Abstract
Deseasin MCP-01 is a bacterial collagenolytic serine protease. Its catalytic domain alone can degrade collagen, and its C-terminal PKD domain is a collagen-binding domain (CBD) that can improve the collagenolytic efficiency of the catalytic domain by an unknown mechanism. Here, scanning electron microscopy (SEM), atomic force microscopy (AFM), zeta potential, and circular dichroism spectroscopy were used to clarify the functional mechanism of the PKD domain in MCP-01 collagenolysis. The PKD domain observably swelled insoluble collagen. Its collagen-swelling ability and its improvement to the collagenolysis of the catalytic domain are both temperature-dependent. SEM observation showed the PKD domain swelled collagen fascicles with an increase of their diameter from 5.3 μm to 8.8 μm after 1 h of treatment, and the fibrils forming the fascicles were dispersed. AFM observation directly showed that the PKD domain bound collagen, swelled the microfibrils, and exposed the monomers. The PKD mutant W36A neither bound collagen nor disturbed its structure. Zeta potential results demonstrated that PKD treatment increased the net positive charges of the collagen surface. PKD treatment caused no change in the content or the thermostability of the collagen triple helix. Furthermore, the PKD-treated collagen could not be degraded by gelatinase. Therefore, though the triple helix monomers were exposed, the PKD domain could not unwind the collagen triple helix. Our study reveals the functional mechanism of the PKD domain of the collagenolytic serine protease MCP-01 in collagen degradation, which is distinct from that of the CBDs of mammalian matrix metalloproteases.
Keywords: Collagen, Enzyme Catalysis, Enzyme Mechanisms, Protein Degradation, Serine Protease, C-terminal PKD, Collagen Swelling, Collagen Triple Helix, Collagenolytic Serine Protease, Deep Sea
Introduction
The polycystic kidney disease (PKD)3 domain is an 80–90-amino acid module originally found in the human PKD1 gene encoding the cell surface glycoprotein polycystin-1 (1), and later, in many surface layer proteins of archaebacteria (2, 3). In addition to cell surface proteins, the PKD domain is found in many biopolymer hydrolases, such as chitinases (4, 5), celluloses (6), and proteases (7–9), suggesting that it may play an important role in biopolymer degradation. The structures of three PKD domains have been solved, which show that, though their sequences are different, they all adopt a β-helix fold and a conserved sequence area with two Trp residues in the hydrophobic core (2, 3). However, the functional mechanism of the PKD domain in biopolymer hydrolases is largely unknown, except that the PKD domains in chitinase ChiA and collagenolytic serine protease deseasin MCP-01 are both reported to function as a binding domain (5, 10). Genome sequence analysis and experimental results show that the PKD domain is common in the hydrolases of marine heterotrophic bacteria. For example, Gramella forsetii has 14 exported proteins with PKD domains (11). Chitin-binding protease AprIV, deseasin MCP-01, and many other marine deseasins contain one or two PKD domains (5, 9). Therefore, elucidating the function and the mechanism of action of the PKD domains in these hydrolases would have universal importance, especially for clarifying the degradation mechanism of marine biopolymers.
Collagen, the most abundant protein in mammals, is water-insoluble and has hierarchical structure. The triple-helical monomer (15 Å in diameter) of collagen is a rod-like structure composed of three left-handed helix polypeptide chains that coil around each other into a right-handed triple helix. Monomeric collagen molecules axially polymerize to form a microfibril (35 Å in diameter). Then, further aggregate structures, the fibril (50–500 nm in diameter), fascicle (50–300 μm in diameter), and tendon (100–500 μm in diameter) are successively formed (12–14). Because of its special triple-helical structure, only a limited number of collagenolytic proteases can degrade collagen.
Collagenolytic proteases include mammalian matrix metalloproteases (MMPs) and bacterial collagenases, consisting of collagenolytic metalloproteases and collagenolytic serine proteases. Collagenolytic proteases usually have a C-terminal collagen-binding domain (CBD). Most MMPs have a hemopexin-like C domain functioning as a CBD (15–19); the collagenolytic metalloproteases ColG and ColH from Clostridium histolyticum both have CBDs (20); and the collagenolytic serine protease from Geobacillus collagenovorans MO-1 has a collagen-binding segment (21). The C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 also functions as a CBD (10). Deseasin MCP-01, secreted by the deep sea bacterium Pseudoalteromonas sp. SM9913, is a new multidomain protease of the S8 family with collagenolytic activity, which is suggested to play an important role in the degradation of deep sea sedimentary particulate organic nitrogen (9, 10).
