers appear to be weaker transcriptional activators than either CREB or certain forms of CREM because ATF-1-mediated activation is enhanced by heterodimerization with either CREB or CREM (15). ATF2, whose known target genes include interferon-γ, tumor necrosis factor-α, and cyclin A (16–18), is activated upon its phosphorylation by stress-activated kinases in response to stress and cytokine stimuli (19, 20). These are just several examples of the complex regulatory control possible for genes whose promoters contain CRE motifs.
Very little, if anything, is known regarding the transcriptional regulation of Cd39. Because a CRE-like regulatory element lies only 210 bp upstream of the Cd39 transcriptional start point, we hypothesized that this might be a major site of transcriptional regulatory control for CD39 expression. It would be logical, given the critical and often parallel vascular homeostatic roles for cAMP and CD39, for cAMP to regulate CD39 expression. This work shows for the very first time that cAMP, working through this CRE-like motif, is indeed an extremely powerful regulator of Cd39 transcription. Furthermore, these results demonstrate that these changes are functionally significant, where the enzymatic activity of CD39 is driven in parallel to the cAMP-mediated regulation of Cd39 transcription. This type of regulatory control could have profound implications for vascular diseases ranging from myocardial infarction to stroke, wherein CD39 expression can mitigate the adverse consequences of interrupted blood flow.
MATERIALS AND METHODS
Cell Culture
All chemical reagents were ordered from Sigma unless otherwise specifically noted. RAW cells (a transformed murine macrophage cell line) were obtained from ATCC and grown in RPMI 1640 medium with 10% fetal calf serum (Invitrogen) and penicillin/streptomycin (50 units/ml and 5 μg/ml, respectively; Invitrogen). When RAW cells achieved confluence, experiments were performed by pretreating them for 20 min with either vehicle (DMSO) or 20 μm H89 (an inhibitor of the cAMP-dependent protein kinase (PKA)), 50 μm LY294002 (an inhibitor of phosphoinositide 3-kinase (PI3K)), wortmannin (an inhibitor of PI3K), or PD98059 (an inhibitor of ERK). Immediately after, the membrane-permeable cAMP analog 8-bromo-cyclic AMP (8-Br-cAMP; 250 μm) was added to the culture dishes for the indicated times.
Isolation of a Mouse Cd39 Genomic Clone
A 4555-bp fragment containing 4474 bp of the 5′-flanking (−4474) region and 81 bp of the untranslated first exon (+81) of mouse Cd39 was isolated from RAW cell total genomic DNA using PCR. The primers (5′-CACTTACTCGGGCATGCTGTTGAAC-3′ and 5′-CCCCCTGAAGGGTTTGGATCAAATCAGTTCTGG-3′) were designed by analysis of the sequence of the mouse Cd39 gene, 5′ flank (GenBankTM accession number NM_009848). The 4555-bp insert was subcloned into pCR-XL-TOPO vector (Invitrogen) using the TOPO XL PCR cloning kit (Invitrogen) and sequenced to confirm identity.
Construction of Mouse Cd39 Promoter-Luciferase Reporter Plasmids and Site-directed Mutagenesis
PCR was used to amplify mouse Cd39 promoter-luciferase reporter constructs representing 5′-deleted Cd39 upstream sequences, and these PCR products were subcloned into the pGL3-basic vector (Promega) using a T4 ligase and Quick Ligasing buffer (Promega). In these PCRs, the Cd39 genomic clone (described above) was used as a template. The 5′-deletion constructs of the Cd39 promoter represent the spans −250 to +32 (pCD39/250) and −52 to +32 (pCD39/52). The Cd39 promoter −220 to −189 fragment (containing a CRE element) was cloned into pCD39/52 to make the deletion construct pCD39/ΔCRE. For mutants, oligonucleotides used for priming were synthesized based on 5′-flanking region sequence of the mouse Cd39 gene. The site-directed mutants of the pCD39/CREmut constructs were generated using QuikChange site-directed mutagenesis kits (Stratagene). In these PCRs, the 5′-deletion construct, pCD39/250, was used as a template. Two mutated priming oligonucleotides representing overlapping sense and antisense sequences of the mutant site were used to amplify the entire pGL3 plasmid and insert. The mutated primers are shown as follows (target regions are indicated by underlines and alterations are indicated by boldface type): 5′-GAGTTTTGAACACATACTACCACAAGCCTAGAAAAAAG-3′ and 5′-CTTTTTTCTAGGCTTGTGGTAGTATGTGTTCAAAACTC-3′. Forward and reverse DNA sequencing of inserts confirmed the sequences of all constructs employed for transfection assays.
Construction of Mouse Creb1, Atf2, and Dominant Negative Creb Overexpression Plasmids
The cDNA clones of mouse Creb1 and Atf2 were ordered from ATCC. Creb1 expression plasmid (pCREB1) and Atf2 expression plasmid (pATF2) were constructed by first using PCR to amplify the open reading frame of the Creb1 and Atf2 cDNAs. The open reading frames of Creb1 and Atf2 cDNA were each cloned into pcDNA3.1 (Invitrogen) expression vectors. The primers used for PCR to amplify the open reading frame of the Creb1 and Atf2 cDNAs are as follows: Creb1, 5′-GGAGAAGCTTGTACCACCGGTAACTAAATGACC-3′ and 5′-CACTCGAGAACTTAAATCCCAAATTAATCTG-3′; Atf2, 5′-AACAGGTACCTGTGGAATATGAGTGATG-3′ and 5′-GCACTCGAGGTTTTAATCAACTTCCTGAGG-3′.
The site-directed mutants of the dominant negative Creb overexpression plasmid (pCREB/Ser133Mut) constructs were generated using QuikChange site-directed mutagenesis kits (Stratagene). In these PCRs, pCREB1 was used as a template. The mutated priming oligonucleotides represented overlapping sense and antisense sequences of the mutant site that amplified the entire pGL3 plasmid and insert. The mutated primers are shown as follows (target regions are indicated by underlines, and alterations are indicated by boldface type): 5′-CCTTTCAAGGAGGCCTGCCTACAGGAAAATTTTG-3′ and 5′-CAAAATTTTCCTGTAGGCAGGCCTCCTTGAAAGG-3′.
Transient Transfections
RAW cells were co-transfected using SuperFect transfection reagent (Qiagen). The day before transfection, RAW cells were plated on 12-well plates (Costar) with 1 × 105 cells/well and grown in RPMI 1640 (Invitrogen) containing 10% fetal calf serum (Invitrogen). Cultures were incubated at 37 °C in a humidified atmosphere containing 5% CO2 until they reached 50–80% confluence. The cells were co-transfected using 0.75 μg of Cd39 promoter-pGL3 reporter plasmid and 0.75 μg of control plasmid, pCMV/β-galactosidase, which served as an internal control to normalize transfection efficiency, as well as 7.5 μl of SuperFect transfection reagent in 75 μl of serum-free RPMI 1640 medium. This was followed by incubation at room temperature for 10 min, after which the complexes were mixed with 0.4 ml of RPMI 1640 medium with 10% FBS. After incubating at 37 °C and 5% CO2 for 3 h, the cells were washed 3 times with HBSS (Invitrogen) and then incubated with RPMI with 10% FBS. After 24–48 h of incubation, the cell medium was changed to RPMI 1640, and then the cells were treated with 250 μm of 8-Br-cAMP or RPMI 1640 only (control) for 5 h. Luciferase reporter assays were performed using a luciferase reporter assay system (Promega). Transfected cells were washed two times with cold phosphate-buffered saline and then harvested, lysed, and assayed for luciferase activity by a VICTOR LIGHT luminometer (PerkinElmer Life Sciences). β-Galactosidase enzyme activity was measured using a β-galactosidase enzyme assay system (Promega). Fifty μl of the sample was mixed with 50 μl of 2× buffer containing 200 mm sodium phosphate, pH 7.3, 2 mm MgCl2, 100 mm β-mercaptoethanol, and 1.33 mg/ml o-nitrophenyl-β-galactopyranoside in a 96-well plate. After incubating for 30 min at 37 °C, the absorbances of the samples were read at 420 nm in a VersaMax tunable microplate reader (Molecular Devices). For the overexpression experiments, the RAW cells were co-transfected with pCREB1, pATF2, or empty vector (pcDNA3.1) with pCD39/250 and pCMV/β-galactosidase vector. The method of transfection and reporter assay used are described above.
Stable Transfections
The day before transfection, RAW cells were plated on 6-well plates (Costar) and grown in RPMI 1640 (Invitrogen) containing 10% fetal calf serum (Invitrogen). Cultures were incubated at 37 °C in a humidified atmosphere containing 5% CO2 until they reached 50–80% confluence. RAW cells were transfected with a mouse Cd39 short hairpin RNA (shRNA) construct (5′-CAGGAACAGAGTTGGCTAAGCCTCATCTC-3′, OriGene (Rockville, MD)) or a control pRS shRNA vector (OriGene), using SuperFect transfection reagent (Qiagen). The cells were transfected using 2 μg of Cd39 shRNA construct, as well as 10 μl of SuperFect transfection reagent in 100 μl of serum-free RPMI 1640 medium. This was followed by incubation at room temperature for 10 min, after which the complexes were mixed with 0.6 ml of RPMI 1640 medium containing 10% FBS. After incubating at 37 °C and 5% CO2 for 3 h, the cells were washed three times with Hanks' balanced salt solution (Invitrogen) and then incubated with RPMI containing 10% FBS. After 24–48 h of incubation, the transfected cells were cultured with RPMI containing 10% FBS and 3 μg/ml puromycin to select stable transfected cells. After transfected cells were established (>2 weeks in culture), membrane protein was isolated, and the CD39 protein expression was measured using Western blot.
Murine Peritoneal Macrophage Isolation
Peritoneal macrophages were isolated as described previously (21). In brief, 10-week-old male C57Bl6/J mice were injected intraperitoneally with 3.0 ml of a 5% thioglycollate solution (BD Biosciences). Four days later, macrophages were isolated by peritoneal lavage and placed in ice-cold phosphate-buffered saline. The cells were plated in 6-well plates at a concentration of 1 × 106 cells/well for total RNA isolation or 150-mm dishes for membrane protein extraction.
