Abstract
PIASy is a small ubiquitin-related modifier (SUMO) ligase that modifies chromosomal proteins in mitotic Xenopus egg extracts and plays an essential role in mitotic chromosome segregation. We have isolated a novel SUMO-2/3-modified mitotic chromosomal protein and identified it as poly(ADP-ribose) polymerase 1 (PARP1). PARP1 was robustly conjugated to SUMO-2/3 on mitotic chromosomes but not on interphase chromatin. PIASy promotes SUMOylation of PARP1 both in egg extracts and in vitro reconstituted SUMOylation assays. Through tandem mass spectrometry analysis of mitotically SUMOylated PARP1, we identified a residue within the BRCA1 C-terminal domain of PARP1 (lysine 482) as its primary SUMOylation site. Mutation of this residue significantly reduced PARP1 SUMOylation in egg extracts and enhanced the accumulation of species derived from modification of secondary lysine residues in assays using purified components. SUMOylation of PARP1 did not alter in vitro PARP1 enzyme activity, poly-ADP-ribosylation (PARylation), nor did inhibition of SUMOylation of PARP1 alter the accumulation of PARP1 on mitotic chromosomes, suggesting that SUMOylation regulates neither the intrinsic activity of PARP1 nor its localization. However, loss of SUMOylation increased PARP1-dependent PARylation on isolated chromosomes, indicating SUMOylation controls the capacity of PARP1 to modify other chromatin-associated proteins.
Keywords: ADP-ribosylation, Cell/Mitosis, Chromatin, Protein/Post-translational Modification, Sumoylation, PIASy, SUMO
Introduction
Small ubiquitin-related modifiers (SUMOs)3 are proteins found in all eukaryotes that become covalently conjugated to other cellular proteins in a manner similar to ubiquitination (1, 2). This modification (SUMOylation) regulates many aspects of interphase nuclear function and structure (1, 3, 4) as well as events during mitosis (5). Consistent with the important and broad roles of the SUMOylation pathway, hundreds of SUMOylation targets have been identified in proteomic screens (6–9). Three major SUMO family proteins, called SUMO-1, SUMO-2, and SUMO-3, are expressed in vertebrates. SUMO-2 and SUMO-3 are closely related in their primary sequence (95% identical), whereas SUMO-1 is around 45% identical to either of the other two paralogues. In contexts where SUMO-2 cannot be functionally distinguished from SUMO-3, we will refer to these paralogues collectively as SUMO-2/3. All vertebrate paralogues are around 50% identical to the single SUMO protein found in budding yeast, Smt3p (2). SUMO conjugation to cellular substrates is carried out in a manner similar to ubiquitin. The conjugation process requires three sequential enzymes: an activating (E1) enzyme, a conjugating (E2) enzyme, and usually a SUMO ligase (E3 enzyme). All SUMOylation utilizes the same E1 and E2 enzymes, which are called Uba2/Aos1 and Ubc9, respectively. However, there are a number of different E3 enzymes that show a high degree of specificity for particular conjugation targets (1).
All eukaryotes possess E3 enzymes with variant RING-finger like domains (SP-RINGs) (1). These proteins are called Siz (SAP and miz-finger domain) proteins in yeast and PIAS (protein inhibitor of activated STAT) proteins in vertebrates. The five vertebrate PIAS proteins (PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy) are involved in a wide variety of processes, including signal transduction, gene expression, and genome maintenance (10). We have previously shown that PIASy-mediated conjugation of SUMO-2/3 to mitotic chromosomal proteins is essential for proper chromosomal segregation at anaphase in Xenopus egg extract (XEE) cell free assays (11). One of the major targets for PIASy-directed SUMOylation is DNA topoisomerase IIα (TopoIIα) (12). PIASy-deficient mice are viable (13), suggesting PIASy-catalyzed SUMOylation is not essential during anaphase in mice. However, siRNA-mediated PIASy knockdown causes mitotic arrest with aberrant chromosome cohesion in HeLa cells (14). Moreover, the yeast Siz1p and Siz2p proteins are essential for accurate segregation of chromosomes in yeast (15). It, thus, appears that PIASy-mediated SUMOylation plays an essential role in regulating mitotic chromosomal structure and progression of mitosis in many cellular contexts.
Poly-ADP-ribosylation (PARylation) of proteins is another major post-translational modification of many nuclear proteins, catalyzed by poly(ADP-ribose) polymerases (PARPs) (16–18). All PARPs share a conserved catalytic domain that interacts with NAD+, which is the donor molecule of poly(ADP-ribose) (19). The importance of PARylation during mitosis has been demonstrated; inhibition of PARylation compromises spindle formation in XEEs (20). PARylation mediated by the enzyme Tankyrase-1 has been particularly implicated the correct assembly of spindle poles (21). Another PARylation enzyme, PARP1, has been shown to interact with centromeres and could mediate PARylation of centromeric proteins (22, 23). PARP1 interacts with the mitotic chromosomal protein kinase, Aurora B, and could mediate PARylation of Aurora B to regulate its activity (24). Although the mitotic function of PARP1 has not been clearly demonstrated, the increasing evidence that PARP1 plays a role in chromatin structure suggests the potential relevance of PARP1 activity to chromosomal structure and function in mitosis (18, 25).
We identified PARP1 as a SUMO-2/3-modified substrate associated with mitotic chromosomes. PARP1 was robustly conjugated to SUMO-2/3 on mitotic chromatin but not on interphase chromatin. PIASy promoted PARP1 SUMOylation both in XEEs and in vitro SUMOylation assays using purified, recombinant proteins. Through tandem mass spectrometry analysis of mitotically SUMOylated PARP1, we identified a residue within the BRCA1 C-terminal domain of PARP1 (lysine 482) as its primary SUMOylation site. Mutation of this residue significantly reduced PARP1 SUMOylation in XEEs and enhanced the accumulation of species derived from modification of secondary lysine residues in assays using purified components. SUMOylation of PARP1 did not alter PARP1 auto-PARylation activity in vitro, nor did inhibition of SUMOylation of PARP1 alter the accumulation of PARP1 on mitotic chromosomes in XEEs, suggesting that SUMOylation regulates neither the intrinsic activity of PARP1 nor its localization. However, loss of SUMOylation increased PARP1-dependent PARylation on isolated chromosomes, indicating that SUMOylation controls the capacity of PARP1 to modify other chromatin-associated proteins.