Our previous study showed that the collagen binding of the PKD domain of deseasin MCP-01 improves the collagenolytic efficiency of its catalytic domain (10). This suggests that, besides binding collagen, the PKD domain of deseasin MCP-01 may have some unknown function that helps MCP-01 digest collagen. In this study, using the techniques of atomic force microscopy (AFM), circular dichroism (CD) spectroscopy, scanning electron microscopy (SEM), and zeta potential, we studied the changes in collagen caused by the binding of the PKD domain of deseasin MCP-01 to collagen to clarify the function of the PKD domain in MCP-01 collagenolysis. We found that the PKD domain of MCP-01 did not unwind the collagen triple helix, but did swell collagen to expose the triple helix monomers, potentially facilitating their hydrolysis by the MCP-01 catalytic domain.
EXPERIMENTAL PROCEDURES
Materials
Deseasin MCP-01 was purified from Pseudoalteromonas sp. SM9913 using the method previously described (9). The enhanced green fluorescent protein (EGFP) and the fusion protein EGFP-PKD, as well as its mutant EGFP-W36A, were expressed in Escherichia coli BL21 (DE3) and purified using methods previously described (10). Acid-soluble collagen (1 mg/ml, dissolved in 0.1 m acetic acid) and 3-aminopropyl triethoxysilane (APTES) were purchased from Sigma. Insoluble type I collagen (bovine tendon collagen) was purchased from Worthington Biochemical. Pepsin-soluble collagen was prepared using bovine tendon collagen, and then dialyzed against 20 mm borate buffer (pH 8.5) containing 1% NaCl at 4 °C. Chymotrypsin was purchased from Klontech Industrial Sales Inc.
Protein Determination and Enzyme Assays
Protein concentration was assayed by the method of Lowry (22). The activities of chymotrypsin against insoluble type I collagen, thermally denatured collagen and PKD-pretreated collagen were measured at 37 °C with a previously described method to assay collagenase activity against collagen (10). Thermally denatured collagen was prepared by heating insoluble type I collagen at 65 °C for 20 min. PKD-pretreated collagen was prepared by incubating insoluble type I collagen with 3 μm EGFP-PKD at 20 °C for 1 h.
Swelling of Insoluble Type I Collagen by the PKD Domain and Urea
A total of 10 mg of insoluble type I collagen in 2 ml of 20 mm borate buffer (pH 8.5) was incubated at 20 °C for 1 h with 6 μm EGFP-PKD, 6 μm EGFP, 6 μm EGFP-W36A, or 6 m urea. To investigate the effect of temperature on collagen swelling caused by the PKD domain, 10 mg insoluble type I collagen in 2 ml of 20 mm borate buffer (pH 8.5) was incubated with 6 μm EGFP-PKD at 4 °C, 10 °C, 20 °C, and 30 °C for 1 h each.
Atomic Force Microscopy
Acid-soluble collagen was diluted with 50 mm Tris-HCl (pH 8.5) at a ratio of 1:3 (v/v) at 20 °C to self-assemble. After 6 h in suspension, collagen was incubated with EGFP, EGFP-PKD, or EGFP-W36A at a ratio of 1:4 (v/v) at 30 °C (or a given temperature) for 1 h. Then, 10 μl of the sample was spread on the surface of freshly cleaved mica for at least 1 min to be adsorbed. Afterward, to observe the binding of the proteins to collagen, the mica surface was gently rinsed with deionized water to remove the samples that did not adsorb firmly to the substrate and then air-dried. To observe the collagen-swelling effect of the PKD domain, the mica surface was rinsed more thoroughly with deionized water to remove both the unadsorbed samples and EGFP-PKD bound to collagen. All AFM images were performed in tapping mode using a Multimode Nanoscope IIIa (Digital Instruments/Veeco, Santa Barbara, CA) with a scan rate of 1 Hz. Cantilevers with a length of 100 μm and a spring constant of 5.5–22.5 N/m was used.
Scanning Electron Microscopy
A total of 10 mg of type I insoluble collagen was mixed with 1 ml of 20 mm borate buffer (pH 8.5) containing 3 μm EGFP-PKD or EGFP-W36A. The samples were incubated at 20 °C for 1 h with continuous stirring. The samples were observed using scanning electron microscopy (Hitachi S-570) by Usha and Ramasami's method (23).
Circular Dichroism Spectroscopy
Collagen solution (0.2 μm) in 20 mm borate buffer (pH 8.5) was incubated with 0.4 μm EGFP-PKD or EGFP-W36A in the same buffer at 20 °C for 1 h. Collagen solution in 20 mm borate buffer (pH 8.5) without any other proteins was used as control. The triple-helical conformation of the samples was investigated using a J-810 circular dichroism spectropolarimeter (JASCO, Japan) equipped with an F-25 (Julabo, Germany) waterbath thermal unit. All samples were in a 1-mm path length cuvette. Spectra were recorded from 190 to 250 nm, with a bandwidth of 4 nm, a response time of 1 s, a scanning rate of 200 nm/min, and an accumulation of three scans. The spectra were reported in terms of molar ellipticity.