Quantitative Reverse Transcription-PCR (qRT-PCR)
RAW cells were transfected with the pCREB1 or pATF2 overexpression constructs to induce CREB1 or ATF2 protein expression. The empty vector, pcDNA3.1, was used as a control. RAW cells were plated onto 6-well plates (Costar). After RAW cells reached 80% confluence, the cells were transfected using 2 μg of expression or control vectors and 10 μl of Superfect transfection reagent. After 24 h of transfection, the cells were treated with 250 μm 8-Br-cAMP for the indicated durations. Total RNA was then isolated from the transfected RAW cells using RNA Easy Kits (Qiagen), and qRT-PCR was used to assay the level of Cd39, Creb1, or Atf2 mRNA. A real-time PCR assay was carried out with the 7000 sequence detection system (Applied Biosystems) using the One-Step RT-PCR master mix (Applied Biosystems) according to the manufacturer's instructions. The primers and probes sets were purchased from Applied Biosystems. All qRT-PCRs were done in triplicate. Cd39, Creb1, or Atf2 mRNA was normalized to β-actin levels and reported as relative -fold increase compared with vector-transfected controls, determined by the 2−ΔΔCt method (22, 23).
Membrane Protein Isolation
After treatment with 250 μm 8-Br-cAMP for the indicated times, the RAW cells were washed twice with cold phosphate-buffered saline (Invitrogen), scraped off the plate with a rubber policeman, and suspended in ice-cold hypotonic lysis buffer containing 50 mm sucrose, 10 mm Hepes, pH 7.4, 1 μg/ml aprotinin, 1 μg of leupeptin, and 1 mm phenylmethylsulfonyl fluoride. The cells were homogenized by 25 strokes of a Dounce homogenizer (Kontes Glass) with a tight pestle. Following the addition of preparation buffer containing 264 μl of 65% (w/w) sucrose (in 10 mm Hepes, pH 7.4), 4 μl of 0.5 m MgCl2 was added into 2 ml of extraction buffer, and the homogenate was subjected to two 20-min spins at 2000 × g at 4 °C to pellet nuclei, mitochondria, and unlysed cells. Crude membranes were pelleted from the resulting supernatant by centrifugation at 100,000 × g for 30 min at 4 °C, washed quickly in 2 ml of hypotonic lysis buffer, and resuspended in the preparation buffer. The membranes were flash-frozen in aliquots and stored at −80 °C. Protein concentrations were determined by a Bio-Rad protein assay (Bio-Rad).
Flow Cytometric Analysis
Surface expression of CD39 on RAW cells was assessed following 8-Br-cAMP treatment or vehicle treatment. Cells were stained for 30 min at 4 °C with either a monoclonal anti-mouse CD39 antibody conjugated to phycoerythrin (eBioscience) and 7-aminoactinomycin D to exclude nonviable cells or a rat IgG2a isotype control antibody (eBioscience). Immunostained cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences) using CellQuestTM software (BD Biosciences). At a minimum, 10,000 events/sample were collected for analysis.
Immunofluorescence
RAW cells were plated on 4-well chamber slides (LabTek). After plating, wells were treated for 16 h with one of the following: 250 μm 8-Br-cAMP or RPMI 1640 only as a negative control. The RAW cells were then fixed for 15 min with 1% paraformaldehyde and then incubated for 16 h with anti-CD39 antibody (1:50) at 4 °C and then with fluorescein isothiocyanate-conjugated goat anti-rat IgG (1:100) for 1 h at room temperature. Photomicrographs were taken using a Nikon Eclipse TE2000-E microscope.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed using an EZ ChIPTM chromatin immunoprecipitation kit (Upstate Biotechnology). RAW cells were cross-linked at 37 °C for 10 min in 1% formaldehyde. Cells were then sonicated in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin) 12 times at 4 °C using 15 sonicating pulses at 30% output. The supernatant was divided into three tubes for subsequent immunoprecipitation. Samples were precleared in immunoprecipitation buffer (0.01% SDS, 1.1% Triton, 1.2 mm EDTA, 16.7 mm Tris, pH 8.1, 167 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin) and added to protein A-Sepharose beads. The mixture was placed on a rotator for 1 h at 4 °C, after which beads were spun out and discarded (this step was to clear the mixture of excess IgG). To the supernatant, 5 μg of anti-phosphorylated CREB1 (Ser133), phosphorylated ATF2 (Thr71) antibody (Santa Cruz), or rabbit IgG were then added, and the mixture was placed on a rotator for 16 h at 4 °C. Immunoprecipitates were recovered by adding protein A-Sepharose beads, which were washed for 5 min with low salt immune complex wash buffer (0.1% SDS, 1% Triton, 2 mm EDTA, 20 mm Tris, pH 8.1, and 150 mm NaCl) and then for 5 min with high salt immune complex wash buffer (0.1% SDS, 1% Triton, 2 mm EDTA, 20 mm Tris, pH 8.1, and 500 mm NaCl), 5 min with LiCl immune complex wash buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, and 10 mm Tris, pH 8.1) and twice for 3 min with TE buffer (10 mm Tris, pH 8.0, and 1 mm EDTA, pH 8.0). Immune complexes were eluted during rotation with 1% SDS and 0.1 m NaHCO3 for a total of 30 min at room temperature. Immunoprecipitates and inputs were reverse cross-linked with 200 mm NaCl at 65 °C for 16 h. Samples were then incubated 1 h at 45 °C with 10 mm EDTA, 40 mm Tris, pH 6.5, and 40 μg/ml proteinase K. DNA was extracted by the Qiaquick PCR purification kit (Qiagen). The input DNA and ChIP DNA were amplified by qRT-PCR. Primers used for PCR correspond to the mouse Cd39 region (−239 to −172; 5′-primer, 5′-GGGAAGGAGAGAGTGAGTTTTGAA-3′; 3′-primer, 5′-CACCGGGTTGTAATTTCTTTTTTC-3′; probe, ACATACTACGTCAAGCCT-3′). Ct values of ChIP DNA were normalized to the Ct values of input DNA.
mRNA Degradation
Cells were exposed to 250 μm 8-Br-cAMP for 4 h, and then the RNA transcriptional inhibitor actinomycin D was added to a final concentration of 5 μg/ml. The RNA was then harvested at 0, 0.5, 1, 2, and 4 h after actinomycin D addition, and the mRNA levels were measured by real-time RT-PCR.
TLC Analysis of CD39 Activities
For assessment of enzymatic function, cell membrane proteins of either CD39 shRNA-transfected or vector-transfected RAW cells were mixed with 1.0 mm [8-14C]ATP (MP Biomedicals), 286 μm AOPCP (Sigma), or 1 mm ouabain (Sigma) in RPMI 1640 (Invitrogen) and incubated at 37 °C for 20 min. The reaction was stopped using 8 m formic acid (52), and the reaction mixture was spotted on silica gel/TLC plates (Fluka, Invitrogen) along with a ladder (a mixture of [8-14C]AMP (GE Healthcare), [8-14C]ADP, and [8-14C]ATP). The nucleotides were separated by TLC with isobutyl alcohol/isoamyl alcohol/2-ethoxyethanol/ammonia/H2O (9:6:18:9:15) as described previously (10, 24, 51). TLC plates were then exposed to a phosphorimaging screen (Eastman Kodak Co.) and then analyzed using a Typhoon Trio+ Variable Mode Imager (GE Healthcare).
Platelet-rich Plasma Isolation and Platelet Aggregometry
Mouse blood was drawn from the inferior vena cava using a 20-gauge needle containing 3.2% sodium citrate. One volume of HT Buffer (137 mm NaCl, 2.68 mm KCl, 12 mm NaHCO3, 0.36 mm NaH2PO4, 10 mm HEPES, 0.2% bovine serum albumin) was added and spun at 50 × g for 10 min at room temperature. The citrated blood was brought back to the original volume with HT Buffer and spun at 50 × g for 10 min at room temperature. Platelet-rich plasma was collected. Platelet-rich plasma aggregometry was then performed using a Chrono-log 560CA with the Aggro/Link 810. 1 × 108 platelets in 500 μl of HT buffer were mixed with 20 μg of membrane protein of either 250 μm 8-Br-cAMP-treated or non-treated peritoneal macrophages before stimulation with 1 μm ADP (Chrono-log).
Western Blotting Assay
Nuclear, membrane, or total protein (prepared as described above) was added to 4× sample buffer and 10× reducing agent (Invitrogen), boiled for 3 min, separated by 10% SDS-PAGE, and electrophoretically transferred onto nitrocellulose membranes (Invitrogen). The membranes were incubated with anti-CD39, anti-CREB1, or anti-ATF2 antibody (Santa Cruz Biotechnology) and autoradiographed using the enhanced chemiluminescence method (ECL detection system, Amersham Biosciences).
PKA Activity Assay
RAW cells were treated with 20 μm H89, 50 μm LY294002, 10 μm wortmannin, or 50 μm PD98059 for 20 min, suspended in PKA extraction buffer (25 mm Tris-HCl, pH 7.4, 0.5 mm EDTA, 0.5 mm EGTA, 10 mm β-mercaptoethanol, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 μm phenylmethylsulfonyl fluoride), and then homogenized using a Dounce homogenizer. PKA activity was measured using the PepTag® assay for non-radioactive detection of cAMP-dependent protein kinase (Promega). According to the kit's instructions, 10 μl of the lysates were mixed with 0.08 μg/μl PepTag® A1 peptide, a brightly colored, fluorescent peptide substrate, 20 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 1 mm ATP, and 1 μm cAMP. After incubation for the indicated times at room temperature, the mixtures were heated to 95 °C for 10 min to stop the reaction and then run on a 0.8% agarose gel. The negatively charged phosphorylated species migrated toward the positive electrode, whereas the positively charged non-phosphorylated substrate migrated toward the negative electrode. The negatively charged phosphorylated bands were excised from the gel using a razor blade, placed into a 1.5-ml graduated microcentrifuge tube, and heated to 95 °C until the gel slice melted. The melted gel was added to 75 μl of gel solubilization solution and 50 μl of glacial acetic acid, quickly vortexed, and then transferred to a 96-well plate. The sample's fluorescent absorbance was ascertained using an excitation/emission of 540 nm/590 nm by a SpectraMax M5 microplate reader.
Statistics
Statistical analysis was performed using analysis of variance to detect differences between groups, using Prism® software. Differences were considered significant if p was <0.05.