EXPERIMENTAL PROCEDURES
Recombinant Protein Expression, Cloning, Site-directed Mutagenesis, and Antibodies
cDNAs of Xenopus PARP1 were cloned from Xenopus tadpole cDNA (kindly provided by T. Amano and Y. B. Shi, NICHD, National Institutes of Health) using PCR amplification with the addition of the BglII recognition sequence at the 5′ end and SalI recognition sequence at the 3′ end. Amplified cDNA was subcloned into pEGFP-C1 plasmids using BglII and SalI sites and was verified by DNA sequencing. The full-length cDNAs were subcloned into pET28 using BamH1 and XhoI restriction sites. cDNA fragments encoding Xenopus PAPR1 (amino acids 1–150 (PARP1xl-N) and 500–650 (PARP1xl-M)) were amplified from a pEGFP-PARP1construct and subcloned into pGEX4T-1 (GE Healthcare) and pET28a (Novagen) using BamHI and XhoI restriction sites. The recombinant proteins encoded by these cDNAs were expressed in Escherichia coli and purified by tag-based purification according to the manufacture's protocol followed by conventional ion-exchange chromatography. The lysine to arginine substitution of PARP1 was produced by site-directed mutagenesis (QuikChange, Stratagene) according to the manufacturer's protocol.
Polyclonal antibodies against PARP1 and TopoIIα were generated in rabbits by injection with recombinant fragments of His6-T7-PAPR1xl-N, His6-T7-PAPR1xl-M, and His6-T7-TopoIIαxl C terminus (amino acids 1359–1579). The rabbit antisera were subjected to affinity purification on NHS-Sepharose (GE Healthcare) columns with their covalently bound antigens. The anti-SUMO-2/3 polyclonal guinea pig antibody was raised and purified similar to previous polyclonal rabbit antibody (12). The anti-PAR monoclonal antibody and anti-T7 tag antibody conjugated with horseradish peroxidase were purchased from Trevigen and EMD Bioscience, respectively. Unless otherwise specified, other reagents were obtained from Sigma.
Protein Purification and in Vitro SUMOylation Assay
The recombinant E1 complex (Aos1/Uba2 heterodimer) was purified by co-expression His6-Aos1 and Uba2 (kindly provided by Dr. F. Melchior) in E. coli as described previously (26). The bacterially expressed E1 complex was purified with Talon affinity resin (Clontech) purification followed by SUMO affinity and conventional chromatography as previously described (27). Purification of recombinant SUMO-2, wild type Ubc9, dnUbc9, and PIASy were as previously described (11, 12).
Recombinant PARP1-wt and PARP1K482R proteins were expressed in BL21(DE3) at 15 °C in the presence of 5% glycerol, 2.5% ethanol, and 0.1 mm ZnCl2. The expressed proteins were purified from the soluble fraction of the bacterial lysate using Talon affinity resin (Clontech) followed by Sephacyryl-300 gel filtration and SP-Sepharose column chromatography (GE Healthcare). Purified PARP1 proteins were concentrated up to 2∼3 mg/ml by centrifugal concentrator (Amicon Ultra-4, 30 kDa, Millipore) then snap-frozen by liquid N2 in small aliquots.
In vitro SUMOylation assay was performed with 15 nm E1, 5 μm SUMO-2, 2.5 mm ATP, and 500 nm PARP1 in reaction buffer (20 mm Tris (pH 7.5), 50 mm NaCl, 10 mm MgCl2, 0.05% Tween 20, and 0.5 mm dithiothreitol) together with the indicated Ubc9 (E2) and PIASy (E3) concentrations. Reactions were incubated at 25 °C for the indicated periods.
Xenopus Egg Extracts
Sperm chromatin and low speed extracts of Xenopus eggs arrested in metaphase by cytostatic factor (CSF) were prepared as described (28). Where required, CSF extracts were driven into interphase by the addition of 0.6 mm CaCl2. For mitotic chromosome assembly or interphase nuclear assembly, demembranated sperm chromatin were added to CSF or interphase extracts and incubated for 40∼60 min at room temperature. Chromatin isolation was performed as previously described (12). Unless otherwise specified, all reactions contained 5000 sperm nuclei per μl.
Immunodepletion and Immunofluorescence
The immunodepletion experiment with XEE was performed as previously described (11) with an affinity-purified anti-PARP1 antibody. For add-back experiments, recombinant PARP1 proteins were added at an endogenous concentration of 4 μg/ml.
The immunofluorescence analysis in Fig. 5C was demonstrated as previously described (11). In brief, 100 μl of CSF extracts were released into interphase by a 0.6 mm CaCl2 addition. 200 sperm/μl were added 5 min after the CaCl2, and the extracts were incubated at 23 °C for 55 min to complete DNA replication. Reentry into mitosis was induced by the addition of 50 μl of fresh CSF extract. In the case of inhibition of mitotic SUMOylation, dnUbc9 (150 ng/μl final concentration) was also added at this point. The reactions were incubated for 40 min after the addition of fresh CSF extract to reach metaphase. Then 50 μl were removed from each reaction, diluted with 200 μl of buffer A (0.8× CSF-XB buffer containing 10 mm β-glycerol phosphate and 250 mm sucrose), and incubated for 0.5 min at 23 °C. 300 μl of buffer A containing 4% paraformaldehyde was added to this sample and incubated for an additional 10 min. Samples were spun onto coverslips through a 35% glycerol cushion. Chromosomal samples on coverslips were post-fixed by 1.6% paraformaldehyde and processed for immunostaining with anti-PARP1 (rabbit polyclonal), anti-SUMO-2/3 (guinea pig polyclonal), and anti-CENP-A (chicken polyclonal). Anti-rabbit Alexa 568, anti-guinea pig Alexa 684, and anti-chicken Alexa 488 were used as secondary antibodies respectively. Specimens were observed by Nikon TE2000-U microscope with Plan Apo 100×/1.40 objective. Images were taken with Retiga SRV CCD camera (QImaging) operated by Volocity software (Improvision).