Thermal Unfolding of Collagen
Pepsin-soluble collagen (2 μm) in 20 mm boric acid buffer (pH 8.5) was incubated with an equimolar concentration of EGFP-PKD or EGFP-W36A or 6 m urea for 2 h. Pepsin-soluble collagen in 20 mm boric acid buffer (pH 8.5) was used as control. The melting curves of the treated and control collagen were measured with the Jasco J810 circular dichroism spectropolarimeter at a rate of 1 °C/min by recording the molar ellipticity at 222 nm. The thermal unfolding curves and their midpoints were obtained from the data.
Zeta Potential Measurements
Pepsin-soluble collagen (1 μm) in 50 mm boric acid buffer (pH 8.5) was incubated with an equimolar concentration of EGFP-PKD or EGFP-W36A for 1 h. Then the zeta potential of the treated and untreated collagens was measured at 25 °C using a zeta potential analyzer (Zetapals, Brookhaven Instruments Corporation).
RESULTS
The PKD Domain of MCP-01 Swells Insoluble Collagen
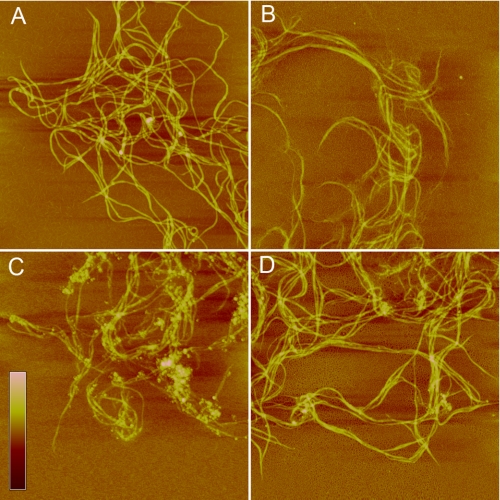
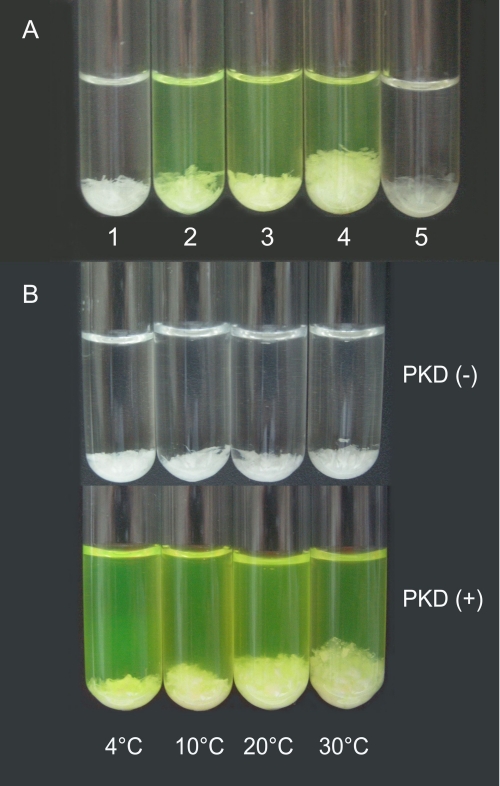
Our previous study showed that the recombinant EGFP-fused PKD domain of MCP-01, EGFP-PKD, can bind to insoluble collagen and that EGFP has no collagen binding ability, indicating that the PKD domain of MCP-01 has collagen binding ability (10). AFM observation directly showed that EGFP-PKD bound to self-assembled collagen (Fig. 1C), while EGFP (Fig. 1B) and the mutant EGFP-W36A (Fig. 1D) had little collagen binding ability. Viewed macroscopically, when EGFP-PKD was incubated with insoluble type I collagen at 20 °C for 1 h, insoluble collagen was swollen, and its volume increased by more than 2-fold, while EGFP and EGFP-W36A did not cause collagen swelling (Fig. 2A). This result indicates that, after binding insoluble collagen, the PKD domain can cause large-scale collagen swelling.
FIGURE 1.
AFM observation of the binding of the PKD domain to collagen. A, self-assembled collagen in 50 mm Tris-HCl (pH 8.5) incubated at 30 °C for 1 h. B, collagen incubated with EGFP at 30 °C for 1 h. C, collagen incubated with EGFP-PKD at 30 °C for 1 h. D, collagen incubated with EGFP-W36A at 30 °C for 1 h. Scale bar: 30 nm. Scan size: A, 6 μm; B–D, 5 μm.
FIGURE 2.
The collagen-swelling effect of the PKD domain. A, collagen swollen by the PKD domain or urea at 20 °C for 1 h. A total of 10 mg of insoluble type I collagen was incubated in 2 ml of 20 mm borate buffer (pH 8.5) (1), 20 mm borate buffer containing 6 μm EGFP (2), 20 mm borate buffer containing 6 μm EGFP-W36A (3), 20 mm borate buffer containing 6 μm EGFP-PKD (4), or 20 mm borate buffer containing 6 m urea (5). B, collagen swollen by the PKD domain for 1 h at different temperatures. A total of 10 mg of insoluble type I collagen in 2 ml of 20 mm borate buffer (pH 8.5) was incubated with 6 μm EGFP-PKD at 4, 10, 20, and 30 °C for 1 h each.