RESULTS
Effects of cAMP on Macrophage Cd39 mRNA Expression
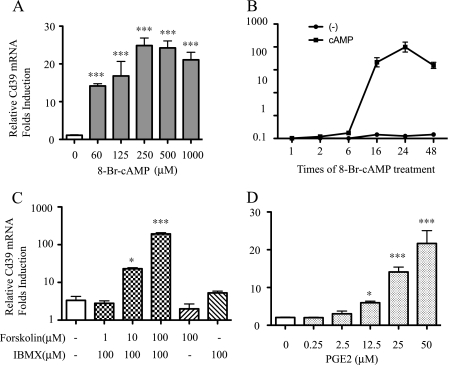
To characterize the role of cAMP pathway stimulation on Cd39 mRNA expression in macrophages, a murine macrophage cell line (RAW 264.7) was grown to confluence and treated with a membrane-permeable cAMP analogue, 8-Br-cAMP, and dose-response and time course studies were conducted. Total RNA was isolated, and Cd39 mRNA levels were measured using qRT-PCR, with results normalized to Cd39 expression for non-treatment control. When cells were treated for 6 h, results showed that Cd39 mRNA levels increased 12.8-fold following 60 μm 8-Br-cAMP treatment and that there was a maximal 22.5-fold increase following 250 μm 8-Br-cAMP (p < 0.001, respectively, compared with non-treatment control) (Fig. 1A). Time course studies showed that, at a fixed dose of 250 μm 8-Br-cAMP, Cd39 mRNA levels increased 10.4-fold by 2 h of treatment and continued to increase up to a peak level of 39.7-fold when treatment persisted for 24 h, finally dropping down to 20-fold when treated for 48 h (p < 0.001 compared with non-treated controls; Fig. 1B). To confirm that intracellular cAMP stimulates Cd39 mRNA expression in macrophages, RAW cells were treated with forskolin, an adenylyl cyclase agonist, and isobutylmethylxanthine, a nonspecific phosphodiesterase inhibitor. A second group of cells were treated for 16 h with prostaglandin E2 (PGE2), an intracellular cAMP stimulator, to increase intracellular cAMP. Results showed that Cd39 mRNA levels were significantly increased 6.8- and 57.8-fold in those cells treated with 10 and 100 μm forskolin, respectively, compared with the non-treated cells (Fig. 1C; p < 0.05 and p < 0.001, respectively). Cd39 mRNA expression in RAW cells also increased 6.8- and 10.5-fold after treatment with 25 and 50 μm PGE2, respectively, compared with the non-treated control group (Fig. 1D; both p < 0.001) (25).
FIGURE 1.
Effect of cAMP on macrophage Cd39 mRNA expression. RAW cells were treated with 8-Br-cAMP for the indicated times and dosages, and total RNA was isolated. qRT-PCR was used to detect Cd39 mRNA expression in dose- and time-dependent manners (A and B). RAW cells were treated with forskolin and isobutylmethylxanthine or PGE2 at the indicated dosages for 16 h, and total RNA was isolated. Quantitative RT-PCR was used to detect Cd39 mRNA expression (C and D). *, p < 0.05; ***, p < 0.001 compared with non-treatment controls. ###, p < 0.001 compared with maximum Cd39 mRNA expression.
Effect of cAMP on Macrophage Cd39 mRNA Degradation Rates
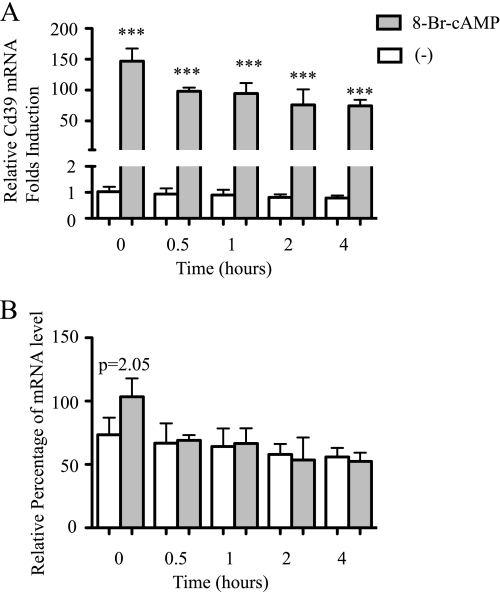
To characterize the effect of cAMP on Cd39 mRNA degradation in macrophages, RAW cells were treated with 8-Br-cAMP for 4 h before adding 5 μg/ml actinomycin D, a mRNA transcription blocker. The cells were harvested at 0, 0.5, 1, 2, and 4 h after the addition of actinomycin D. Total RNA was isolated, and Cd39 mRNA levels were measured using qRT-PCR. The results showed that cAMP does not affect degradation rates of Cd39 mRNA in macrophages. The values of relative quantities (RQ) were normalized to the values of RQ for the 0 min actinomycin D-treated cells. The levels of Cd39 mRNA in the 8-Br-cAMP-treated cells are higher than in the non-treated cells after 0–4 h of actinomycin D treatment (Fig. 2A, p < 0.001). In order to compare the degradation rates of Cd39 mRNA in 8-Br-cAMP-treated cells to that in non-treated cells, the values of RQ for 0.5, 2, and 4 h actinomycin D plus 8-Br-cAMP-treated cells were normalized to the 0 min actinomycin D plus 8-Br-cAMP-treated cells and shown as percentages (Fig. 2B) (26).
FIGURE 2.
Effect of cAMP on Cd39 mRNA stability in macrophages. RAW cells were treated with 8-Br-cAMP for 4 h and then incubated with 5 μg/ml of actinomycin D for the indicated times. Total RNA was isolated, and then qRT-PCR was used to detect Cd39 mRNA expression as described under “Materials and Methods.” The values of RQ were normalized to the values of RQ for the 0 min actinomycin D-treated control (non-cAMP-treated) cells (A). In order to compare the relative rates of RNA degradation, the values of RQ for actinomycin D and cAMP co-treated cells were normalized to the 0 min actinomycin D and cAMP-treated cells and shown as percentages (B). In order to assess the relative levels of Cd39 mRNA, the values of actinomycin D and non-cAMP treated samples were normalized to C3 (0 min actinomycin D-treated normoxic cells) (B) (26). ***, p < 0.001. 0, 0.5, 1, 2, or 4 h of 8-Br-cAMP treatment versus non-cAMP-treated cells, respectively.
Effect of cAMP on Macrophage Plasmalemmal CD39 Expression
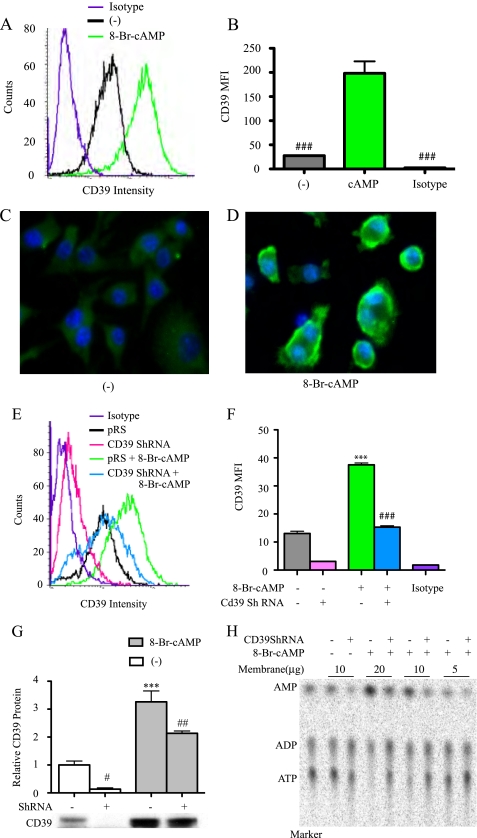
To characterize the effect of cAMP on cell membrane CD39 expression, RAW cells were grown to confluence and then treated with 8-Br-cAMP (250 μm for 16 h). The expression of CD39 on RAW cell membranes was measured by FACS and Western blot. Immunostaining for CD39 was also performed on cells that were plated and then fixed. 8-Br-cAMP treatment resulted in a consistent and striking increase in CD39 antigen compared with non-treatment controls (Fig. 3, A–D). Even under experimental conditions in which a Cd39 shRNA was used to silence expression of native Cd39, 8-Br-cAMP (250 μm for 5 h) was still able to increase CD39 expression, albeit largely dampened by the Cd39 silencing construct (Fig. 3, E–G). Enzymatic activity of CD39 was then used as a functional correlate of CD39 antigen expression. To measure CD39 activity, the isolated membrane proteins were incubated with [8-14C]ATP, after which phosphohydrolysis was assessed by a TLC assay (Fig. 3H). CD39 activity was increased by 40% in 8-Br-cAMP-treated cells compared with the non-treated control (Fig. 3H). CD39 activity was reduced by 55% in Cd39 shRNA-transfected cells compared with cells transfected with the vector alone (Fig. 3H).
FIGURE 3.
Effect of cAMP on macrophage CD39 protein expression. RAW cells were treated with 250 μm 8-Br-cAMP for 16 h, and CD39 protein expression was measured by FACS (Isotype, rat IgG2a isotype control; MFI, mean fluorescence intensity) (A and B) and immunofluorescence staining (C and D). RAW cells were stably transfected with Cd39 shRNA or pRS shRNA control vector. The transfected cells were treated with 250 μm 8-Br-cAMP for 5 h, CD39 protein expression was measured by FACS (E and F), and membrane protein was isolated for Western blot (G). Membrane proteins were also used to perform TLC assays to assess CD39 enzyme activity (H). ***, p < 0.001 compared with non-cAMP-treated controls. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 compared with the cAMP-treated group.
Transcriptional Regulation of Cd39 by cAMP
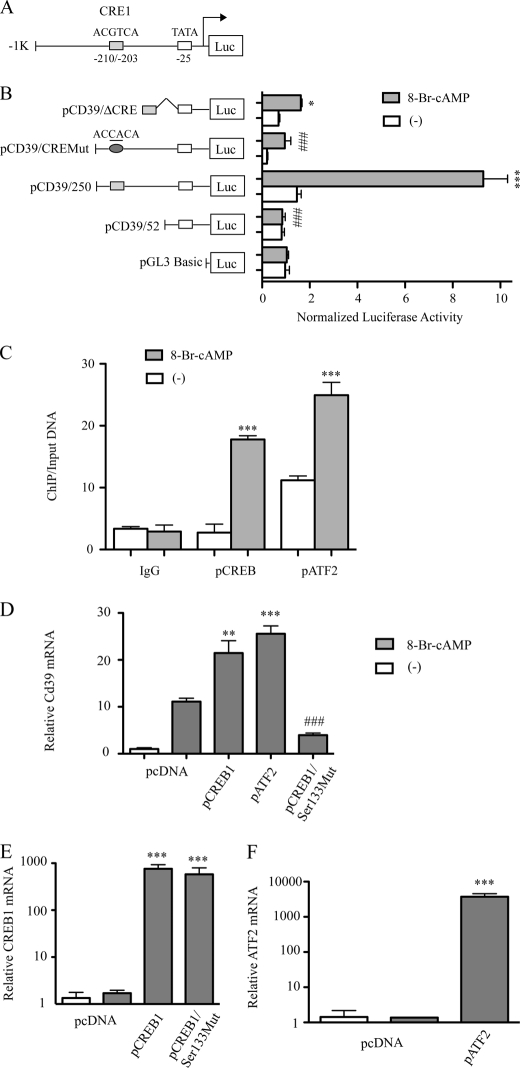
To elucidate mechanisms driving transcription of Cd39 in response to cAMP stimulation, we cloned a 1037-bp genomic fragment that contains 1000 bp of the 5′-flanking region upstream of the mouse Cd39 gene (Fig. 4A) and proceeded to perform a series of promoter truncation and promoter mutation experiments. Note that this cloned version of the native promoter sequence contained a CRE-like motif in reverse orientation at −210 to −203 bp upstream from the transcription start point as well as a TATA box site to which RNA polymerase II typically binds. To delineate promoter elements that regulate Cd39 transcription and may be responsive to cAMP, a series of luciferase reporter gene constructs was generated in a pGL3 basic vector that represents a 5′ deletion of the 5′-flanking region of the Cd39 gene (Fig. 4B). The resulting chimeric constructs (termed pCD39/250 and pCD39/52) were transfected into RAW 264.7 cells to measure their transcriptional rate in the presence or absence of 8-Br-cAMP.