FIGURE 5.
Inhibition of SUMOylation on mitotic chromosomes alters chromosomal PARylation. A, SUMOylated PARP1 retains robust enzymatic activity of poly-ADP-ribosylation in vitro. An in vitro SUMOylation reaction was performed in the presence of the indicated SUMO-2 proteins as in Fig. 3. After the SUMOylation reaction, NAD+ was added to the reaction mixture, and then the reaction mixture was incubated for the indicated periods. Poly(ADP-ribosyl) chain formation was analyzed by a monoclonal anti-PAR antibody (Trevigen). PARP1 was detected with an anti-T7 antibody. B, SUMOylation affects PARylation on chromosomes prepared from XEE. Mitotic chromosomes were isolated from the extracts that were manipulated as indicated. Isolated chromosomal fractions were incubated with NAD+-containing buffer to promote PARylation on chromosomal proteins, and then the reactions were terminated with the addition of SDS-PAGE sample buffer. The obtained chromosomal samples were analyzed by immunoblotting with the indicated antibodies. The bracket indicates the area of PARylation signals affected by the inhibition of SUMOylation. C, inhibition of SUMOylation does not affect localization of PARP1 on mitotic chromosomes significantly. CSF extracts were released into interphase by CaCl2 addition. 200 sperm/μl were added 5 min after CaCl2, and the extract was incubated at 23 °C for 55 min. Reentry into mitosis was induced by the addition of fresh CSF extract (50% of reaction volume) with or without dnUbc9 (150 ng/μl final concentration). Chromosomes from each reaction were spun onto coverslips and analyzed by immunofluorescence with antibodies against SUMO-2/3, PARP1, and CENP-A as described under “Experimental Procedures.” Each signal was pseudocolored as indicated at the top of the panels. DNA was visualized with Hoechst 33342 staining. Insets are magnified images around the centromere region.
PARylation Assay
To measure PARylation activity of in vitro SUMOylated PARP1, reactions of in vitro SUMOylation were incubated for 60 min, and then the reactions were diluted twice with the reaction buffer (same as in vitro SUMOylation reaction buffer) containing 2 mm NAD+. The reactions were incubated further at 25 °C for indicated periods. Termination of reaction was done by the addition of SDS-PAGE sample buffer to the reaction mixture. For analysis of PARylation of chromosomal proteins, the mitotic chromosomes, assembled in an XEE reaction, were suspended into 30 μl of the reaction buffer containing 1 mm NAD+ and incubated at room temperature for 10 min. The reactions were centrifuged, and chromosomal pellets were boiled in SDS-PAGE sample buffer. PARylation of proteins in SDS-PAGE samples were determined by immunoblotting with anti-PAR monoclonal or polyclonal antibodies (Trevigen).
RESULTS
PIASy Mediates SUMO-2/3 Conjugation of PARP1 on Mitotic Chromosomes
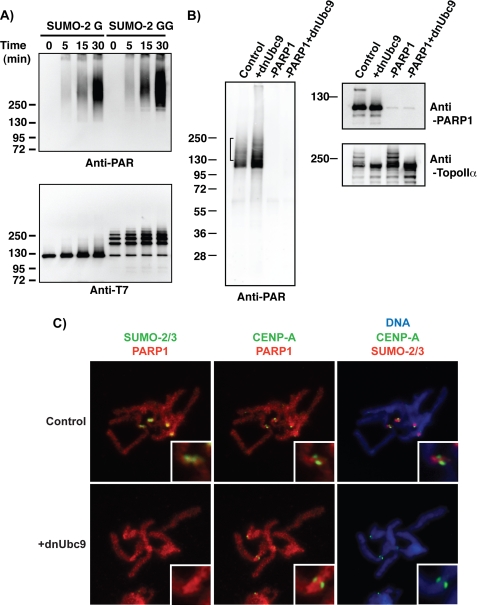
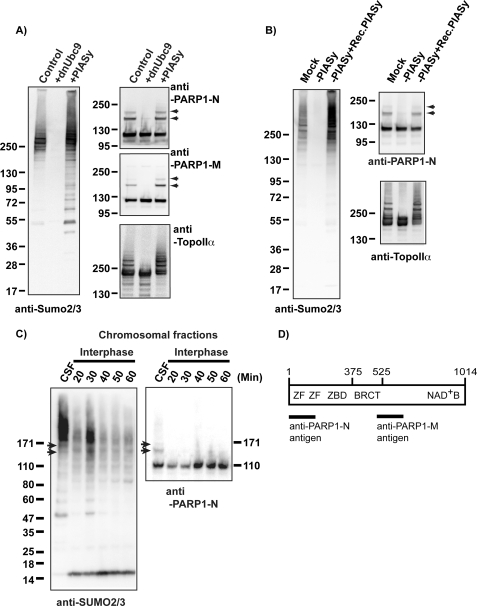
PIASy-mediated SUMO-2/3 modification of chromosomal proteins is essential for faithful anaphase chromosome segregation in the XEE in vitro system (11). There are a number of readily detectable SUMOylated proteins on mitotic chromosomes in XEEs whose modification requires PIASy, including TopoIIα (11). To identify the full spectrum of mitotic SUMOylation substrates for PIASy in XEEs, we added hexahistidine (His6)-tagged SUMO-2 protein to XEEs arrested in meiotic M phase through the action of cytostatic factor (CSF XEEs) in the presence of demembranated sperm chromatin. We purified a chromosomal fraction from these reactions and isolated the SUMOylated proteins by Co2+ affinity chromatography under denaturing conditions (supplemental Fig. 1) in a manner analogous to earlier studies (12). We observed a prominent SUMOylated species with a molecular mass around 140 kDa and subjected it to LC (liquid chromatography)-tandem mass spectometry analysis for protein identification at the Harvard Microchemistry and Proteomics Facility (Cambridge, MA). The results indicated that this species was SUMO-2-conjugated PARP1. To confirm this identification, we cloned Xenopus laevis PARP1 and prepared antibodies against fragments of the PARP1 protein (Fig. 1D). Immunoblotting analysis of mitotic chromosomes prepared from CSF XEEs clearly showed more slowly migrating forms of PARP1 protein, which were abolished when a dominant negative form of Ubc9 (dnUbc9 (11)) was added to the reactions or increased in abundance upon the addition of exogenous PIASy (Fig. 1A).