Urea is a known collagen-swelling agent (23). Compared with urea, the collagen-swelling ability of 6 μm PKD was stronger than 6 m urea, based on the volume change of collagen (Fig. 2A). In addition, insoluble collagen became semitransparent after urea treatment, while PKD did not cause such a change in insoluble collagen (Fig. 2A), which suggests that the collagen-swelling mechanism of PKD may be different from that of urea.
When EGFP-PKD was incubated with insoluble collagen at different temperatures from 4 to 30 °C, the collagen-swelling effect of PKD varied: its collagen-swelling ability increased with temperature between 4 °C and 30 °C (Fig. 2B). Correspondingly, improvement of the collagenolytic efficiency of the catalytic domain caused by PKD pretreatment also increased with temperature between 10 and 40 °C (supplemental Fig. S1). These results suggest that collagen swelling caused by the PKD domain facilitates collagen hydrolysis by the MCP-01 catalytic domain.
The PKD Domain of MCP-01 Swells the Aggregate Structures of Collagen and Exposes Collagen Monomers
To study the collagen-swelling mechanism of the PKD domain, we observed the changes in the aggregate structures of collagen caused by the PKD domain using SEM and AFM.
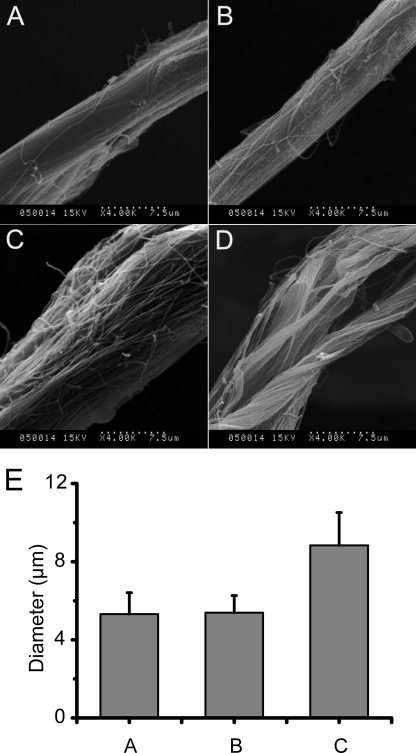
SEM observation showed that after insoluble type I collagen was treated with EGFP-PKD for 0.5–1 h, collagen fascicles were swollen (Fig. 3, A, C, and D). Statistical analysis showed that the diameters of the fibrils increased from 5.3 ± 1.1 μm to 8.8 ± 1.7 μm (Fig. 3E). Moreover, the fibrils that form the fascicles were dispersed. In contrast, EGFP-W36A had no such effect on collagen fascicles and fibrils (Fig. 3B).
FIGURE 3.
SEM observation of collagen fascicles swollen by the PKD domain. A total of 10 mg of type I insoluble collagen in 1 ml of 20 mm borate buffer (pH 8.5) was incubated at 20 °C with 3 μm EGFP-PKD or EGFP-W36A. The samples were observed using scanning electron microscopy (Hitachi S-570) by Usha and Ramasami's method (23). A, collagen fascicle incubated at 20 °C for 1 h. B, collagen fascicle incubated with W36A-EGFP at 20 °C for 1 h. C, collagen fascicle incubated with EGFP-PKD at 20 °C for 0.5 h. D, collagen fascicle incubated with EGFP-PKD at 20 °C for 1 h. E, diameter of untreated and EGFP-W36A- and EGFP-PKD-treated collagen fascicles. Columns in E show the diameter of: 1) untreated collagen fascicles, 2) EGFP-W36A-treated collagen fascicles, and 3) EGFP-PKD-treated collagen fascicles. All data in E are averages of the data from 50 collagen fascicles measured.
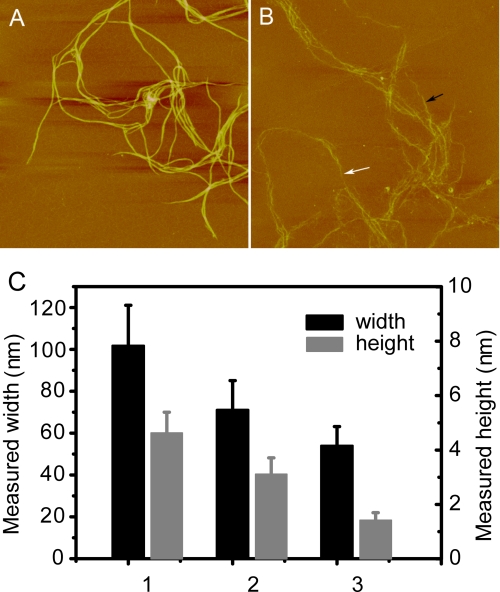
Furthermore, we observed the structural changes of collagen caused by the PKD domain at the microfibril level using AFM. Under AFM, the height of the self-assembled collagen polymers was ∼4.5 nm, indicating that these polymers were collagen microfibrils (Fig. 4). When they were treated with EGFP-PKD at 30 °C for 1 h, the collagen microfibrils swelled and became loose (Fig. 4A). The height of PKD-treated collagen microfibrils markedly decreased to 3.8–1.2 nm, indicating that the tight collagen microfibrils were disjoined (Fig. 4B). It is noteworthy that the diameter of many PKD-treated collagen microfibrils was ∼1.4 nm (Fig. 4C), consistent with that of a collagen triple helix monomer, which is reported to be 15 Å, considering the compression effect of atomic force microscopy during scanning (24). Therefore, the PKD domain exposed the collagen triple helix monomers that had originally formed tight microfibrils.