FIGURE 4.
cAMP-induced CD39 expression results from transcriptional activation at CRE-like sites; deletional analysis of the Cd39 promoter. A, schematic of the 5′-flanking region of the mouse Cd39 gene with its CRE-like binding sites. Transient co-transfection of RAW cells was performed using either pCD39/250, pCD39/52, or pCD39/D206; pCD39/CRE1mut; and pCMV/β-galactosidase. Cultures were transfected with each of the indicated constructs using the SuperFect procedure, and then cells were treated with 250 μm 8-Br-cAMP (B). Luciferase activities were then determined with a luciferase reporter assay system. Relative firefly luciferase activity is normalized to control pCMV/β-galactosidase activity (***, p < 0.001 compared with non-treatment controls; ###, p < 0.001 compared with pCD39/250-transfected and cAMP-treated groups). C, ChIP of CREB1 or ATF2 interaction with Cd39 promoter in RAW cells. RAW cells were treated with 250 μm 8-Br-cAMP for 30 min. qRT-PCR products targeting −239 to −172 of the Cd39 promoter are shown. Ct values of ChIP DNA were normalized to the Ct values of the input DNA. ***, p < 0.001 compared with non-treatment control. D–F, effect of CREB1 or AFT2 overexpression on Cd39 mRNA expression. RAW cells were transfected either with a pCREB1 overexpression construct, a pATF2 overexpression construct, a dominant negative “phosphorylation-resistant” CREB1 overexpression construct (pCREB/Ser133Mut), or vector alone. After 48 h, transfected cells were treated with 250 μm 8-Br-cAMP for 5 h, total RNA was extracted for reverse transcription, and quantitative PCR was done to evaluate Cd39 (D), Creb1 (E), or Atf2 (F) mRNA levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with non-treated controls. ###, p < 0.001 compared with pCREB1-transfected and 8-Br-cAMP-treated groups.
After treatment with 8-Br-cAMP (250 μm for 6 h), luciferase activity was increased in RAW cells transfected with pCD39/250 (5.94-fold, p < 0.0001) compared with untreated controls (Fig. 4B). When truncation was performed closer (3′) to the transcription start site (at −52 bp), a complete loss of the cAMP effect on transcription was observed. cAMP failed to induce luciferase activity in RAW cells transfected with pCD39/52 plasmids; these treated cells demonstrated low rates of Cd39 transcription no different from those of RAW cells transfected with the same plasmid but incubated without the cAMP analog (Fig. 4B). Together, these truncation construct data indicate that cAMP controls Cd39 transcription in RAW cells by its effects on a segment of the 5′-flanking DNA located between −250 and −52 bp upstream of the transcription start site of the Cd39 gene.
To predict the cAMP-activated transcription factor binding sites between −250 and −52 bp upstream of the transcription start site of the mouse Cd39 gene, we searched for sequences similar to known transcription factor binding sequences. This search revealed a potential transcription factor binding sequence, a CRE, in the −210/−203 region of Cd39 promoter (which we designated CRE1; Fig. 4A).
To determine whether the CRE1 binding sites are functionally important for inducing Cd39 transcription by cAMP, we generated reporter constructs that contained the CRE1 (pCD39/ΔCRE1) binding site fused to a short promoter segment 52 bp upstream of the transcription start site (Fig. 4B). After treatment with 8-Br-cAMP for 6 h, luciferase activity was increased in RAW cells transfected with pCD39/ΔCRE1 (2.14-fold increase, p < 0.01) compared with untreated control cells (Fig. 4B). We also generated reporter constructs from the −250/+52 DNA segment (pCD39/CREMut) that contained mutated CRE binding sites (Fig. 4B). As a control, a wild-type construct (pCD39/250) was used. After treatment with 8-Br-cAMP for 6 h, the RAW cells transfected with mutant CRE site constructs (pCD39/CRE1Mut) exhibited an 86.6% (p < 0.001) decrease in transcriptional activity compared with wild-type constructs (Fig. 4B). These data suggest that the CRE binding site within the Cd39 promoter plays an important role in the transcriptional activation of Cd39 expression by cAMP.
Binding of CREB/ATF to Cd39 Promoter
To demonstrate that CREB1 and ATF2 can bind to the mouse Cd39 promoter in the native macrophage chromatin environment, ChIP assays were performed using RAW cells. RAW cells were treated with 8-Br-cAMP, and cross-linked RAW cell lysates were then immunoprecipitated with anti-phosphorylated CREB1 (Ser133) or anti-phosphorylated ATF2 (Thr71) antibodies. qRT-PCR was used to amplify a 68-bp fragment of the mouse Cd39 promoter (−239 to −172). These ChIP data show that the binding of phosphorylated CREB (Ser133) and phosphorylated ATF2 (Thr71) to the mouse Cd39 promoter was increased 6.48- and 2.3-fold, respectively, in 8-Br-cAMP-treated cells compared with the non-treated control cells (Fig. 4C; both p < 0.0001). These results indicate that the transcriptional increase of Cd39 observed in 8-Br-cAMP-treated RAW cells is associated with the binding of phosphorylated CREB and phosphorylated ATF2 to the CRE binding site in authentic cellular chromatin.
To characterize the effect that phosphorylation of CREB (Ser133) had on Cd39 mRNA levels in macrophages, RAW cells were transfected with a pCREB1 overexpression construct or one in which the phosphorylatable amino acid (Ser133) in CREB was mutated to a non-phosphorylatable amino acid (Ala133). Transfection of RAW cells was then performed with vector alone, pCREB1 overexpression construct, or this dominant negative “phosphorylation-resistant” CREB overexpression construct (pCREB/Ser133Mut). After 48 h, transfected cells were treated with 8-Br-cAMP, and then total RNA was extracted for reverse transcription and quantitative PCR analysis, which was performed to evaluate levels of Cd39 mRNA (Fig. 4D) and Creb1 mRNA levels measured (Fig. 4E). Levels of Cd39 mRNA, which were markedly increased (11.1-fold, p < 0.01) by cAMP treatment of empty vector transfectants (Fig. 4D), were increased yet further in pCREB1/cAMP-treated cells. In sharp contrast, Cd39 mRNA levels decreased 82% in pCREB/Ser133Mut-transfected cells despite concurrent cAMP treatment compared with pCREB-transfected/cAMP-treated cells (p < 0.001, Fig. 4D). In pCREB1-transfected or pCREB/Ser133Mut-transfected cells, Creb1 mRNA expression was increased more than 1000-fold compared with controls (p < 0.0001; Fig. 4E).
These experiments show that the transfection strategy was equally efficacious between the native pCREB1 (Ser133) and the mutated CREB1 (Ala133) transfectants, suggesting that phosphorylation status drives the differences in Cd39 transcription. These data show that the ATF-2 transfection strategy was highly efficacious. When all of these transfection data are considered together, they show that cAMP stimulates CREB and that Ser133 represents the critical phosphorylatable residue in CREB that drives Cd39 transcription. To assess transfection efficiency in experiments in which pATF-2 overexpression constructs were employed, relative Cd39 mRNA and ATF-2 mRNA levels were measured after cAMP treatment. ATF-2 has a role in the response of CD39 to cAMP, similar to that of CREB (Fig. 4, D and F), which is not surprising given their functional homology in the same transcription factor family.
Intracellular Second Messenger Signaling Effects on cAMP-induced Macrophage CD39 Expression
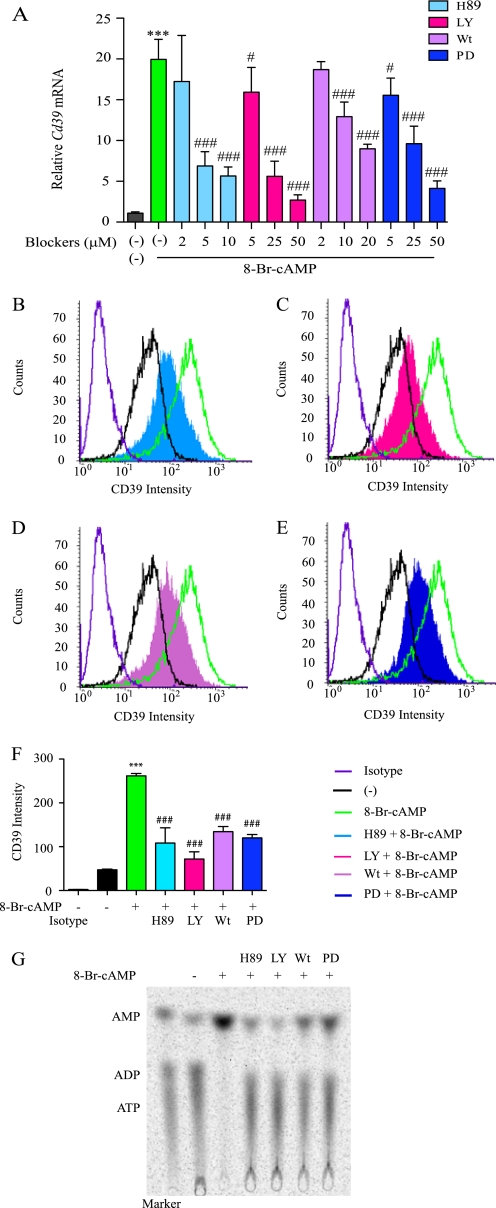
Because the cAMP analogue 8-Br-cAMP was a potent inducer of both CD39 antigen expression and CD39 activity levels, three intracellular second messenger-dependent signaling kinase cascades were examined; PKA, PI3K, and ERKs (27, 28). RAW cells were pretreated with different dosages of the PKA inhibitor H89 (10, 5, and 2 μm), the PI3K inhibitor LY294002 (50, 25, and 5 μm), wortmannin (20, 10, and 2 μm), or the ERK inhibitor PD98059 (50, 25, and 5 μm) for 20 min and then treated with 250 μm 8-Br-cAMP for 4 h. Cd39 mRNA levels were measured by quantitative real time RT-PCR. The results show that Cd39 mRNA was increased 20-fold in 8-Br-cAMP-treated cells in comparison with non-treated cells. When comparing Cd39 mRNA levels of those cells treated with 8-Br-cAMP alone with those cells treated with a combination of 8-Br-cAMP and either 10 or 5 μm H89, we observed that Cd39 mRNA induction was reduced to 71 and 65% respectively (p < 0.0001). The addition of either 50 or 25 μm LY294002 reduced Cd39 mRNA induction to 86 and 71%, respectively, compared with 8-Br-cAMP alone (p < 0.0001). Similarly, 20 and 10 μm wortmannin reduced Cd39 mRNA induction to 55 and 35%, respectively (p < 0.001). Finally, 50 and 25 μm PD98059 reduced Cd39 mRNA induction to 79 and 51%, respectively (p < 0.0001) (Fig. 5A).