FIGURE 1.
PARP1 is a PIASy-mediated SUMOylated protein on mitotic chromosomes. A, PARP1 is SUMOylated on mitotic chromosomes. Chromosomal fractions were prepared in CSF extracts in the presence of either dnUbc9 or PIASy as described under “Experimental Procedures.” Isolated chromosomes were analyzed by immunoblotting with indicated antibodies. Arrows indicate the position of SUMOylated PARP1, which were diminished when SUMOylation was inhibited by dnUbc9. B, PIASy is required for PARP1 SUMOylation. CSF extracts were subjected to immunodepletion with anti-PIASy antibody (−PIASy), and then a purified recombinant PIASy was supplemented to PIASy-depleted extracts (−PIASy+Rec.PIASy). Chromosomal fractions were prepared in the indicated conditions of CSF extracts, then the SUMOylation status of isolated chromosomes was analyzed with the indicated antibodies. Arrows indicate the position of SUMOylated PARP1. C, PARP1 SUMOylation is specific in mitosis. CSF extracts were released into interphase by addition of CaCl2. Sperm nuclei were added to either CSF or interphase extracts. After incubation of the indicated periods with interphase extracts or 40 min with CSF extracts, the chromatin fractions were isolated as described under “Experimental Procedures.” Isolated chromatin fractions were subjected to immunoblotting with the indicated antibodies. Arrows indicate SUMOylated PARP1. D, representation of functional domains of PARP1 (adjusted from Ref. 18) is shown. Lines indicate the fragments of PARP1 that were used as antigens. BRCT, BRCA1 C-terminal domain.
To test directly whether the SUMOylation of PARP1 requires PIASy, we depleted PIASy from CSF XEEs (Fig. 1B); PIASy depletion eliminated the slowly migrating SUMOylated forms of PARP1. These forms of PARP1 were restored upon the addition of a recombinant PIASy to the depleted CSF XEEs, confirming that they arise in a PIASy-dependent manner. To test whether SUMOylation of PARP1 was sensitive to the cell cycle status of the XEE, we compared chromatin fractions isolated from CSF XEEs to chromatin fractions from interphase XEEs (Fig. 1C). Immunoblotting analysis with anti-PARP1 and anti-SUMO-2/3 antibodies showed robust SUMOylation in the CSF XEEs (indicated with arrows) as expected, but no SUMOylated PARP1 could be detected in the interphase chromatin fractions. Together, these results show that PARP1 is a target of mitotic, PIASy-mediated SUMO-2/3 conjugation that is associated with mitotic chromosomes in XEEs. Notably, this pattern was closely similar to the SUMOylation of TopoIIα, an established PIASy substrate in this system (12).
In Vitro Reconstitution of PARP1 SUMOylation by PIASy
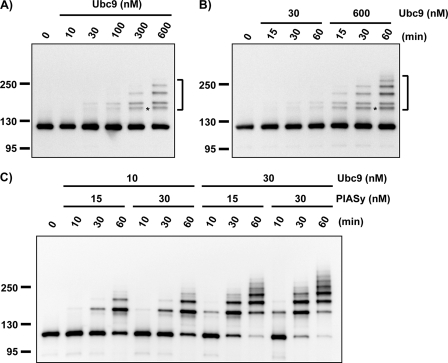
To examine PARP1 SUMOylation by PIASy further, we reconstituted this modification in an in vitro system using entirely purified components that were expressed in bacteria and prepared as described under “Experimental Procedures.” We found that PARP1 could be modified by SUMO-2/3 in the absence of PIASy in reactions containing a very high concentration of Ubc9 (600 nm) (Fig. 2A indicated with brackets). These concentrations significantly exceeded the physiological concentration of Ubc9 in XEEs, which was determined to be around 30 nm through quantitative immunoblotting of XEE in comparison to purified recombinant Ubc9 (data not shown). In reactions containing 30 nm Ubc9, SUMOylated PARP1 was hardly detected (Fig. 2B), even with prolonged incubation. Notably, strong SUMOylation of PARP1 was observed when PIASy was added to reactions containing near-physiological concentrations of Ubc9 (Fig. 2C). Modified forms of PARP1 were evident in reactions containing 10 nm Ubc9 and 15 nm PIASy. Such SUMOylation was enhanced in reactions containing 30 nm PIASy. Increasing the concentration of Ubc9 to 30 nm provided a robust boost in PARP1 SUMOylation at both concentrations of PIASy.
FIGURE 2.
PIASy stimulates PARP1 modification by SUMO-2/3 in reconstituted in vitro SUMOylation reaction. All recombinant proteins, E1, E2, E3, PARP1, and SUMO, were expressed in E. coli and purified as described under “Experimental Procedures.” In vitro SUMOylation reaction was performed with 15 nm E1, 500 nm PARP1, and 6 μm SUMO at 25 °C. Reactions were terminated at the indicated times by the addition of SDS-PAGE sample buffer. SUMOylation of PARP1 was analyzed by immunoblotting with horseradish peroxidase-conjugated anti-T7 antibody (Novagen). The bracket indicates the position of SUMOylated PARP1. A, Ubc9 dose dependence of PARP1 SUMOylation without E3 is shown. B, kinetics of Ubc9-dependent PARP1 SUMOylation are shown. The SUMOylation reaction was performed with the indicated concentration of Ubc9 without PIASy. The reaction mixtures were incubated for 60 min in A and for the indicated time periods in B. C, PIASy-dependent SUMOylation of PARP1 is shown. The reaction mixtures containing the indicated concentration of Ubc9 and PIASy were incubated for the indicated times.