FIGURE 4.
A, AFM micrograph of collagen microfibrils incubated at 30 °C for 1 h. B, AFM micrograph of collagen microfibrils incubated with EGFP-PKD at 30 °C for 1 h. The black arrow shows the relatively thicker collagen microfibrils, and the white arrow shows the relatively thinner collagen microfibrils after incubation with EGFP-PKD. Scan size is 5 μm in both A and B. C, height and width of the PKD-untreated and PKD-treated collagen microfibrils calculated from AFM micrographs A and B. Columns in C show the height and width of: 1) the PKD-untreated collagen microfibrils in AFM micrograph A, 2) the thicker collagen microfibrils in AFM micrograph B, and 3) the thinner collagen microfibrils in AFM micrograph B. All data in C are averages of the data from 20 sites measured.
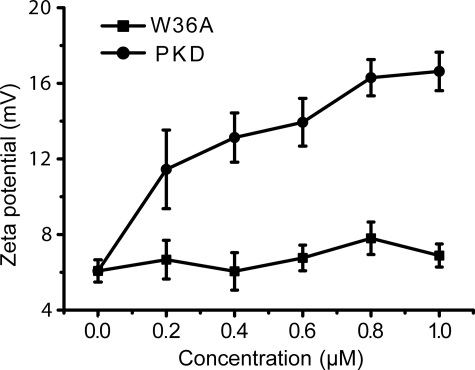
To probe the mechanism of PKD domain to swell collagen, the effect of PKD treatment on the collagen surface charges was investigated by measuring zeta potential. The zeta potential of type I collagens was 6.1 ± 0.6 mV, showing that the collagens bear net positive charges. After treatment with EGFP-PKD, the zeta potential of collagen increased in a PKD concentration-dependent manner. At 1.0 μm EGFP-PKD, the zeta potential increased to 16.6 ± 1.0 mV. In contrast, when treated with EGFP-W36A mutant, the zeta potential of collagen did not change (Fig. 5). Therefore, after PKD treatment, collagen exposed more internal positive charges to the solution, suggesting that binding of the PKD domain to collagen induced structural changes in collagen.
FIGURE 5.
Zeta potential of collagen treated with different concentrations of EGFP-PKD or EGFP-W36A at 20 °C for 1 h.
Binding of the PKD Domain to Collagen Fibers Does Not Unwind the Collagen Triple Helix
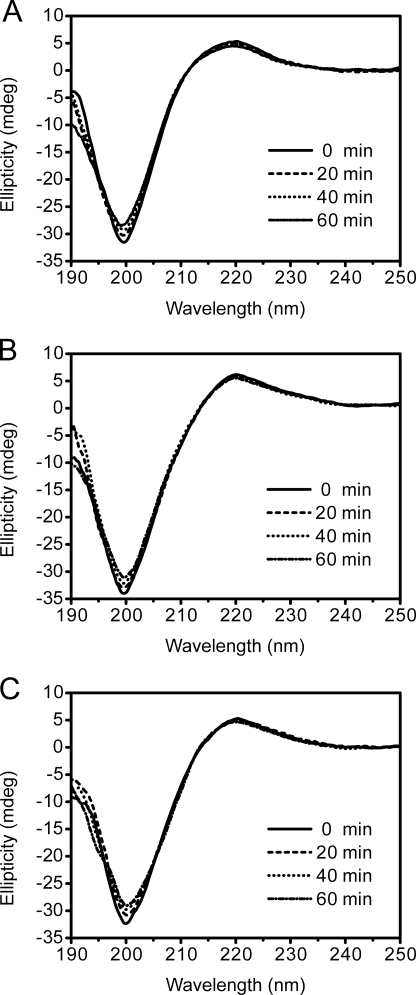
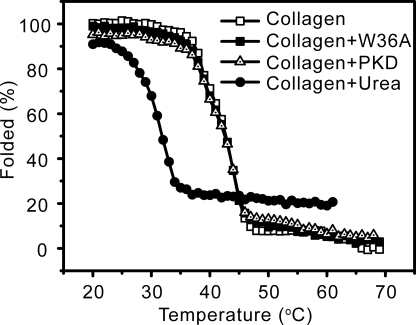
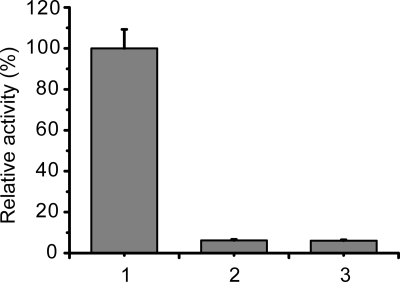
The collagen triple helix structure displays a typical spectrum when detected by CD (25). To investigate whether the binding of the PKD domain to collagen unwinds its triple helix, the changes in collagen structure caused by the PKD domain were detected using CD spectroscopy. The collagen treated with EGFP-PKD (Fig. 6B) or EGFP-W36A (Fig. 6C) displayed similar CD spectra to the untreated collagen (Fig. 6A), showing that the binding of the PKD domain to collagen fibers caused no change in the secondary structure of collagen. This result indicates that the PKD domain of MCP-01 has no helicase activity, and therefore cannot unwind the collagen triple helix. To confirm this, we compared the thermal unfolding temperature (Tm) of collagen treated with or without EGFP-PKD or EGFP-W36A. The treated and untreated collagen had the same Tm values (Fig. 7), indicating that the PKD domain binding to collagen did not affect the thermostability of the collagen triple helix. In contrast, urea treatment led to a significant decrease in the Tm value of collagen. This result also indicates that the functional collagen-interacting mechanisms of the MCP-01 PKD domain and urea are different. In addition, chymotrypsin, a gelatinase that is able to hydrolyze denatured collagen, could not hydrolyze the PKD-swelled collagen (Fig. 8), suggesting that the triple helix of this collagen was not unwound. Taken together, the above results show that the PKD domain of MCP-01 cannot unwind the collagen triple helix.
FIGURE 6.