FIGURE 5.
Intracellular second messenger signaling effects on macrophage CD39 expression. A, RAW cells were treated with H89, LY294002 (LY), wortmannin (Wt), or PD98059 (PD) for 20 min and then treated with 250 μm 8-Br-cAMP for 4 h. Cd39 mRNA levels were then measured using qRT-PCR. B–F, RAW cells were pretreated with 20 μm H-89, 50 μm LY294002, 20 μm wortmannin, or 50 μm PD98059 for 20 min and then treated with 250 μm 8-Br-cAMP for 16 h. CD39 protein expression was then detected by FACS. MFI, mean fluorescence intensity. ***, p < 0.001 compared with non-treated controls. ##, p < 0.01; ###, p < 0.001 compared with 8-Br-cAMP-treated group. G, after 16 h of 8-Br-cAMP treatment, RAW cell membrane protein was extracted, and CD39 enzymatic activity was measured by a TLC assay.
To measure the CD39 protein expression changes effected by PKA, PI3K, or ERK, RAW cells were pretreated with PKA inhibitor, H89 (10 μm), PI3K inhibitor LY294002 (50 μm) or wortmannin (20 μm), or ERK inhibitor PD98059 (50 μm) for 20 min and then treated with 250 μm 8-Br-cAMP for 16 h, followed by CD39 quantification by FACS and Western blot. The data show (Fig. 5, B–F) clearly that CD39 protein levels increase on the membrane of cells treated with 8-Br-cAMP alone and do not increase in other conditions (DMSO alone or 8-Br-cAMP with H89, LY294002, wortmannin, or PD98059). CD39 protein function was next tested by a TLC assay. The results show that H89, LY294002, wortmannin, and PD98059 all block cAMP-induced CD39 activity on the cell membrane (Fig. 5G). These data indicate, therefore, that PKA, PI3K, and ERK are necessary for the 8-Br-cAMP-stimulated increase in Cd39 mRNA and protein expression.
Intracellular Second Messenger Signaling Effects on Transcriptional Regulation of Cd39 by cAMP
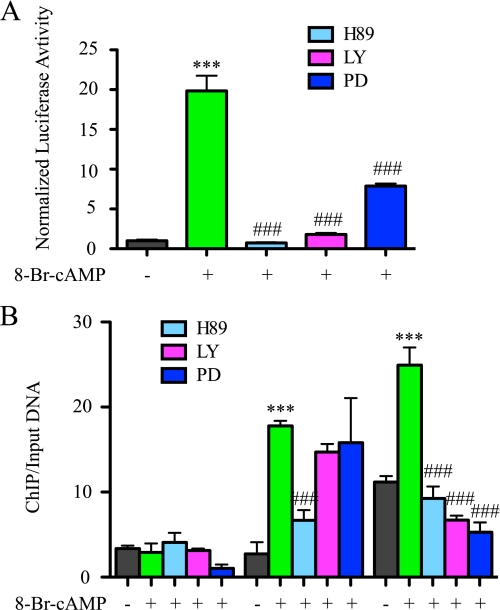
To characterize the mechanism(s) by which PKA, PI3K, and ERK augment CD39 expression, a Cd39 promoter-luciferase construct (pCD39/1K) was transfected into RAW 264.7 cells to measure the effect of PKA, PI3K, and ERK on Cd39 gene transcription. For these experiments, an inhibitor of either PKA (H89, 20 μm), PI3K (LY294002, 50 μm), or ERK (PD98059, 50 μm) was added 24 h following transfection, 20 min after which a cAMP analogue (250 μm 8-Br-cAMP) was added. The experiment was designed such that the downstream signaling cascades of cAMP would be blocked regardless of the presence of cAMP. When examined after 7 h, cells treated with 8-Br-cAMP alone demonstrated a 19-fold increase in the transcriptional activity of the Cd39 promoter compared with vehicle-treated controls (p < 0.001) (Fig. 6A). When pretreated with a combination of 8-Br-cAMP and either H89, LY294002, or PD98059, luciferase activity decreased by 96% (p < 0.0001), 90% (p < 0.0001), and 60% (p < 0.0001), respectively, compared with 8-Br-cAMP treatment alone (Fig. 6A). These data confirm the requirement of intact intracellular signaling cascades in order to transduce the cAMP signal increased expression of CD39.
FIGURE 6.
Intracellular second messenger signaling effects on the transcriptional activation of CRE-like sites. A, RAW cells were co-transfected with pCD39/1K and pCMV/β-galactosidase. After 24 h of transfection, 20 μm H89, 50 μm LY294002 (LY), or 50 μm PD98059 (PD) was added. After 20 min, 250 μm 8-Br-cAMP was added. Cells were lysed after 7 h of treatment, and luciferase reporter assays were performed. B, ChIP of CREB1 or ATF2 binding of the Cd39 promoter in RAW cells. RAW cells were treated with 20 μm H89, 50 μm LY294002, or 50 μm PD98059 for 20 min and then treated with 250 μm 8-Br-cAMP for 30 min. qRT-PCR products targeting the Cd39 promoter (−239 to −172) are shown. Ct values of ChIP DNA were normalized to the Ct values of input DNA. ***, p < 0.01 compared with non-treated control; ###, p < 0.001 compared with the 8-Br-cAMP-treated group.
To characterize the mechanism(s) by which PKA, PI3K, and ERK affect the ability of CREB1 and ATF2 to bind to the mouse Cd39 promoter in a macrophage chromatin environment, ChIP assays were performed using RAW cells. RAW cells were pretreated with 20 μm H89, 50 μm LY294002, and 50 μm PD98059 for 20 min and then incubated with 250 μm 8-Br-cAMP for 30 min. Cross-linked RAW cell lysates were then immunoprecipitated with anti-phosphorylated CREB1 (Ser133) or anti-phosphorylated ATF2 (Thr71) antibodies, and qRT-PCR was used to amplify a 68-bp fragment of mouse Cd39 promoter (−239 to −172). These ChIP data show that the binding of phosphorylated CREB to the CRE binding site decreases 62% (p < 0.0001) in cells treated with both H89 and 8-Br-cAMP, compared with cells that only received 8-Br-cAMP. The binding of phosphorylated ATF2 to the CRE binding site decreased 62% (p < 0.0001), 72% (p < 0.0001), and 79% (p < 0.0001) in cells treated with 8-Br-cAMP and either H89, LY294002, or PD98059, compared with cells that were only treated with 8-Br-cAMP (Fig. 6B). These results indicate that the binding of phosphorylated CREB and phosphorylated ATF2 to the CRE binding site in 8-Br-cAMP-treated RAW cells closely involves the activities of PKA, PI3K, and ERK.
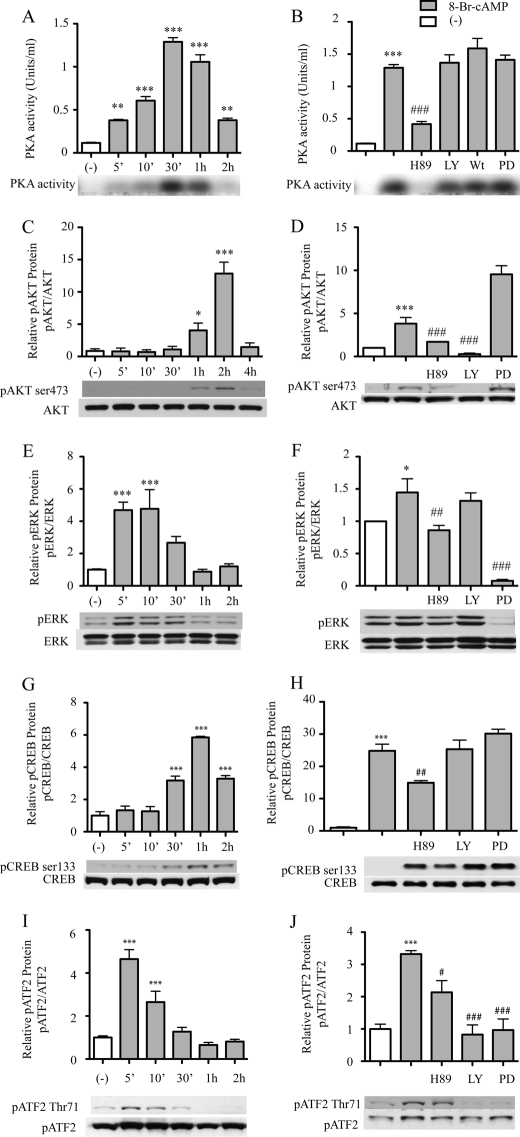
Effect of cAMP on Activation of PKA, PI3K, and ERK in Macrophages
Because inhibiting the activity of PKA, PI3K, and ERK led to a decrease in cAMP-stimulated CD39 expression on macrophages, we aimed to determine whether cAMP regulates PKA activity and the phosphorylation-dependent activation of Akt and ERK. To measure PKA activity, RAW cells were treated with H89, LY294002, wortmannin, or PD98059 for 20 min, after which, PKA activity was measured using the PepTag® non-radioactive protein kinase assay kit. The data showed that cAMP increased the PKA activity of RAW cell in a time-dependent manner (Fig. 7A). After 5 min of 8-Br-cAMP treatment, PKA activity was increased 3.27-fold compared with the non-treated control group (p < 0.01), with maximal activation at 30 min, at which time there was an 11-fold increase compared with non-treated controls (p < 0.0001). Pretreatment with the PKA inhibitor H89 blocked cAMP induced PKA activity at the 30 min time point by 68% compared with those cells treated with cAMP alone (p < 0.0001). PKA activity was not, however, blocked by the PI3K inhibitors LY294002 and wortmannin or by the ERK inhibitor PD98059 (Fig. 7A). Western blot analyses revealed that the phosphorylation of Akt and ERK also occurred in a time-dependent manner. Compared with the non-treated control, phosphorylation of Akt was increased 4.67-fold after 1 h of cAMP treatment (p < 0.05) and peaked at 2 h after cAMP treatment, when there was a 14-fold increase (p < 0.0001; Fig. 7B). cAMP-induced phosphorylation of Akt was reduced 93% by pretreatment with the PI3K inhibitor LY294002, compared with the cells with only cAMP treatment. Pretreatment with the PKA inhibitor H89 partially (55%, p < 0.05) blocked the cAMP-induced phosphorylation of Akt compared with the peak level Akt phosphorylation. The ERK inhibitor PD98059 did not block cAMP-induced phosphorylation of Akt (Fig. 7D). The maximum phosphorylation of ERK was seen at 5 min after cAMP treatment, at which point there was a 4.7-fold increase compared with the non-treated control (Fig. 7D, p < 0.001). Compared with this maximum, co-treatment with PD98059 or H89 reduced phosphorylation by 94.6% (p < 0.0001) and 40.5% (p < 0.01), respectively; LY294002 treatment, however, did not show a reduction (Fig. 7E).