Our results collectively showed that PIASy stimulates SUMOylation of PARP1 at physiological concentrations of Ubc9. A subtler but equally important finding from these in vitro reactions was that the pattern of SUMOylated PARP1 bands differed between the reactions with and without PIASy. Without PIASy, we observed a SUMOylated form of PARP1 with an aberrant migration on SDS-PAGE that did not correspond to the bands observed in CSF XEEs (indicated with the asterisk in Fig. 2, A and B). This form was dramatically decreased in PIASy-dependent reactions (compare Fig. 2, B and C). In other words, the pattern of SUMOylated PARP1 species found in the PIASy-dependent in vitro reactions (Fig. 2C) was much closer to the pattern observed within the chromatin fractions of complete CSF XEEs.
Lysine 482 Is a Primary SUMOylation Site of PARP1
To determine the function of PARP1 SUMOylation, we mapped its SUMOylation site(s). We isolated SUMOylated PARP1 by immunoprecipitation with anti-PARP1 antibodies under denaturing conditions from a mitotic chromatin fraction made from CSF XEEs (supplemental Fig. 2A) (29). Both SUMOylated and non-SUMOylated PARP1 were subjected to tandem mass spectometry analysis for mapping of SUMOylation site(s). The SUMOylation site was identified by mass spectrometry after a double digestion with trypsin and chymotrypsin. Chymotrypsin generated a signature QQQTGG tag on the modified lysine of PARP1. With >70% sequence coverage of PARP1, this analysis indicated that lysine 482 was a candidate SUMOylation site on PARP1. Moreover, alignment of X. laevis PARP1 primary sequence to Homo sapiens PARP1 (supplemental Fig. 2B) indicated that lysine 482 is conserved to a recently reported SUMOylation site of human PARP1, which was found by mutational analysis (30).
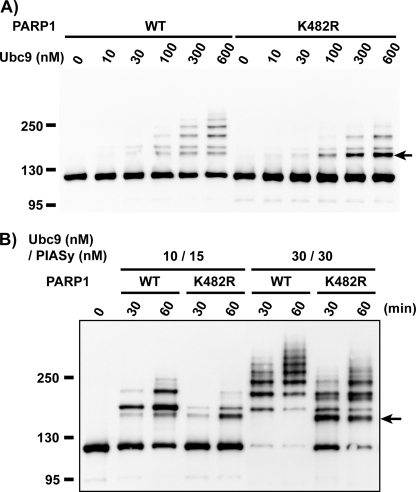
To confirm this mapping result, we introduced a mutation at lysine 482 to encode an arginine at this position (PARP1-K482R). We compared the SUMOylation patterns of wild type PARP1 (PARP1-wt) and PARP1-K482R within SUMOylation assays using recombinant proteins. In the absence of PIASy, PARP1-wt and PARP1-K482R became SUMO-2/3 conjugated to similar extents and showed similar dependence upon the concentration of Ubc9 (Fig. 3A). However, PARP1-K482R showed a much greater accumulation of the aberrant SUMOylation product that was preferentially observed in PIASy-independent reactions (Fig. 3A, indicated with an arrow; compare with Fig. 2).
FIGURE 3.
The substitution of lysine 482 to arginine causes an alteration in the efficiency and specificity of the SUMOylation of PARP1 in in vitro SUMOylation assay. We have introduced a mutation at lysine 482 to arginine (PARP1 K482R) to confirm the mapping result. Purified PARP1 proteins were subjected to in vitro SUMOylation assay as in Fig. 3. A, shown is the Ubc9 dose-dependent reaction without PIASy. The SUMOylation reaction was performed with the indicated concentration of Ubc9 without PIASy. The reaction mixtures were incubated for 60 min. B, shown is the PIASy-dependent reaction. The reaction mixtures containing the indicated concentration of Ubc9 and PIASy were incubated for the indicated times. The arrow indicates the aberrant molecular mass shifted form that is more abundant in K482R mutant than the wild type.
In reactions containing PIASy, there were two important differences between PARP1-K482R and PARP1-wt (Fig. 3B). First, PARP1-K482R showed a significantly lower efficiency of SUMOylation than PARP1-wt. For instance, more than 50% of PARP1-wt became SUMOylated within 60 min at the lowest Ubc9 and PIASy concentrations (10 and 15 nm, respectively), whereas the extent of PARP1-K482R SUMOylation was much less under the same conditions. In the presence of 30 nm Ubc9 and 30 nm PIASy, the decreased efficiency of PARP1-K482R SUMOylation was even more obvious; PARP1-wt was almost 100% SUMOylated within 30 min, whereas PARP1-K482R was only about 50% SUMOylated in the same interval. Second, aberrantly migrating SUMOylated species were more abundant in reactions containing PARP1-K482R (Fig. 3B, indicated by an arrow). As before, these forms were largely suppressed by the presence of PIASy in reactions containing PARP1-wt (Figs. 2C and 3B). Collectively, these results indicate that lysine 482 is a primary SUMOylation site of PARP1 for PIASy-mediated SUMO-2/3 modification in vitro. Mutation of this residue is accompanied by the enhanced modification of other lysines within PARP1, leading to the formation of species with electrophoretic motilities that differ from the primary SUMOylation products observed in intact CSF XEEs.