CD spectra of collagen at 20 °C for 1 h (A), collagen incubated with EGFP-PKD at 20 °C for 1 h (B), and collagen incubated with EGFP-W36A (C) at 20 °C for 1 h.
FIGURE 7.
Thermal unfolding curves of untreated collagen and collagens treated with EGFP-PKD, EGFP-W36A, or urea at 20 °C for 1 h.
FIGURE 8.
Activities of chymotrypsin against thermally denatured collagen (1), PKD-untreated collagen (2), and PKD-treated collagen (3). Thermally denatured collagen was prepared by heating insoluble type I collagen at 65 °C for 20 min. PKD-treated collagen was prepared by incubating insoluble type I collagen with 3 μm PKD at 20 °C for 1 h. Chymotrypsin activity was measured at 37 °C by a previously described method (10).
DISCUSSION
Collagenases have different kinds of CBDs. The CBD of MMPs is a hemopexin-like C domain composed of ∼200 amino acid residues (15, 20). The CBD of deseasin MCP-01 is a PKD domain of ∼90 residues (10). The collagenases from Clostridium histolyticum also have small CBDs of ∼100 residues (20). However, the CBD of the collagenases from G. collagenovorans MO-1 is as large as 35 kDa (21). The function of the CBD of MMPs in collagenolysis has been intensively studied. Most studies show that MMPs unwind triple-helical collagen prior to peptide bond hydrolysis because the catalytic sites of MMPs are too narrow to accommodate the triple-helical structure of collagen (16, 26, 27). The CBD of MMPs has triple-helicase activity and induces local structural disruption of the collagen triple helix. Then the catalytic domain of MMPs cleaves the three chains one by one (27). Another hypothesis for the action of MMP CBDs on triple-helical collagen posits that the binding of the CBD can stabilize partially unfolded conformers so that the catalytic domain can recognize the unwound part and initiate collagen degradation (28). Although the structure and collagen-binding mechanism of the CBDs of some bacterial collagenases have been studied (20, 21), it is unclear whether the binding of these CBDs can potentiate collagenolysis by modifying collagen structure.
We have demonstrated that the C-terminal PKD domain of deseasin MCP-01 from the deep sea bacterium Pseudoalteromonas sp. SM9913 functions as a CBD, and its binding facilitates the hydrolysis by the catalytic domain of insoluble collagen (10). To study how the PKD domain facilitates the collagenolysis of the catalytic domain, in this study we analyzed the changes in collagen structure caused by the binding of the PKD domain of deseasin MCP-01 to collagen using SEM, AFM, and CD. We directly observed under AFM that this PKD domain bound to collagen. Viewed macroscopically, PKD binding caused insoluble collagen to swell at a large scale, a phenomenon that has never been reported for other collagenase CBDs. SEM observation showed that collagen fascicles were swollen by the PKD domain and that the fibrils in the collagen fascicle were dispersed and also swollen. AFM images further showed that the PKD domain swelled collagen at the microfibril level and that the collagen monomers in the microfibril were dispersed and exposed. CD spectroscopy analysis and thermal unfolding experiments showed that the PKD domain did not unwind collagen triple helices. Therefore, the PKD domain of deseasin MCP-01 can destroy the aggregate structures of collagen but has no triple-helicase activity, making its mechanism of action different from that of the hemopexin-like C domains of MMPs.