FIGURE 7.
Effect of PKA activity and phosphorylation of CREB/ATF on macrophage CD39 expression. A and B, RAW cells were pretreated with 20 μm H89, 50 μm LY294002 (LY), 20 μm wortmannin (Wt), or 50 μm PD98059 (PD) for 20 min and then treated with 1 μm 8-Br-cAMP for the indicated time, after which the cell lysates were used to measure PKA activity. C–J, RAW cells were pretreated with 20 μm H89, 50 μm LY294002, or 50 μm PD for 20 min and then treated with 250 μm 8-Br-cAMP for the indicated times. Total protein was extracted, and phospho-AKT (pAKT; Ser473), total AKT (C and D), phospho-ERK (pERK), total ERK (E and F), phospho-CREB1 (pCREB; Ser133), total CREB1 (G and H), phospho-ATF2 (pATF2; Thr71), or total ATF2 (I and J) were detected by Western blot. *, p < 0.05; ***, p < 0.001 compared with non-treated controls. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 compared with the 8-Br-cAMP-treated group.
Effect of Phosphorylation of CREB/ATF on Macrophage CD39 Expression
Phosphorylation of CREB and ATF by cAMP may drive Cd39 transcription. To detect an effect of cAMP on phosphorylation status of CREB and ATF, nuclear extracts were obtained from RAW cells treated with 8-Br-cAMP with or without co-treatment with either H89, LY294002, or PD98059. Extracted proteins were then immunoblotted with anti-phospho-CREB1 (Ser133) or anti-phospho-ATF2 (Thr71) antibodies. The results show that cAMP induces CREB1 and ATF2 phosphorylation in a time-dependent manner. Phosphorylated CREB1 was increased 3.17-fold after 30 min of cAMP treatment, with maximal activation at 1 h when there was a 5.85-fold increase compared with non-treated controls (both p < 0.0001; Fig. 7, F and G).
Furthermore, this phosphorylation is blocked by H89 but not by LY294002, wortmannin, or PD98059 (Fig. 7G). Phosphorylation of ATF-1 (detected with the anti-CREB1 phosphospecific antibody) paralleled phospho-CREB-1 status, with up-regulation by cAMP and suppression by PKA blockade. In contrast, although phospho-ATF2 was also induced by cAMP, its phosphorylation appeared to be diminished by PKA, PI3K, or ERK blockade (Fig. 7, H and I). These results demonstrated that the cAMP effect on CREB1 (Ser133) phosphorylation involves only PKA, whereas the cAMP effect on ATF2 (Thr71) phosphorylation involves PKA, PI3K, and ERK.
Effect of cAMP on Cd39 mRNA Expression in Murine Peritoneal Macrophages
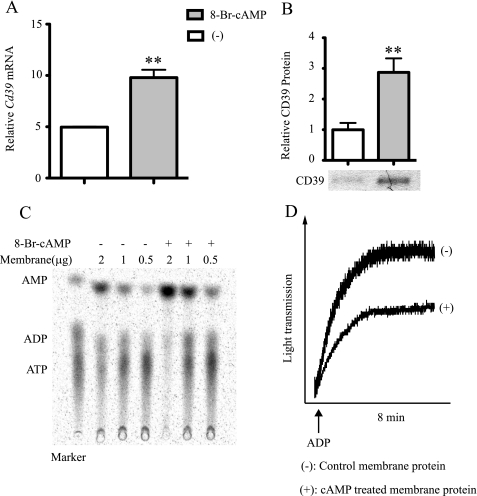
To verify that the cAMP-driven Cd39 mRNA and protein expression and the subsequently increased enzymatic activity of CD39 observed in RAW cells was not cell line-dependent, primary murine peritoneal macrophages were isolated. Peritoneal macrophages were treated with 8-Br-cAMP, and total RNA and membrane protein were isolated. From these samples, Cd39 mRNA levels were measured using qRT-PCR. The data showed that in cAMP-treated cells, Cd39 mRNA increased by 2-fold, relative to non-cAMP-treated controls (p < 0.01, Fig. 8A). We then measured CD39 expression on the cell membrane by Western blot. cAMP increased CD39 protein expression by 2.87-fold compared with the non-treated control (p < 0.01). The functional enzymatic activity of CD39 on peritoneal macrophages was measured using a TLC assay and a platelet aggregation assay. The result showed that the conversion of ATP to AMP was increased by 2.5-fold and that platelet aggregation was reduced by 46% in 8-Br-cAMP-treated macrophages compared with non-treated controls (Fig. 8, C and D). These primary cell data confirm the results obtained from the cAMP-treated RAW cells and show that, indeed, cAMP strongly induces Cd39 expression in primary murine macrophages.
FIGURE 8.
Effect of cAMP on CD39 expression in primary macrophage cultures. 10-week-old male C57Bl6/J mice were injected intraperitoneally with 3.0 ml of a 5% thioglycollate solution. Four days later, macrophages were isolated by peritoneal lavage, red blood cells were lysed, and remaining cells were plated. After 24 h, the recovered cells were pretreated with 20 μm H89, 50 μm LY294002, or DSMO (vehicle) for 20 min. To this medium, 250 μm 8-Br-cAMP was added, after which the cells were allowed to incubate for 2 h. Total RNA was then isolated from the plated murine peritoneal macrophages, and qRT-PCR was used to assay levels of Cd39 mRNA (A). Isolated peritoneal macrophages were treated with 250 8-Br-cAMP for 16 h before membrane protein was extracted. CD39 protein expression was measured by Western blot (B), and CD39 enzymatic activity was tested using a TLC assay (C) and a platelet aggregation assay (D). **, p < 0.01 compared with non-treated controls. ###, p < 0.001 compared with the 8-Br-cAMP-treated group.
DISCUSSION
CD39 functions on the surface of vascular cells to decrease platelet activation and recruitment by metabolizing ATP and ADP released from stimulated platelets (1). The critical nature of its expression is manifest in several murine models of ischemia (including cerebral, renal, myocardial, and intestinal) wherein CD39-deficient mice had aberrant platelet activation and a concomitant increase in ischemic susceptibility compared with littermate controls. CD39 is known to regulate inflammation based on its known properties to modulate cytokine expression, type II inflammatory responses, homotypic cellular adhesion (29–32), and leukocyte trafficking (24). Although CD39 is expressed on the surface of many cell types, including endothelium, macrophages, and lymphocytes, very little is known about the mechanisms responsible for regulating its expression. Here we present a previously unknown mechanism linking expression of CD39 with a ubiquitous intracellular second messenger, which has, among its other properties, the ability to modulate and, in fact, restore vascular homeostasis when vascular cells are subjected to hypoxic or ischemic stress. These data are the first to show that cAMP, acting through stimulation of PKA, drives CREB to bind to the promoter region of Cd39 to induce its transcription. The implications of this finding are quite broad, especially because it provides new insights into a pathway by which endogenous CD39 levels can be modulated to restore vascular homeostasis under various conditions, including ischemic stress.
Because macrophages have been shown to play a critical role in thrombogenesis in response to hypoxic or ischemic insult (33) and CD39 is itself a major endogenous regulator of blood fluidity and anticoagulation, the current experiments focused on transcriptional regulation of CD39 expression in macrophages. Because CD39 is a critical modulator of vascular homeostasis, and mechanisms regulating expression of this gene in all cell types, including macrophages, are poorly understood, the current studies examined the Cd39 promoter region to elicit any clues into its regulation. Study of this region revealed a proximal site in the Cd39 promoter, which closely matched a known CRE consensus motif. The work presented here provides compelling evidence that not only does cAMP transduce its signal through this sequence element, but also this interaction is functionally important in both the transcriptional regulation and the ultimate activity of Cd39. Lines of proof include the fact that a cAMP analogue (8-Br-cAMP) rapidly and markedly induces macrophage Cd39 mRNA (by more than 40-fold). Concordant with these data, CD39 antigen and activity levels were strongly up-regulated by cAMP, an up-regulation that was diminished by transfection of cells with Cd39 shRNA. The up-regulation of Cd39 by 8-Br-cAMP was both exposure time- and dose-dependent. Furthermore, results from both the TLC and platelet aggregation assays demonstrated a marked increase in CD39 activity with 8-Br-cAMP treatment. As a whole, these data show that cAMP elicits a brisk and profound increase in Cd39 mRNA, protein expression, and enzymatic function. In the current work, signaling mechanisms underlying transcriptional activation of the Cd39 gene by cAMP were also elucidated, showing that PKA, PI3K/AKT, ERK, and CREB/ATF signaling pathways contribute to Cd39 gene transcription, in both an independent and potentially cooperative manner (Fig. 9).
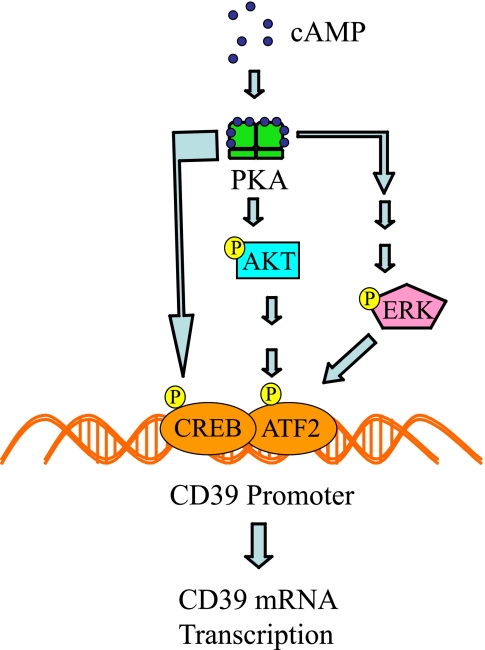
FIGURE 9.
cAMP signaling promotes transcription of mouse Cd39. Shown is a scheme outlining the effects of cAMP-PKA activity on the phosphorylation of CREB1; activation of the PI3K/AKT and ERK pathways (27) leads to the phosphorylation of ATF2. Phosphorylated CREB and ATF2 bind the Cd39 promoter to drive the transcription of Cd39.