PARP1-K482R Shows Deficient SUMOylation in XEE Assays
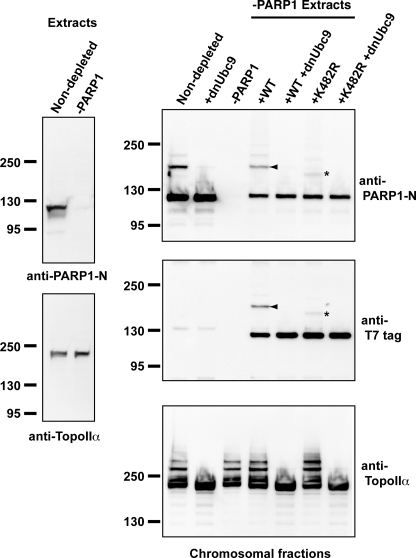
We wished to compare the SUMOylation of PARP1-K482R to PARP1-wt under more physiological conditions in XEE CSF extracts. We added recombinant, T7-tagged PARP1-K482R and PARP1-wt to chromatin-containing CSF XEEs that had been immunodepleted of endogenous PARP1 using anti-PARP1 antibodies (−PARP1) (Fig. 4). The efficiency of depletion was measured by immunoblotting the extracts with an anti-PARP1 antibody (left panel, indicated as Extracts). More than 99% of PARP1 was eliminated from CSF XEEs without detectable changes in the concentration of another PIASy substrate, TopoIIα. After a 60-min incubation, mitotic chromosomes were prepared from these reactions, and the chromosomal proteins were isolated (Fig. 4, right panels). As expected, chromosomes prepared from the PARP1-depleted extract did not contain detectable levels of PARP1 (Fig. 4, −PARP1 lane on the right panels). Reactions reconstituted through the addition of recombinant PARP1-wt to depleted CSF XEEs showed levels of SUMOylation similar to endogenous PARP1 modification in non-depleted CSF XEEs (right panels, arrowheads), and this modification was similarly sensitive to dnUbc9. On the other hand, PARP1-K482R was SUMOylated to a lesser extent, and the resultant species showed greater electrophoretic mobility than the SUMOylated forms of the endogenous protein (Fig. 4, indicated with asterisks), as we had previously observed the assays with fully purified components (compare Fig. 3 and 4). Notably, the formation of these species remained sensitive to dnUbc9. Our findings would be consistent with the utilization of an alternative or secondary SUMOylation site(s) in PARP1 that lacks lysine 482.
FIGURE 4.
Lysine 482 of PARP1 is a primary acceptor site of SUMO-2/3 on mitotic chromosomes. CSF extracts were immunodepleted with an anti-PARP1-M antibody. Either wt or K482R mutant PARP1 (K482R) was added to the PARP1-depleted extracts. Mitotic chromosomes were assembled with these altered CSF extracts, and the chromosomal fractions were isolated. The chromosomal proteins were analyzed by immunoblotting with the indicated antibodies. Exogenous PARP1 is detected with anti-T7 tag antibody (middle right panel). The arrowhead indicates SUMOylated PARP1 corresponding to the endogenous SUMOylated PARP1. The asterisk indicates a shifted PARP1 that does not correspond to the endogenous SUMOylated PARP1 and was observed only with K482R.
SUMOylation Regulates PARP1 Activity on Chromosomes
Lysine 482 is located within the BRCA1 C-terminal domain of PARP1. This domain is predicted to bind to other proteins as well as to mediate auto-PARylation. Therefore, we hypothesized that SUMOylation of Lys-482 may either affect auto-PARylation activity by modulating the conformation of the catalytic domain or affect the PARylation of other substrates by modulating PARP1 protein-protein interactions (18, 19).
To test the first possibility, we examined whether SUMOylation of PARP1 altered auto-PARylation activity in an in vitro reaction. An in vitro SUMOylation reaction was performed in the presence of PIASy as in Fig. 3B using either unconjugate-able SUMO-2 that lacked a C-terminal diglycine motif (SUMO-2-G) or conjugate-able SUMO-2 (SUMO-2-GG). As expected, there was no SUMOylated PARP1 detected in the SUMOylation reaction with SUMO-2-G, whereas the bulk of PARP1 was modified in the reaction with conjugate-able SUMO-2-GG (Fig. 5A, lower panel at t = 0 min) (27). After PARP1 had become SUMOylated, NAD+ was added to the reaction, and further incubation allowed the formation of poly(ADP-ribosyl) chains on PARP1. Poly(ADP-ribosyl) chain formation was analyzed by immunoblotting with a monoclonal anti-PAR antibody (Fig. 5A, upper panel). There was a similar amount of PARylated PARP1 in both SUMO-2-G- and SUMO-2-GG-containing reactions. These results indicate that SUMOylated PARP1 retains robust auto-PARylation activity. Our findings suggest that SUMOylation of PARP1 has neither an inhibitory nor an enhancing effect on the auto-PARylation of PARP1.
To test the second possibility, we examined whether inhibition of SUMOylation in XEE assay affects a PARylation on chromosomal proteins in CSF XEEs (Fig. 5B). We prepared mitotic chromosomes under conditions where SUMOylation occurred normally and under conditions where it was blocked by dnUbc9. We then induced PARylation of chromosomal proteins by incubating the isolated chromosomes with NAD+ as described under “Experimental Procedures.” The PARylation of chromosomal proteins was analyzed by immunoblotting with anti-PAR polyclonal antibody. As shown in Fig. 5B, the chromosomes from the reaction where SUMOylation was inhibited (+dnUbc9 lane) showed more PARylation, and the pattern of PARylated proteins was clearly different from the control reaction (Control lane). To confirm that this PARylation was dependent upon PARP1, we performed the same experiment using CSF XEEs that were depleted of PARP1. The PARP1-depleted chromosomes showed little PARylation, indicating that PARP1 was the primary enzyme that mediated chromosomal PARylation in this assay.
Finally, we investigated whether SUMOylation may control PARylation by modulating PARP1 localization on mitotic chromosomes. Sperm chromatin was allowed to undergo a single round of DNA replication in interphase XEEs. The XEEs were then returned to mitosis through the addition of a fresh aliquot of CSF XEE, allowing the formation of mitotic chromosomes. Individual chromosomes were isolated on coverslips and stained using antibodies against SUMO-2/3, PARP1, and centromeric histone variant CENP-A (Fig. 5C). Consistent with earlier experiments (11), the anti-SUMO2/3 antibody stained the inner centromere region of the chromosomes, and this signal was completely abolished in the presence of dnUbc9 (Fig. 5C, right panels). PARP1 associated broadly on chromosome arms, with slight accumulation on the inner centromere region, partially co-localizing with the region of highest SUMO-2/3 accumulation (Fig. 5C, upper left panel). As shown in the magnified insets, red (PARP1) and green (SUMO-2/3) merged to yellow at the edge of SUMO-2/3 signals, although the majority of SUMO-2/3 signal did not overlap with PARP1. This pattern was not obviously changed by the addition of dnUbc9, arguing against the possibility that SUMO-dependent changes in PARylation by PARP1 are due to its mislocalization on chromosomes. Together, our findings favor the possibility that SUMOylation primarily controls the capacity of PARP1 to act on PARylation of substrates without substantially altering its intrinsic enzymatic activity or subcellular localization.