Efforts on studying collagen of the last couple of decades have made the hierarchical structure of collagen basically clear (12–14, 29). However, the deformation mechanisms under mechanical load, and, in particular, the relationship between those mechanisms and collagen molecular and intermolecular properties, are not well understood. Moreover, the limiting factors of the strength of collagen fibrils and the origins of toughness remain largely unknown (30). New evidence demonstrates that stereoelectronic effects play a key role in the stability of collagen structure (29). Thus, to gain insights into the mechanism of collagen swelling by PKD, we studied the effects of PKD treatment on the collagen surface charges. The result showed that type I collagens bear net positive charges, and PKD treatment resulted in an obvious increase in the surface charges of collagens, indicating that, after PKD treatment, more aggregate structures, such as fibrils, microfibrils, and monomers are exposed, resulting in more charges on the exposed surfaces. The electrostatic repulsive forces produced by these surface charges would prohibit the fibrils/monomers from reassembling to higher level assemblies and promote their dispersion. In addition, these charges may form H-bonds to water and/or solute molecules, which will stabilize the dispersion of fibrils/monomers in the solution. This is consistent with our observations that the PKD-induced swelling of collagen was irreversible (data not shown). Further studies are needed to elucidate the detailed mechanism on the structural changes in collagen induced by the PKD domain, especially to determine the three-dimensional structure of the PKD domain and the PKD-collagen complexes.
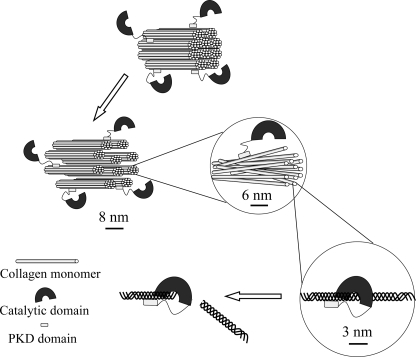
Unlike the catalytic domains of MMPs that have no collagenolytic activity (17, 31, 32), the catalytic domain of deseasin MCP-01 alone can hydrolyze insoluble type I collagen (10), suggesting that the catalytic cavity of the catalytic domain of deseasin MCP-01 can accommodate the triple-helical structure of collagen. Moreover, MCP-01 has various but specific cleavage sites on insoluble type I collagen (10), different from the case in MMPs, which have only one cleavage site on collagens (33). Based on our previous and present results, a collagenolysis model of serine protease MCP-01 is proposed (Fig. 9). The architecture of collagen is notably more resistant to collagenolysis than lone collagen monomers because the tight arrangement of neighboring collagen monomers restricts enzyme access to the cleavage sites. During collagenolysis by MCP-01, the C-terminal PKD domain binds collagen, swells collagen aggregate structures, fascicles, fibrils, and microfibrils, and finally exposes collagen monomers. Then, the N-terminal catalytic domain accommodates the triple-helical collagen monomer in its catalytic cavity and cleaves it. The result that the collagen-swelling ability of the PKD domain and its improvement to the collagenolytic efficiency of the catalytic domain is both temperature dependent also suggests that collagen swelling caused by the PKD domain can exposes more cleavage sites for the catalytic domain. Therefore, our study reveals the functional mechanism of the PKD domain of the collagenolytic serine protease MCP-01 in collagen degradation. As a collagenase CBD, the interaction of the PKD domain with collagen is quite different from that of the CBDs of MMPs. Because PKD domains are common among marine bacterial hydrolases, our study may be helpful in clarifying the degradation mechanism of marine biopolymers.
FIGURE 9.
A schematic diagram of the mechanism of collagenolysis by serine protease MCP-01. During collagenolysis by MCP-01, the C-terminal PKD domain binds collagen, swells collagen aggregate structures, fascicles, fibrils, and microfibrils, and finally exposes collagen triple-helical monomers. Then, the N-terminal catalytic domain accommodates the triple-helical collagen monomer in its catalytic cavity and cleaves it.