Cyclic PKAs are ubiquitous signaling molecules that mediate extracellular signals in eukaryotes. The PKA holoenzyme is a heterotetramer composed of a regulatory subunit homodimer and two associated catalytic subunits. Cooperative binding of two cAMP molecules to each regulatory subunit results in dissociation and consequent activation of the catalytic subunits. These active subunits are then able to phosphorylate target proteins (such as CREB/ATF) on serine or threonine residues. Once phosphorylated, target proteins are activated to either promote or suppress downstream mRNA transcription. In this study, cAMP increased PKA activity, which was only blocked by pretreatment with a PKA inhibitor, H89, but not affected by PI3K or ERK inhibitors. The PKA inhibitor H89 was also employed to test the role of PKA in cAMP-mediated CD39 induction. CD39 protein expression was reduced following pretreatment of cells with H89 compared with cells treated with 8-Br-cAMP alone. Further, pretreatment of cells with H89 blocked 98% of the 8-Br-cAMP-induced Cd39 mRNA expression. Cells transfected with a luciferase reporter linked to the first kilobase of the Cd39 promoter (pCD39/250) lost 65% of cAMP-induced luciferase activity when pretreated with H-89. The results of our ChIP assays showed that H89 did block the binding of phosphorylated CREB (Ser133) and ATF2 (Thr71) to the CRE-like binding site in the Cd39 promoter. Because transcriptional up-regulation could be abrogated by transfection into macrophages of a Creb overexpression mutant construct encoding a nonphosphorylatable residue at the site of Ser133, this provides further evidence for the criticality of interaction between phosphorylated CREB and the CRE-like promoter motif in driving cAMP-mediated Cd39 up-regulation. All of these data demonstrate that cAMP activates PKA phosphorylative activity and that CREB/ATF, when phosphorylated by PKA, increases Cd39 mRNA transcription. Use of the ChIP assay in these experiments increases the likelihood that the observed transcriptional regulation of Cd39 by cAMP in the in vitro promoter construct experiments is biologically relevant in the actual chromatin environment in living cell nuclei.
Activation of second messenger pathways triggers multiple tertiary or amplifying pathways to effect a biological outcome. For the studies shown in this paper, a hypothetical schema was developed, with supporting evidence, in which cAMP activation of PKA leads to downstream activation of other kinases, such as PI3K and ERK (28, 34). PI3K, a heterodimeric enzyme composed of one 110-kDa catalytic subunit and one 85-kDa regulatory subunit, serves as a major signaling component downstream of growth factor receptor tyrosine kinases (35). The PI3K subunit p85α (serine 83) is phosphorylated in vivo and in vitro by PKA (27). PI3K catalyzes the production of the lipid secondary messenger phosphatidylinositol 3,4,5-triphosphate, which in turn activates a wide range of its own downstream targets, including the serine/threonine kinase Akt (36). The PI3K-Akt pathway regulates multiple cellular processes, including cell proliferation, survival, growth, and motility (35). In this study, we found that phosphorylated Akt expression increased in 8-Br-cAMP-treated RAW cells and was reduced by the PKA inhibitor H89. In addition, the PI3K inhibitor LY294002 blocked 97% of the 8-Br-cAMP-induced Cd39 mRNA and protein expression, a reduction confirmed as well in its ability to inhibit cAMP-driven luciferase activity in pCD39/1K-transfected cells. Further confirmation of the important role for PI3K comes from use of an alternate inhibitor of this pathway, wortmannin. Additional ChIP data demonstrated that cAMP activates PI3K and increases interactions of phosphorylated ATF2 with the promoter of Cd39, driving mRNA transcription.
The ERKs are serine/threonine protein kinases. ERKs phosphorylate many substrates, thereby regulating numerous cellular functions, such as gene expression, metabolism, and morphology. Consequently, ERK signaling plays an important role in neuronal development and memory, cardiac muscle hypertrophy, apoptosis, and cancer (37). The ERK cascade is a target of the cAMP- and PKA-mediated regulation of the Ras/Raf/MEK/ERK pathway and can be activated or inhibited by cAMP in a cell-specific manner (38, 39). In cells where cAMP stimulates ERK activity, cAMP activates PKA, which activates Rap1 in a PKA-dependent manner, which then stimulates B-Raf/ERK signaling (34). In this study, we found that 8-Br-cAMP-induced ERK phosphorylation and that PD98059, an ERK inhibitor, blocked 86.8% of the 8-Br-cAMP-induced Cd39 mRNA and protein expression. The PKA inhibitor H89 also blocked 8-Br-cAMP-induced ERK phosphorylation. These reductions were then confirmed by an 8-Br-cAMP-driven promoter-luciferase assay showing a loss of luciferase activity with inhibitor treatment in cells transfected with pCD39/1K. Additional ChIP data demonstrated that 8-Br-cAMP directly activates ERK and increases interactions between phosphorylated ATF2 and the Cd39 promoter, thus driving transcription. PD98059 also reduced the 8-Br-cAMP-stimulated increase in the phosphohydrolytic activity of CD39 to convert ATP to AMP, which supports the role of ERK as a necessary part of the pathway initiated by cAMP toward the regulation of CD39.
The CREB/ATF family of transcription factors resides within the bZIP superfamily, whose members are defined by their ability to heterodimerize with each other (40). Hagiwara et al. (41) showed, using quantitative analysis of nuclear extracts, that CREB is expressed at about 50,000 molecules/cell, with the majority bound to chromatin. CREB may constitutively bind to the target gene's CRE site (TGACGTCA) (40, 42, 43). Up to half of all of cellular CREB protein is phosphorylated at the Ser133 position in response to maximal cAMP stimulation (41). It is possible and perhaps even likely that if basal cAMP levels in cells could drive some level of phosphorylated CREB binding to CRE elements, thus resulting in the tonic low level expression of CD39 in the absence of cAMP analogue treatment. The phosphorylation of CREB at Ser133 results in the recruitment of CREB to bind CREB-binding protein (44), which leads to the activation of RNA polymerase II to initiate the transcription of target genes (45, 46), such as Egr-1 (early growth response factor-1). The binding of phosphorylated CREB (Ser133) to proximal CRE sites located in the Egr-1 promoter is necessary for the induction of Egr-1 transcription by exendin-4 (47). For example, CRE has been shown to be necessary but not sufficient for the activation of calcitonin gene-related peptide by nerve growth factor (48).
The mere presence of a CRE element in a gene promoter is insufficient to ascribe functional significance to it. For instance, CRE was found to be unimportant in the transcription of rat tyrosine hydrolase (49). ATF2 also binds with high affinity to CRE sites. Under uninduced conditions, the ATF2 activation domain specifically binds to the bZIP DNA binding domain to inhibit ATF2 transcriptional activation. This inhibition of the ATF2 activation domain does not involve bZIP DNA binding (50). Interruption of the binding of ATF2 activation and bZIP domains driving ATF2 transcription activities requires phosphorylation of ATF2 on threonines 69 and 71 that is mediated by the c-Jun N-terminal kinase (JNK), p38/mitogen-activated protein kinase (MAPK), or ERK pathway (51–53). For its transcription program, ATF2 heterodimerizes with other bZIP family members, including Fos, Fra2, c-Jun, CREB, or ATF1. ATF2 heterodimeric partners are also activated in a stimulus-specific manner. Therefore, different stimuli can lead to different ATF2 complexes and thereby activate specific subsets of target genes (54, 55).
By in silico analysis, we noted Cd39 to have a CRE site, but there had not been prior information as to the importance of this site in Cd39 regulation. In the work shown here, we critically evaluated, tested, and confirmed the importance of this CRE site within the mouse Cd39 promoter for CD39 regulation because mutation of this serine 133 residue resulted in the abrogation of the ability of cAMP to drive Cd39 transcription. Furthermore the results of our ChIP analyses showed that the binding of phosphorylated CREB (Ser133) and ATF2 (Thr71) to the CRE element in the Cd39 promoter was increased via the cAMP/PKA pathway, and phosphorylated ATF2 (Thr71) binding to the CRE element was increased via the cAMP/PKA/PI3K or the cAMP/PKA/ERK pathway. Together, our results show that Cd39 expression is up-regulated by the increased activity of these transcriptional complexes.
This work intersects with our previous observations on the role of cAMP in cardiac preservation for transplantation and the maintenance of normal vascular homeostatic mechanisms. Hypoxia and reoxygenation increase endothelial cell permeability, induce procoagulant activity, and alter endothelial cell/leukocyte interactions, with a parallel reduction in endothelial cAMP and nitric oxide levels. Under conditions of oxygen deprivation, cAMP promotes endothelial cell antithrombogenicity, maintenance of barrier function, and inhibition of leukocyte/endothelial cell interactions, although the exact mechanisms remain unclear (56). Our data lead us to hypothesize that enhancement by cAMP of vascular anti-thrombotic properties could be in large measure due to the cAMP-stimulated induction of CD39. Both adenine nucleotide phosphohydrolysis data and platelet aggregation data, demonstrating that cAMP up-regulates CD39 enzyme activity and platelet-inhibitory function, lend credence to this hypothesis. Supporting this, in a rat heterotopic cardiac transplant model, grafts stored with the membrane-permeable cAMP analogues dibutyryl-cAMP or 8-Br-cAMP demonstrated a 5.5-fold increase in blood flow and a 3.2-fold decrease in neutrophil infiltration after transplantation (56). Provision of cAMP also enhances lung preservation in an orthotopic rat left lung transplant model. Furthermore, a cAMP pulse during cardiac preservation reduces the incidence and severity of transplant-associated coronary artery disease (57). Given the protective effects of CD39 in cerebral, myocardial, renal, and intestinal ischemia reperfusion models, our data suggest a potential pharmacological means to bolster endogenous CD39 expression, by provision of a cAMP analogue, phosphodiesterase inhibitors that augment endogenous cAMP levels, or perhaps even another agent that activates CRE sites.
As a whole, this work demonstrates a novel cAMP-dependent mechanism by which CD39 is potently regulated on the surface of macrophages. Furthermore, these data demonstrate that cAMP drives CD39 expression through PKA phosphorylation of CREB and PI3K, ERK, and ATF2 (Fig. 9). Given the ubiquitous nature of cAMP signaling in the vasculature and the critical role CD39 plays in limiting thrombosis, it is likely that cAMP-driven up-regulation of CD39 may be fundamental to the maintenance of vascular homeostasis.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL085149, R01HL086676, R01HL055397, and P01HL089407. This work was also supported by a Taubman Medical Research Institute Scholarship, the Ruth Professorship, and the Scleroderma Research Foundation.