DISCUSSION
We have identified PARP1 as a novel mitotic SUMO-2/3-modified chromosomal substrate in M-phase X. laevis egg extracts (CSF XEEs). In a manner similar to a previously identified SUMO-2/3 conjugation target, DNA topoisomerase IIα, PARP1 modification in CSF XEEs required PIASy, the presence of chromatin, and the mitotic phase of the cell cycle (11, 12). We have mapped the SUMOylated residues of PARP1 that were purified from mitotic chromosomes and found that lysine at 482 was a primary SUMOylation site. We have reconstituted SUMOylation of PARP1 using purified components and shown that PIASy plays two important roles in its conjugation; it enhances both the efficiency of SUMOylation and the accuracy of site selection. At high concentrations of Ubc9, PARP1 could be SUMOylated in the absence of PIASy. At more physiological concentrations of Ubc9, however, PARP1 SUMOylation became almost entirely PIASy-dependent. The aberrant migration of the modified species in reactions lacking PIASy suggested that the secondary or tertiary SUMOylation site(s) on PARP1 was being used at a greater frequency than when it was present. Similar species were observed even in the presence of PIASy when the reactions were performed with a PARP1 mutant, PARP1-K482R, that lacked the major SUMOylation site that we had mapped.
Control of Chromosomal PARylation by SUMOylation
We did not find that PARP1 activity in auto-PARylation assays was substantially dependent upon its SUMOylation status (Fig. 5A), arguing against the idea the SUMOylation of PARP1 changes its intrinsic enzymatic activity. On the other hand, we observed that PARP1 was the major PARylation enzyme associated with mitotic chromosomes in XEEs, as this modification was essentially abolished in its absence, and that its capacity to mediate PARylation of other chromosomal proteins was enhanced under conditions where SUMOylation was suppressed (Fig. 5B).
These data are consistent with the idea that SUMOylation of PARP1 substantially decreases its capacity to recognize or modify other proteins. Because lysine 482 is located within the BRCA1 C-terminal domain of PARP1, we speculate that SUMOylation of this residue may substantially change PARP1 association with other proteins (31). Such interactions did not appear to be important for targeting of PARP1 onto mitotic chromosomes (Fig. 5C) but may disrupt its capacity to PARylate its chromatin-bound substrates. An alternative explanation for our findings is that SUMOylation inhibits PARylation through modification of PARP1 target proteins. The dnUbc9 would, thus, increase PARylation by relieving such inhibition. It has not been possible to restore chromosomal PARylation to PARP1-depleted XEEs with recombinant PARP1, possibly because immunodepletion of PARP1 co-precipitates other factors necessary for its activity (data not shown), so it has not been possible to rigorously test these two alternatives; notably, they are not mutually exclusive, so that SUMOylation of both PARP1 and its targets could modulate the level of chromosomal PARylation.
Further investigation will be required to identify the substrates of PARP1-mediated PARylation in XEEs and the role of this modification in altering their function. Consistent to our finding that a portion of PARP1 localizes on inner centromere region with XEE assay (Fig. 5C), PARP1 is localized to the centromeres of mammalian cells (22, 32). It has been shown to be involved in the regulation of chromatin structure (33) and to promote the PARylation of several centromeric proteins (23). One attractive possibility is that these aspects of PARP1 function might be regulated through SUMOylation. In this context, the Aurora B kinase is a particularly intriguing potential target for PARP1; it interacts with the BRCA1 C-terminal domain of PARP1 and can be PARylated by PARP1 (24). Moreover, the fact that PARP1 associated with the inner centromere region might be consistent with this possibility (Fig. 5C). However, it was not possible for us to validate this notion because we did not find evidence that Aurora B becomes PARylated in XEEs nor that such a modification could be modulated by changes in SUMOylation (data not shown). It also remains possible that PARP1 itself is a major PARylation substrate on mitotic chromosomes. In this case the capacity of chromosome-bound PARP1 to catalyze auto-PARylation may be more regulated through SUMOylation than isolated PARP1 protein in our purified assays (Fig. 5A).
Role of PIASy in PARP1 SUMOylation
SUMOylation sites of many targets lie within a preferred consensus motif, ΨKX(D/E) (Ψ is an aliphatic branched amino acid; X is any amino acid). Ubc9 can directly bind this motif and conjugate the lysine residue within it in an E3-independent manner (34). Lysine 482 of PARP1 lies within such a consensus motif (supplemental Fig. 2), and Ubc9 interacts PARP1 in yeast two-hybrid assays (35). Consistent with these facts, we found that PARP1 could be SUMOylated in the absence of PIASy (Fig. 2). We note, however, that Ubc9 was also able to recognize other lysine residues of PARP1 in these E3-independent reactions, as we continued to observe SUMOylation in reactions with a mutant form of PARP1 that lacked this residue (PARP1-K482R). Notably, direct recognition by Ubc9 is relatively inefficient, and we observed little PARP1 SUMOylation at physiological concentrations of Ubc9, even for PARP1 possessing a wild type lysine at residue 482.
Recent structural analysis on a fragment of Siz1p, a SIZ/PIAS family SUMO ligase from budding yeast, showed that it binds the thioester-linked Ubc9·Smt3p complex in such a way as to properly configure it for catalysis and to promote correct interactions between the conjugation target and Ubc9's active site (36). PIASy dramatically increased the extent of PARP1 SUMOylation at physiological concentration of Ubc9 (Fig. 2C). This increased SUMOylation is likely to reflect the capacity of PIASy to recognize both Ubc9·SUMO-2 and PARP1, analogous to the interactions formed by Siz1p. This binding would bring them into a single complex, elevating the effective concentration of Ubc9·SUMO-2 as well as increasing the efficiency of SUMO-2 transfer. Notably, Siz1p also directs the transfer of Smt3p to the appropriate lysine residue of the target protein (36). We find that PIASy can similarly bias conjugation toward a preferred residue, lysine 482. The preference for this residue is not absolute, as we find that alternative residues can be utilized when it is mutated either in purified assays (Fig. 3B) or in XEEs (Fig. 4), albeit inefficiently.