Supplementary Material
This work was supported by National Natural Science Foundation of China (30770040 and 50925205), Hi-Tech Research and Development program of China (2007AA091903), and COMRA Program (DYXM-115-02-2-6).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PKD
- polycystic kidney disease
- CBD
- collagen-binding domain
- SEM
- scanning electron microscopy
- AFM
- atomic force microscopy
- CD
- circular dichroism
- MMPs
- matrix metalloproteases
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1.Gluecksmann-Kuis M. A., Tayber O., Woolf E. A., Bougueleret L., Deng N., Alperin G. D., Iris F., Hawkins F. (1995) Cell 81, 289–2987736581 [Google Scholar]
- 2.Bycroft M., Bateman A., Clarke J., Hamill S. J., Sandford R., Thomas R. L., Chothia C. (1999) EMBO J. 18, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jing H., Takagi J., Liu J. H., Lindgren S., Zhang R. G., Joachimiak A., Wang J. H., Springer T. A. (2002) Structure 10, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 4.Perrakis A., Tews I., Dauter Z., Oppenheim A. B., Chet I., Wilson K. S., Vorgias C. E. (1994) Structure 2, 1169–1180 [DOI] [PubMed] [Google Scholar]
- 5.Orikoshi H., Nakayama S., Hanato C., Miyamoto K., Tsujibo H. (2005) J. Appl. Microbiol. 99, 551–557 [DOI] [PubMed] [Google Scholar]
- 6.Ahsan M. M., Kimura T., Karita S., Sakka K., Ohmiya K. (1996) J. Bacteriol. 178, 5732–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oda K., Ito M., Uchida K., Shibano Y., Fukuhara K., Takahashi S. (1996) J. Biochem. 120, 564–572 [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto K., Nukui E., Itoh H., Sato T., Kobayashi T., Imada C., Watanabe E., Inamori Y., Tsujibo H. (2002) J. Bacteriol. 184, 1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X. L., Xie B. B., Lu J. T., He H. L., Zhang Y. Z. (2007) Microbiology 153, 2116–2125 [DOI] [PubMed] [Google Scholar]
- 10.Zhao G. Y., Chen X. L., Zhao H. L., Xie B. B., Zhou B. C., Zhang Y. Z. (2008) J. Biol. Chem. 263, 36100–36107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer M., Kube M., Teeling H., Richter M., Lombardot T., Allers E., Würdemann C. A., Quast C., Kuhl H., Knaust F., Woebken D., Bischof K., Mussmann M., Choudhuri J. V., Meyer F., Reinhardt R., Amann R. I., Glöckner F. O. (2006) Environ. Microbiol. 8, 2201–2213 [DOI] [PubMed] [Google Scholar]
- 12.Kadler K. (1994) Protein Profile 1, 519–638 [PubMed] [Google Scholar]
- 13.Kadler K. E., Holmes D. F., Trotter J. A., Chapman J. A. (1996) Biochem. J. 316, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perumal S., Antipova O., Orgel J. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomis-Rüth F. X., Gohlke U., Betz M., Knäuper V., Murphy G., López-Otín C., Bode W. (1996) J. Mol. Biol. 264, 556–566 [DOI] [PubMed] [Google Scholar]
- 16.Tam E. M., Wu Y. I., Butler G. S., Stack M. S., Overall C. M. (2002) J. Biol. Chem. 277, 39005–39014 [DOI] [PubMed] [Google Scholar]
- 17.Clark I. M., Cawston T. E. (1989) Biochem. J. 263, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knäuper V., Cowell S., Smith B., López-Otin C., O'Shea M., Morris H., Zardi L., Murphy G. (1997) J. Biol. Chem. 272, 7608–7616 [DOI] [PubMed] [Google Scholar]
- 19.Monaco S., Sparano V., Gioia M., Sbardella D., Di-Pierro D., Marini S., Coletta M. (2006) Protein Sci. 15, 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson J. J., Matsushita O., Okabe A., Sakon J. (2003) EMBO J. 22, 1743–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoi Y., Horinaka M., Tsujimoto Y., Matsui H., Watanabe K. (2006) J. Bacteriol. 188, 6572–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 23.Usha R., Ramasami T. (2002) J. App. Poly. Sci. 84, 975–982 [Google Scholar]
- 24.Poenitzsch V. Z., Musselman I. H. (2006) Microsc. Microanal. 12, 221–227 [DOI] [PubMed] [Google Scholar]
- 25.Ikoma T., Kobayashi H., Tanaka J., Walsh D., Mann S. (2003) Int. J. Biol. Macromol. 32, 199–204 [DOI] [PubMed] [Google Scholar]
- 26.Chung L., Dinakarpandian D., Yoshida N., Lauer-Fields J. L., Fields G. B., Visse R., Nagase H. (2004) EMBO J. 23, 3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam E. M., Moore T. R., Butler G. S., Overall C. M. (2004) J. Biol. Chem. 279, 43336–43344 [DOI] [PubMed] [Google Scholar]
- 28.Nerenberg P. S., Salsas-Escat R., Stultz C. M. (2008) Proteins 70, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 29.Shoulders M. D., Raines R. T. (2009) Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buehler M. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12285–12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy G., Allan J. A., Willenbrock F., Cockett M. I., O'Connell J. P., Docherty A. J. P. (1992) J. Biol. Chem. 267, 9612–9618 [PubMed] [Google Scholar]
- 32.Overall C. M. (2002) Mol. Biotechnol. 22, 51–86 [DOI] [PubMed] [Google Scholar]
- 33.De Souza S. J., Pereira H. M., Jacchieri S., Brentani R. R. (1996) FASEB J. 10, 927–930 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.