- CRE
- cAMP-response element
- PKA
- cAMP-dependent protein kinase
- ATF
- activating transcription factor
- PI3K
- phosphoinositide 3-kinase
- Akt/PKB
- protein kinase B
- ERK
- extracellular signal-regulated kinase
- bZIP
- basic leucine zipper
- FBS
- fetal bovine serum
- shRNA
- short homologous RNA
- RT
- reverse transcription
- qRT
- quantitative reverse transcription
- ChIP
- chromatin immunoprecipitation
- RQ
- relative quantities
- FACS
- fluorescence-activated cell sorting
- 8-Br-cAMP
- 8-bromo-cyclic AMP.
REFERENCES
- 1.Marcus A. J., Broekman M. J., Drosopoulos J. H., Olson K. E., Islam N., Pinsky D. J., Levi R. (2005) Semin. Thromb. Hemost. 31, 234–246 [DOI] [PubMed] [Google Scholar]
- 2.Marcus A. J., Broekman M. J., Drosopoulos J. H., Islam N., Pinsky D. J., Sesti C., Levi R. (2003) J. Pharmacol. Exp. Ther. 305, 9–16 [DOI] [PubMed] [Google Scholar]
- 3.Marcus A. J., Broekman M. J., Drosopoulos J. H., Islam N., Pinsky D. J., Sesti C., Levi R. (2003) J. Thromb. Haemost. 1, 2497–2509 [DOI] [PubMed] [Google Scholar]
- 4.Atkinson B., Dwyer K., Enjyoji K., Robson S. C. (2006) Blood Cells Mol. Dis. 36, 217–222 [DOI] [PubMed] [Google Scholar]
- 5.Gerlo S., Verdood P., Hooghe-Peters E. L., Kooijman R. (2006) Cell Mol. Life Sci. 63, 92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohler D., Eckle T., Faiglem M., Grenz A., Mittelbronn M., Laucher S., Hart M. L., Robson S. C., Muller C. E., Eltzschig H. K. (2007) Circulation 116, 1784–1794 [DOI] [PubMed] [Google Scholar]
- 7.Marcus A. J., Broekman M. J., Drosopoulos J. H., Pinsky D. J., Islam N., Maliszewsk C. R. (2001) Ital. Heart J. 2, 824–830 [PubMed] [Google Scholar]
- 8.Pinsky D. J., Broekman M. J., Peschon J. J., Stocking K. L., Fujita T., Ramasamy R., Connolly E. S., Jr., Huang J., Kiss S., Zhang Y., Choudhri T. F., McTaggart R. A., Liao H., Drosopoulos J. H., Price V. L., Marcus A. J., Maliszewski C. R. (2002) J. Clin. Invest. 109, 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperlágh B., Mergl Z., Jurányi Z., Vizi E. S., Makara G. B. (1999) J. Endocrinol. 160, 343–350 [DOI] [PubMed] [Google Scholar]
- 10.Montminy M. (1997) Annu. Rev. Biochem. 66, 807–822 [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez G. A., Montminy M. R. (1989) Cell 59, 675–680 [DOI] [PubMed] [Google Scholar]
- 12.Montminy M. R., Sevarino K. A., Wagner J. A., Mandel G., Goodman R. H. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6682–6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr B., Montminy M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto K. K., Gonzalez G. A., Biggs W. H., 3rd, Montminy M. R. (1988) Nature 334, 494–498 [DOI] [PubMed] [Google Scholar]
- 15.Lee K. A., Masson N. (1993) Biochim. Biophys. Acta 1174, 221–233 [DOI] [PubMed] [Google Scholar]
- 16.Falvo J. V., Parekh B. S., Lin C. H., Fraenkel E., Maniatis T. (2000) Mol. Cell. Biol. 20, 4814–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura T., Okuyama S., Okamoto S., Nakajima T., Sekiya S., Oda K. (1995) Exp. Cell Res. 216, 422–430 [DOI] [PubMed] [Google Scholar]
- 18.Tsai E. Y., Yie J., Thanos D., Goldfeld A. E. (1996) Mol. Cell. Biol. 16, 5232–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P., Angel P. (1995) EMBO J. 14, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S., Campbell D., Dérijard B., Davis R. J. (1995) Science 267, 389–393 [DOI] [PubMed] [Google Scholar]
- 21.Liao H., Hyman M. C., Lawrence D. A., Pinsky D. J. (2007) FASEB J. 21, 935–949 [DOI] [PubMed] [Google Scholar]
- 22.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 23.Schmittgen T. D., Zakrajsek B. A., Mills A. G., Gorn V., Singer M. J., Reed M. W. (2000) Anal. Biochem. 285, 194–204 [DOI] [PubMed] [Google Scholar]
- 24.Hyman M. C., Petrovic-Djergovic D., Visovatti S. H., Liao H., Yanamadala S., Bouïs D., Su E. J., Lawrence D. A., Broekman M. J., Marcus A. J., Pinsky D. J. (2009) J. Clin. Invest. 119, 1136–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujino H., Salvi S., Regan J. W. (2005) Mol. Pharmacol. 68, 251–259 [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Narayan V. M., Juedes N., Patel J. M. (2009) Int. J. Clin. Exp. Med. 2, 87–94 [PMC free article] [PubMed] [Google Scholar]
- 27.Cosentino C., Di Domenico M., Porcellini A., Cuozzo C., De Gregorio G., Santillo M. R., Agnese S., Di Stasio R., Feliciello A., Migliaccio A., Avvedimento E. V. (2007) Oncogene 26, 2095–2103 [DOI] [PubMed] [Google Scholar]
- 28.De Gregorio G., Coppa A., Cosentino C., Ucci S., Messina S., Nicolussi A., D'Inzeo S., Di Pardo A., Avvedimento E. V., Porcellini A. (2007) Oncogene 26, 2039–2047 [DOI] [PubMed] [Google Scholar]
- 29.Kansas G. S., Wood G. S., Tedder T. F. (1991) J. Immunol. 146, 2235–2244 [PubMed] [Google Scholar]
- 30.Mizumoto N., Kumamoto T., Robson S. C., Sévigny J., Matsue H., Enjyoji K., Takashima A. (2002) Nat. Med. 8, 358–365 [DOI] [PubMed] [Google Scholar]
- 31.Langston H. P., Ke Y., Gewirtz A. T., Dombrowski K. E., Kapp J. A. (2003) J. Immunol. 170, 2962–2970 [DOI] [PubMed] [Google Scholar]
- 32.Dombrowski K. E., Ke Y., Brewer K. A., Kapp J. A. (1998) Immunol. Rev. 161, 111–118 [DOI] [PubMed] [Google Scholar]
- 33.Lawson C. A., Yan S. D., Yan S. F., Liao H., Zhou Y. S., Sobel J., Kisiel W., Stern D. M., Pinsky D. J. (1997) J. Clin. Invest. 99, 1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vuchak L. A., Tsygankova O., Prendergast G., Meinkoth J. (2009) Mol. Pharmacol. 76, 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen K., Iribarren P., Gong W., Wang J. M. (2005) Cell Mol. Immunol. 2, 241–252 [PubMed] [Google Scholar]
- 36.Carnero A., Blanco-Aparicio C., Renner O., Link W., Leal J. F. (2008) Curr. Cancer Drug Targets 8, 187–198 [DOI] [PubMed] [Google Scholar]
- 37.Shaul Y. D., Seger R. (2007) Biochim. Biophys. Acta 1773, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 38.Stork P. J., Schmitt J. M. (2002) Trends Cell Biol. 12, 258–266 [DOI] [PubMed] [Google Scholar]
- 39.Dumaz N., Marais R. (2005) FEBS J. 272, 3491–3504 [DOI] [PubMed] [Google Scholar]
- 40.Meyer T. E., Habener J. F. (1993) Endocr. Rev. 14, 269–290 [DOI] [PubMed] [Google Scholar]
- 41.Hagiwara M., Brindle P., Harootunian A., Armstrong R., Rivier J., Vale W., Tsien R., Montminy M. R. (1993) Mol. Cell. Biol. 13, 4852–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cha-Molstad H., Keller D. M., Yochum G. S., Impey S., Goodman R. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13572–13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaywitz A. J., Greenberg M. E. (1999) Annu. Rev. Biochem. 68, 821–861 [DOI] [PubMed] [Google Scholar]
- 44.Chrivia J. C., Kwok R. P., Lamb N., Hagiwara M., Montminy M. R., Goodman R. H. (1993) Nature 365, 855–859 [DOI] [PubMed] [Google Scholar]
- 45.Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. (1994) Nature 370, 223–226 [DOI] [PubMed] [Google Scholar]
- 46.Ferreri K., Gill G., Montminy M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1210–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang J. H., Kim M. J., Jang H. I., Koh K. H., Yum K. S., Rhie D. J., Yoon S. H., Hahn S. J., Kim M. S., Jo Y. H. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E215–E222 [DOI] [PubMed] [Google Scholar]
- 48.Freeland K., Liu Y. Z., Latchman D. S. (2000) Biochem. J. 345, 233–238 [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon S. O., Chikaraishi D. M. (1992) Neuron 9, 55–67 [DOI] [PubMed] [Google Scholar]
- 50.Li X. Y., Green M. R. (1996) Genes Dev. 10, 517–527 [DOI] [PubMed] [Google Scholar]
- 51.Morton S., Davis R. J., Cohen P. (2004) FEBS Lett. 572, 177–183 [DOI] [PubMed] [Google Scholar]
- 52.Buschmann T., Yin Z., Bhoumik A., Ronai Z. (2000) J. Biol. Chem. 275, 16590–16596 [DOI] [PubMed] [Google Scholar]
- 53.Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., Davis R. J. (1996) Mol. Cell. Biol. 16, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kerppola T. K., Curran T. (1993) Mol. Cell. Biol. 13, 5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dam H., Castellazzi M. (2001) Oncogene 20, 2453–2464 [DOI] [PubMed] [Google Scholar]
- 56.Pinsky D., Oz M., Liao H., Morris S., Brett J., Sciacca R., Karakurum M., Van Lookeren Campagne M., Platt J., Nowygrod R. (1993) J. Clin. Invest. 92, 2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C. Y., Aronson I., Takuma S., Homma S., Naka Y., Alshafie T., Brovkovych V., Malinski T., Oz M. C., Pinsky D. J. (2000) Circ. Res. 86, 982–988 [DOI] [PubMed] [Google Scholar]