In summary, we have shown that PARP1 is a substrate of PIASy-mediated SUMOylation on mitotic chromosomes in XEEs. This modification regulated its activity in this context, perhaps through its capacity to recognize or modify other chromosomal proteins. PIASy facilitates the modification of PARP1 by increasing both the extent of SUMOylation and by directing the modification to a preferred site on PARP1. In combination with earlier observations showing that PIASy modifies TopoIIα in XEEs (11, 12), our findings suggest that PIASy is responsible for a series of coordinated changes in the activity of chromatin-associated enzymes that contribute to chromosome segregation in this system.
Supplementary Material
Acknowledgments
Protein identification was done at the Harvard Microchemistry and Proteomics Analysis Facility. We appreciate the gifts of the X. laevis cDNA from Drs. Toshikazu Amano and Yun-Bo Shi and plasmids for E1 expression from Dr. Frauke Melchior. We thank Alexei Arnaoutov for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants P20 RR015563 (National Center for Research Resources COBRE-Center for Cancer Experimental Therapeutics) and GM80278 and GM67945 (NIGMS to S. P. G.). This work was also supported in part by a start-up grant from Department of Molecular Biosciences at University of Kansas.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- SUMO
- small ubiquitin-related modifiers
- XEE
- Xenopus egg extract
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- PARylation
- poly-ADP-ribosylation
- CSF
- cytostatic factor
- wt
- wild type
- TopoIIα
- topoisomerase IIα.
REFERENCES
- 1.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 2.Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 3.Seeler J. S., Bischof O., Nacerddine K., Dejean A. (2007) Curr. Top Microbiol. Immunol. 313, 49–71 [DOI] [PubMed] [Google Scholar]
- 4.Heun P. (2007) Curr. Opin. Cell Biol. 19, 350–355 [DOI] [PubMed] [Google Scholar]
- 5.Dasso M. (2008) Cell Div. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. (2005) Mol. Cell. Proteomics 4, 246–254 [DOI] [PubMed] [Google Scholar]
- 7.Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. (2005) J. Biol. Chem. 280, 4102–4110 [DOI] [PubMed] [Google Scholar]
- 8.Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. (2004) J. Biol. Chem. 279, 41346–41351 [DOI] [PubMed] [Google Scholar]
- 9.Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd (2004) J. Biol. Chem. 279, 45662–45668 [DOI] [PubMed] [Google Scholar]
- 10.Palvimo J. J. (2007) Biochem. Soc. Trans. 35, 1405–1408 [DOI] [PubMed] [Google Scholar]
- 11.Azuma Y., Arnaoutov A., Anan T., Dasso M. (2005) EMBO J. 24, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azuma Y., Arnaoutov A., Dasso M. (2003) J. Cell Biol. 163, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong K. A., Kim R., Christofk H., Gao J., Lawson G., Wu H. (2004) Mol. Cell. Biol. 24, 5577–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díaz-Martínez L. A., Giménez-Abián J. F., Azuma Y., Guacci V., Giménez-Martín G., Lanier L. M., Clarke D. J. (2006) PLoS One 1, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi Y., Yong-Gonzalez V., Kikuchi Y., Strunnikov A. (2006) Genetics 172, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 17.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M. Y., Zhang T., Kraus W. L. (2005) Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 19.Amé J. C., Spenlehauer C., de Murcia G. (2004) BioEssays 26, 882–893 [DOI] [PubMed] [Google Scholar]
- 20.Chang P., Jacobson M. K., Mitchison T. J. (2004) Nature 432, 645–649 [DOI] [PubMed] [Google Scholar]
- 21.Chang P., Coughlin M., Mitchison T. J. (2005) Nat. Cell Biol. 7, 1133–1139 [DOI] [PubMed] [Google Scholar]
- 22.Saxena A., Wong L. H., Kalitsis P., Earle E., Shaffer L. G., Choo K. H. (2002) Hum. Mol. Genet 11, 2319–2329 [DOI] [PubMed] [Google Scholar]
- 23.Saxena A., Saffery R., Wong L. H., Kalitsis P., Choo K. H. (2002) J. Biol. Chem. 277, 26921–26926 [DOI] [PubMed] [Google Scholar]
- 24.Monaco L., Kolthur-Seetharam U., Loury R., Murcia J. M., de Murcia G., Sassone-Corsi P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14244–14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caiafa P., Guastafierro T., Zampieri M. (2009) Faseb J. 23, 672–678 [DOI] [PubMed] [Google Scholar]
- 26.Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 27.Azuma Y., Tan S. H., Cavenagh M. M., Ainsztein A. M., Saitoh H., Dasso M. (2001) FASEB J. 15, 1825–1827 [DOI] [PubMed] [Google Scholar]
- 28.Kornbluth S., Yang J., Powers M. (2001) in Current Protocols in Cell Biology (M. Yamada K. ed) pp. 11.11.11–11.11.13, John Wiley & Sons, Inc., New York [Google Scholar]
- 29.Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. (2002) J. Biol. Chem. 277, 22115–22118 [DOI] [PubMed] [Google Scholar]
- 30.Messner S., Schuermann D., Altmeyer M., Kassner I., Schmidt D., Schär P., Müller S., Hottiger M. O. (2009) FASEB J. 23, 3978–3989 [DOI] [PubMed] [Google Scholar]
- 31.Glover J. N., Williams R. S., Lee M. S. (2004) Trends Biochem. Sci. 29, 579–585 [DOI] [PubMed] [Google Scholar]
- 32.Earle E., Saxena A., MacDonald A., Hudson D. F., Shaffer L. G., Saffery R., Cancilla M. R., Cutts S. M., Howman E., Choo K. H. (2000) Hum. Mol. Genet 9, 187–194 [DOI] [PubMed] [Google Scholar]
- 33.Quénet D., El Ramy R., Schreiber V., Dantzer F. (2009) Int. J. Biochem. Cell Biol. 41, 60–65 [DOI] [PubMed] [Google Scholar]
- 34.Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 35.Masson M., Menissier-de Murcia J., Mattei M. G., de Murcia G., Niedergang C. P. (1997) Gene 190, 287–296 [DOI] [PubMed] [Google Scholar]
- 36.Yunus A. A., Lima C. D. (2009) Mol. Cell 35, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.